Identification, Culture and Targeting of Cancer Stem Cells
Abstract
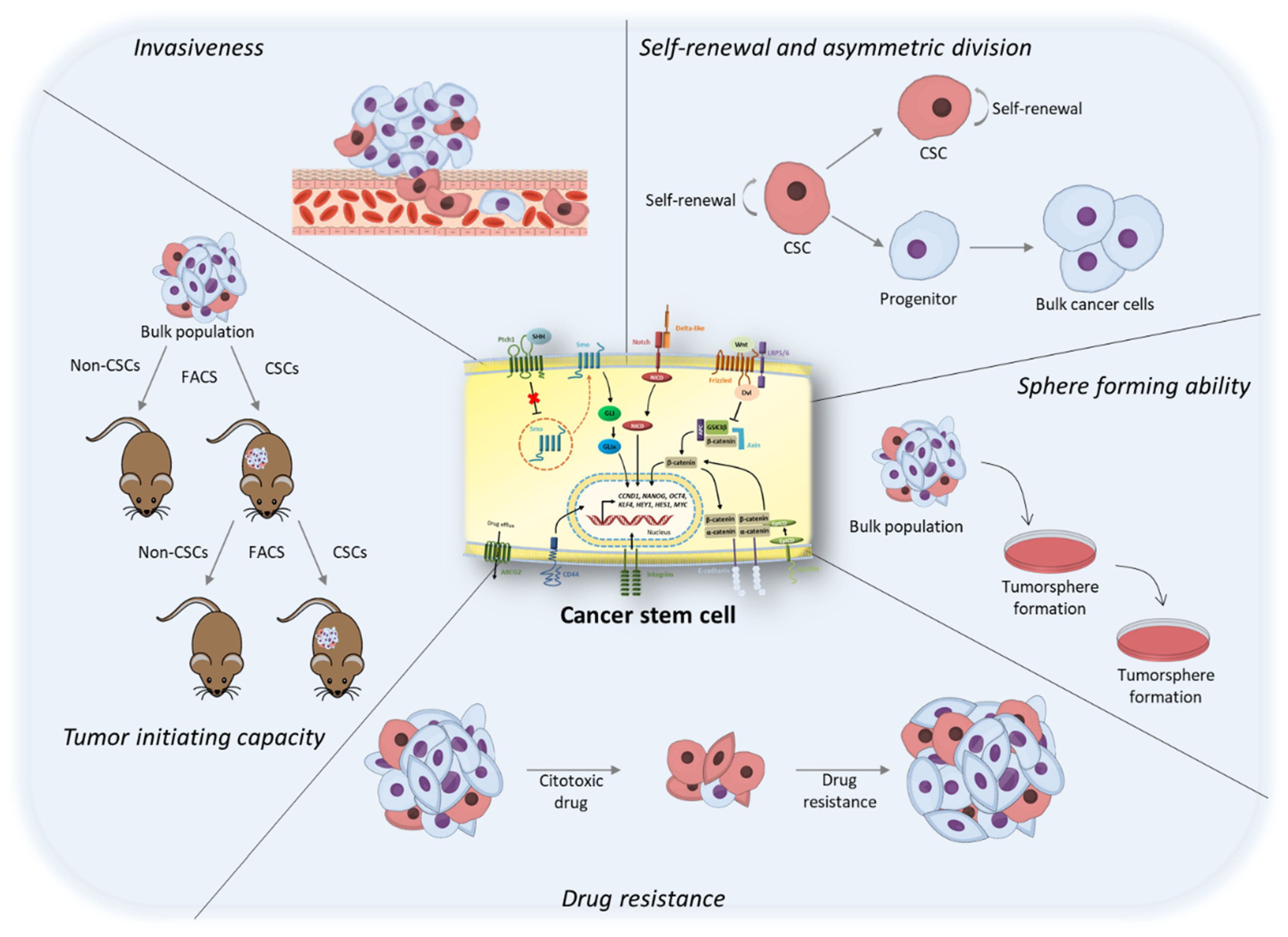
1. Tumor Heterogeneity: The Origin of the “Cancer Stem Cell” Concept
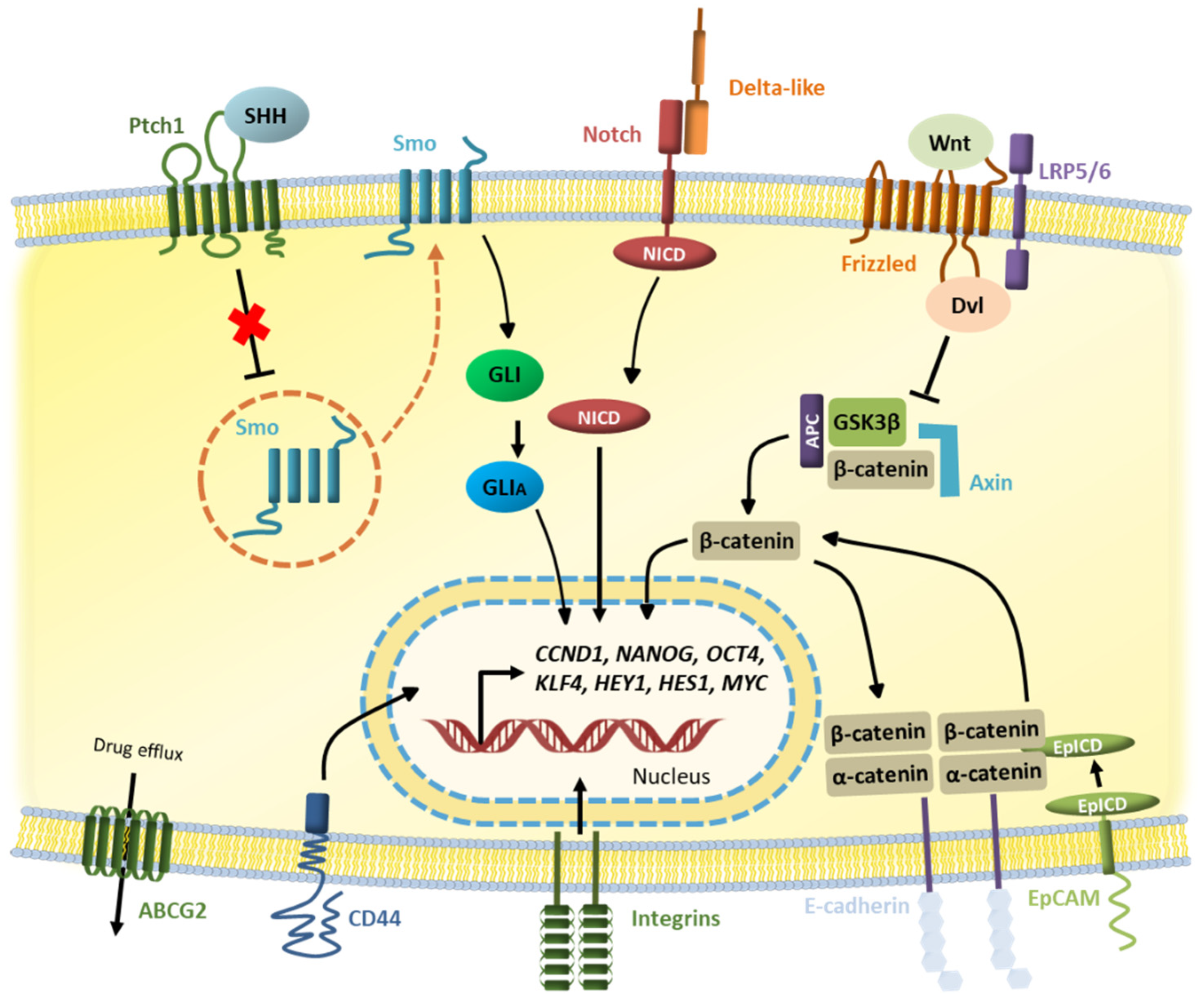
2. Cancer Stem Cells Markers and Pathways
2.1. Notch Pathway
2.2. Wnt Pathway
2.3. Hedgehog Pathway
2.4. Hippo Pathway
2.5. JAK/STAT Pathway
3. Culture of Cancer Stem Cells
4. Targeting of Cancer Stem Cells
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Greaves, M.; Maley, C.C. Clonal Evolution in Cancer. Nature 2012, 481, 306–313. [Google Scholar] [CrossRef]
- Zardavas, D.; Irrthum, A.; Swanton, C.; Piccart, M. Clinical management of breast cancer heterogeneity. Nat. Rev. Clin. Oncol. 2015, 12, 381–394. [Google Scholar] [CrossRef]
- Burrell, R.A.; McGranahan, N.; Bartek, J.; Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013, 501, 338–345. [Google Scholar] [CrossRef]
- Shackleton, M.; Quintana, E.; Fearon, E.; Morrison, S. Heterogeneity in cancer: Cancer stem cells versus clonal evolution. Cell 2009, 138, 822–829. [Google Scholar] [CrossRef]
- Nowell, P.C. The clonal evolution of tumor cell populations. Science 1976, 194, 23–28. [Google Scholar] [CrossRef]
- Wang, Y.; Water, J.; Leung, M.L.; Unruh, A.; Roh, W.; Shi, X.; Chen, K.; Scheet, P.; Vattathil, S.; Liang, H.; et al. Clonal Evolution in Breast Cancer Revealed by Single Nucleus Genome Sequencing. Nature 2014, 512, 155–160. [Google Scholar] [CrossRef]
- Landau, D.-A.; Carter, S.L.; Getz, G.; Wu, C.J. Clonal evolution in hematologic malignancies and therapeutic implications. Leukemia 2014, 28, 34–43. [Google Scholar] [CrossRef]
- Dick, J.E. Stem cell concepts renew cancer research. Blood 2008, 112, 4793–4807. [Google Scholar] [CrossRef]
- Sourisseau, T.; Hassan, K.A.; Wistuba, I.; Penault-Llorca, F.; Adam, J.; Deutsch, E.; Soria, J.C. Lung cancer stem cell: Fancy conceptual model of tumor biology or cornerstone of a forthcoming therapeutic breakthrough? J. Thorac. Oncol. 2014, 9, 7–17. [Google Scholar] [CrossRef]
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar] [PubMed]
- Smalley, M.; Ashworth, A. Stem cells and breast cancer: A field in transit. Nat. Rev. Cancer 2003, 3, 832–844. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.F.B.; Jackson, E.L.; Woolfenden, A.E.; Lawrence, S.; Babar, I.; Vogel, S.; Crowley, D.; Bronson, R.T.; Jacks, T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 2005, 121, 823–835. [Google Scholar] [CrossRef]
- Ricci-Vitiani, L.; Lombardi, D.G.; Pilozzi, E.; Biffoni, M.; Todaro, M.; Peschle, C.; De Maria, R. Identification and expansion of human colon-cancer-initiating cells. Nature 2007, 445, 111–115. [Google Scholar] [CrossRef]
- Li, C.; Heidt, D.G.; Dalerba, P.; Burant, C.F.; Zhang, L.; Adsay, V.; Wicha, M.; Clarke, M.F.; Simeone, D.M. Identification of pancreatic cancer stem cells. Cancer Res. 2007, 67, 1030–1037. [Google Scholar] [CrossRef]
- He, S.; Nakada, D.; Morrison, S.J. Mechanisms of stem cell self-renewal. Annu. Rev. Cell Dev. Biol. 2009, 25, 377–406. [Google Scholar]
- Cheng, L.; Zhang, S.; Davidson, D.D.; Montironi, R.; Lopez-beltran, A. Implications of Cancer Stem Cells for Cancer Therapy. In Stem Cells and Cancer; Springer: Framingham, MA, USA, 2009; pp. 255–262. [Google Scholar]
- Meacham, C.E.; Morrison, S.J. Tumour heterogeneity and cancer cell plasticity. Nature 2013, 501, 328–337. [Google Scholar] [CrossRef]
- Kreso, A.; Dick, J.E. Evolution of the Cancer Stem Cell Model. Cell Stem Cell 2014, 14, 275–291. [Google Scholar] [CrossRef]
- Knoblich, J.A. Mechanisms of asymmetric stem cell division. Cell 2008, 132, 583–597. [Google Scholar] [CrossRef]
- Ragoussis, J. Regulators of Asymmetric Cell Division in Breast Cancer. Trends Cancer 2018, 4, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Seyfrid, M.; Bobrowski, D.; Bakhshinyan, D.; Tatari, N.; Venugopal, C.; Singh, S.K. In Vitro Self-Renewal Assays for Brain Tumor Stem Cells. Methods Mol. Biol. 2019, 1869, 79–84. [Google Scholar] [PubMed]
- Bayik, D.; Lathia, J.D. Cancer stem cell-immune cell crosstalk in tumour progression. Nat. Rev. Cancer 2021, 21, 526–536. [Google Scholar] [CrossRef] [PubMed]
- Moore, N.; Lyle, S. Quiescent, slow-cycling stem cell populations in cancer: A review of the evidence and discussion of significance. J. Oncol. 2011, 2011, 396076. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Dong, J.; Haiech, J.; Kilhoffer, M.-C.; Zeniou, M. Cancer Stem Cell Quiescence and Plasticity as Major Challenges in Cancer Therapy. Stem Cells Int. 2016, 2016, 1740936. [Google Scholar] [CrossRef] [PubMed]
- Moitra, K.; Lou, H.; Dean, M. Multidrug efflux pumps and cancer stem cells: Insights into multidrug resistance and therapeutic development. Clin. Pharmacol. Ther. 2011, 89, 491–502. [Google Scholar] [CrossRef]
- Moitra, K. Overcoming Multidrug Resistance in Cancer Stem Cells. Biomed. Res. Int. 2015, 2015, 635745. [Google Scholar] [CrossRef]
- Dontu, G.; Liu, S.; Wicha, M.S. Stem cells in mammary development and carcinogenesis: Implications for prevention and treatment. Stem Cell Rev. 2005, 1, 207–213. [Google Scholar] [CrossRef]
- Takebe, N.; Miele, L.; Harris, P.J.; Jeong, W.; Bando, H.; Yang, S.X.; Ivy, S.P. Targeting Notch, Hedgehog, and Wnt Pathways in cancer stem cells: Clinical update. Nat. Rev. Clin. Oncol. 2015, 12, 445–464. [Google Scholar] [CrossRef]
- Lambert, A.W.; Weinberg, R.A. Linking EMT programmes to normal and neoplastic epithelial stem cells. Nat. Rev. Cancer 2021, 21, 325–338. [Google Scholar] [CrossRef]
- Medema, J.P. Cancer stem cells: The challenges ahead. Nat. Cell Biol. 2013, 15, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Z.; White, J.; Zhang, L. Lung cancer stem cells and implications for future therapeutics. Cell. Biochem. Biophys. 2014, 69, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Mannelli, G.; Gallo, O. Cancer stem cells hypothesis and stem cells in head and neck cancers. Cancer Treat. Rev. 2012, 38, 515–539. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Wu, S.; Tang, W.; Qian, H.; Zhou, H.; Guo, T. Reduced SLC27A2 induces cisplatin resistance in lung cancer stem cells by negatively regulating Bmi1-ABCG2 signaling. Mol. Carcinog. 2015, 55, 1822–1832. [Google Scholar] [CrossRef]
- Monzani, E.; Facchetti, F.; Galmozzi, E.; Corsini, E.; Benetti, A.; Cavazzin, C.; Gritti, A.; Piccinini, A.; Porro, D.; Santinami, M.; et al. Melanoma contains CD133 and ABCG2 positive cells with enhanced tumourigenic potential. Eur. J. Cancer 2007, 43, 935–946. [Google Scholar] [CrossRef]
- Jia, Q.; Zhang, X.; Deng, T.; Gao, J. Positive correlation of Oct4 and ABCG2 to chemotherapeutic resistance in CD90(+)CD133(+) liver cancer stem cells. Cell Reprogram. 2013, 15, 143–150. [Google Scholar] [CrossRef]
- Herpel, E.; Jensen, K.; Muley, T.; Warth, A.; Schnabel, P.A.; Meister, M.; Herth, F.J.F.; Dienemann, H.; Thomas, M.; Gottschling, S. The cancer stem cell antigens CD133, BCRP1/ABCG2 and CD117/c-KIT are not associated with prognosis in resected early-stage non-small cell lung cancer. Anticancer Res. 2011, 31, 4491–4500. [Google Scholar]
- Margaritescu, C.; Pirici, D.; Cherciu, I.; Barbalan, A.; Cartana, T.; Saftoiu, A. CD133/CD166/Ki-67 triple immunofluorescence assessment for putative cancer stem cells in colon carcinoma. J. Gastrointestin. Liver Dis. 2014, 23, 161–170. [Google Scholar] [CrossRef]
- Yan, M.; Yang, X.; Wang, L.; Clark, D.; Zuo, H.; Ye, D.; Chen, W.; Zhang, P. Plasma Membrane Proteomics of Tumor Spheres Identify CD166 as a Novel Marker for Cancer Stem-like Cells in Head and Neck Squamous Cell Carcinoma. Mol. Cell. Proteom. 2013, 12, 3271–3284. [Google Scholar] [CrossRef]
- Jiao, J.; Hindoyan, A.; Wang, S.; Tran, L.M.; Goldstein, A.S.; Lawson, D.; Chen, D.; Li, Y.; Guo, C.; Zhang, B.; et al. Identification of CD166 as a surface marker for enriching prostate stem/progenitor and cancer initiating cells. PLoS ONE 2012, 7, e42564. [Google Scholar] [CrossRef]
- Koren, A.; Rijavec, M.; Kern, I.; Sodja, E.; Korosec, P.; Cufer, T. BMI1, ALDH1A1, and CD133 Transcripts Connect Epithelial-Mesenchymal Transition to Cancer Stem Cells in Lung Carcinoma. Stem Cells Int. 2016, 2016, 9714315. [Google Scholar] [CrossRef] [PubMed]
- Reuben, J.M.; Lee, B.-N.; Gao, H.; Cohen, E.N.; Mego, M.; Giordano, A.; Wang, X.; Lodhi, A.; Krishnamurthy, S.; Hortobagyi, G.N.; et al. Primary breast cancer patients with high risk clinicopathologic features have high percentages of bone marrow epithelial cells with ALDH activity and CD44(+)CD24lo cancer stem cell phenotype. Eur. J. Cancer 2011, 47, 1527–1536. [Google Scholar] [CrossRef] [PubMed]
- Karimi-Busheri, F.; Zadorozhny, V.; Li, T.; Lin, H.; Shawler, D.L.; Fakhrai, H. Pivotal role of CD38 biomarker in combination with CD24, EpCAM, and ALDH for identification of H460 derived lung cancer stem cells. J. Stem Cells 2011, 6, 9–20. [Google Scholar] [PubMed]
- Boonyaratanakornkit, J.B.; Yue, L.; Strachan, L.R.; Scalapino, K.J.; LeBoit, P.E.; Lu, Y.; Leong, S.P.; Smith, J.E.; Ghadially, R. Selection of tumorigenic melanoma cells using ALDH. J. Investig. Dermatol. 2010, 130, 2799–2808. [Google Scholar] [CrossRef]
- Sullivan, J.P.; Spinola, M.; Dodge, M.; Raso, M.G.; Behrens, C.; Gao, B.; Schuster, K.; Shao, C.; Larsen, J.E.; Sullivan, L.A.; et al. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on Notch signaling. Cancer Res. 2010, 70, 9937–9948. [Google Scholar] [CrossRef]
- Mansour, S.F.; Atwa, M.M. Clinicopathological Significance of CD133 and ALDH1 Cancer Stem Cell Marker Expression in Invasive Ductal Breast Carcinoma. Asian Pac. J. Cancer Prev. 2015, 16, 7491–7496. [Google Scholar] [CrossRef]
- Liu, S.; Liu, C.; Min, X.; Ji, Y.; Wang, N.; Liu, D.; Cai, J.; Li, K. Prognostic Value of Cancer Stem Cell Marker Aldehyde Dehydrogenase in Ovarian Cancer: A Meta-Analysis. PLoS ONE 2013, 8, e81050. [Google Scholar]
- Zhang, S.; Han, Z.; Jing, Y.; Tao, S.; Li, T.; Wang, H.; Wang, Y.; Li, R.; Yang, Y.; Zhao, X.; et al. CD133(+)CXCR4(+) colon cancer cells exhibit metastatic potential and predict poor prognosis of patients. BMC Med. 2012, 10, 85. [Google Scholar] [CrossRef]
- Kahlert, U.D.; Bender, N.O.; Maciaczyk, D.; Bogiel, T.; Bar, E.E.; Eberhart, C.G.; Nikkhah, G.; Maciaczyk, J. CD133/CD15 defines distinct cell subpopulations with differential in vitro clonogenic activity and stem cell-related gene expression profile in in vitro propagated glioblastoma multiforme-derived cell line with a PNET-like component. Folia Neuropathol. 2012, 50, 357–368. [Google Scholar] [CrossRef]
- Erhart, F.; Blauensteiner, B.; Zirkovits, G.; Printz, D.; Soukup, K.; Klingenbrunner, S.; Fischhuber, K.; Reitermaier, R.; Halfmann, A.; Lotsch, D.; et al. Gliomasphere marker combinatorics: Multidimensional flow cytometry detects CD44+/CD133+/ITGA6+/CD36+ signature. J. Cell. Mol. Med. 2019, 23, 281–292. [Google Scholar] [CrossRef]
- Sarvi, S.; Mackinnon, A.C.; Avlonitis, N.; Bradley, M.; Rintoul, R.C.; Rassl, D.M.; Wang, W.; Forbes, S.J.; Gregory, C.D.; Sethi, T. CD133+ cancer stem-like cells in small cell lung cancer are highly tumorigenic and chemoresistant but sensitive to a novel neuropeptide antagonist. Cancer Res. 2014, 74, 1554–1565. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Convery, P.A.; Matsumura, N.; Whitaker, R.S.; Kondoh, E.; Perry, T.; Huang, Z.; Bentley, R.C.; Mori, S.; Fujii, S.; et al. Epigenetic regulation of CD133 and tumorigenicity of CD133+ ovarian cancer cells. Oncogene 2009, 28, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, J.-N.; Zeng, T.-T.; He, F.; Chen, S.-P.; Ma, S.; Bi, J.; Zhu, X.-F.; Guan, X.-Y. CD133+ liver cancer stem cells resist interferon-gamma-induced autophagy. BMC Cancer 2016, 16, 15. [Google Scholar] [CrossRef] [PubMed]
- Todaro, M.; Gaggianesi, M.; Catalano, V.; Benfante, A.; Iovino, F.; Biffoni, M.; Apuzzo, T.; Sperduti, I.; Volpe, S.; Cocorullo, G.; et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell 2014, 14, 342–356. [Google Scholar] [CrossRef]
- Herreros-Pomares, A.; De-Maya-Girones, J.D.; Calabuig-Fariñas, S.; Lucas, R.; Martínez, A.; Pardo-Sánchez, J.M. Lung tumorspheres reveal cancer stem cell-like properties and a score with prognostic impact in resected non-small-cell lung cancer. Cell Death Dis. 2019, 10, 660. [Google Scholar] [CrossRef]
- Yan, Y.; Zuo, X.; Wei, D.; Cells, C.S. Concise Review: Emerging Role of CD44 in Cancer Stem Cells: A Promising Biomarker and Therapeutic Target. Stem Cells Transl. Med. 2015, 4, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Sahlberg, S.H.; Spiegelberg, D.; Glimelius, B.; Stenerlöw, B.; Nestor, M. Evaluation of cancer stem cell markers CD133, CD44, CD24: Association with AKT isoforms and radiation resistance in colon cancer cells. PLoS ONE 2014, 9, e94621. [Google Scholar] [CrossRef]
- de Beca, F.F.; Caetano, P.; Gerhard, R.; Alvarenga, C.A.; Gomes, M.; Paredes, J.; Schmitt, F. Cancer stem cells markers CD44, CD24 and ALDH1 in breast cancer special histological types. J. Clin. Pathol. 2013, 66, 187–191. [Google Scholar] [CrossRef]
- Yoon, C.; Park, D.J.; Schmidt, B.; Thomas, J.; Lee, H.-J.; Kim, T.S.; Janjigian, Y.Y.; Cohen, D.J. CD44 expression denotes a subpopulation of gastric cancer cells in which Hedgehog signaling promotes chemotherapy resistance. Clin. Cancer Res. 2014, 20, 3974–3988. [Google Scholar] [CrossRef]
- Yan, X.; Luo, H.U.; Zhou, X.; Zhu, B.; Wang, Y.; Bian, X. Identification of CD90 as a marker for lung cancer stem cells in A549 and H446 cell lines. Oncol. Rep. 2013, 30, 2733–2740. [Google Scholar] [CrossRef]
- Patriarca, C.; Macchi, R.M.; Marschner, A.K.; Mellstedt, H. Epithelial cell adhesion molecule expression (CD326) in cancer: A short review. Cancer Treat. Rev. 2012, 38, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Cozzi, P.; Hao, J.; Beretov, J.; Chang, L.; Duan, W.; Shigdar, S.; Delprado, W.; Graham, P.; Bucci, J.; et al. Epithelial cell adhesion molecule (EpCAM) is associated with prostate cancer metastasis and chemo/radioresistance via the PI3K/Akt/mTOR signaling pathway. Int. J. Biochem. Cell Biol. 2013, 45, 2736–2748. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.A.; Jiang, W.G. Evaluation of the expression of stem cell markers in human breast cancer reveals a correlation with clinical progression and metastatic disease in ductal carcinoma. Oncol. Rep. 2014, 31, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Hoogland, A.M.; Verhoef, E.I.; Roobol, M.J.; Schroder, F.H.; Wildhagen, M.F.; van der Kwast, T.H. Validation of stem cell markers in clinical prostate cancer: Alpha6-integrin is predictive for non-aggressive disease. Prostate 2014, 74, 488–496. [Google Scholar] [CrossRef]
- Haraguchi, N.; Ishii, H.; Mimori, K.; Ohta, K.; Uemura, M.; Nishimura, J.; Hata, T.; Takemasa, I.; Mizushima, T.; Yamamoto, H.; et al. CD49f-positive cell population efficiently enriches colon cancer-initiating cells. Int. J. Oncol. 2013, 43, 425–430. [Google Scholar] [CrossRef]
- Ajani, J.A.; Song, S.; Hochster, H.S.; Steinberg, I.B. Cancer stem cells: The promise and the potential. Semin. Oncol. 2015, 42 (Suppl. 1), S3–S17. [Google Scholar] [CrossRef]
- Borggrefe, T.; Oswald, F. The Notch signaling pathway: Transcriptional regulation at Notch target genes. Cell. Mol. Life Sci. 2009, 66, 1631–1646. [Google Scholar] [CrossRef]
- Takebe, N.; Nguyen, D.; Yang, S.X. Targeting notch signaling pathway in cancer: Clinical development advances and challenges. Pharmacol. Ther. 2014, 141, 140–149. [Google Scholar] [CrossRef]
- Sun, W.; Gaykalova, D.A.; Ochs, M.F.; Mambo, E.; Liu, Y.; Loyo, M.; Agrawal, N.; Howard, J.; Li, R.; Fertig, E.; et al. Activation of the NOTCH pathway in head and neck cancer. Cancer Res. 2014, 74, 1091–1104. [Google Scholar] [CrossRef]
- Vinson, K.E.; George, D.C.; Fender, A.W.; Bertrand, F.E.; Sigounas, G. The Notch pathway in colorectal cancer. Int. J. Cancer 2016, 138, 1835–1842. [Google Scholar] [CrossRef]
- Xiao, M.J.; Han, Z.; Shao, B.; Jin, K. Notch signaling and neurogenesis in normal and stroke brain. Int. J. Physiol. Pathophysiol. Pharmacol. 2009, 1, 192–202. [Google Scholar] [PubMed]
- Penton, A.L.; Leonard, L.D.; Spinner, N.B. Notch signaling in human development and disease. Semin. Cell Dev. Biol. 2012, 23, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Westhoff, B.; Colaluca, I.N.; D’Ario, G.; Donzelli, M.; Tosoni, D.; Volorio, S.; Pelosi, G.; Spaggiari, L.; Mazzarol, G.; Viale, G.; et al. Alterations of the Notch pathway in lung cancer. Proc. Natl. Acad. Sci. USA 2009, 106, 22293–22298. [Google Scholar] [CrossRef]
- Yu, Z.; Pestell, T.; Lisanti, M.P. Cancer Stem Cells. Int. J. Biochem. Cell. Biol. 2012, 44, 2144–2151. [Google Scholar] [CrossRef] [PubMed]
- Egloff, A.M.; Grandis, J.R. Molecular Pathways: Context-dependent approaches to Notch targeting as cancer therapy. Clin. Cancer Res. 2012, 18, 5188–5195. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, T.C.; Smith, S.D.; Sklar, J. Analysis of DNA surrounding the breakpoints of chromosomal translocations involving the beta T cell receptor gene in human lymphoblastic neoplasms. Cell 1987, 50, 107–117. [Google Scholar] [CrossRef]
- Bhola, N.E.; Jansen, V.M.; Koch, J.P.; Li, H.; Formisano, L.; Williams, J.A.; Grandis, J.R.; Arteaga, C.L. Treatment of Triple-Negative Breast Cancer with TORC1/2 Inhibitors Sustains a Drug-Resistant and Notch-Dependent Cancer Stem Cell Population. Cancer Res. 2016, 76, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Yuan, R.; Ke, J.; Sun, L.; He, Z.; Zou, Y.; He, X.; Chen, Y.; Wu, X.X.; Cai, Z.; Wang, L.; et al. HES1 promotes metastasis and predicts poor survival in patients with colorectal cancer. Clin. Exp. Metastasis 2015, 32, 169–179. [Google Scholar] [CrossRef]
- Kushwah, R.; Guezguez, B.; Lee, J.B.; Hopkins, C.I.; Bhatia, M. Pleiotropic roles of Notch signaling in normal, malignant, and developmental hematopoiesis in the human. EMBO Rep. 2014, 15, 1128–1138. [Google Scholar] [CrossRef]
- Stewart, D.J. Wnt signaling pathway in non-small cell lung cancer. J. Natl. Cancer Inst. 2014, 106, 1–11. [Google Scholar] [CrossRef]
- Allen, T.D.; Rodriguez, E.M.; Jones, K.D.; Bishop, J.M. Activated NOTCH1 induces lung adenomas in mice and cooperates with MYC in the generation of lung adenocarcinoma. Cancer Res. 2011, 71, 6010–6018. [Google Scholar] [CrossRef]
- Clevers, H.; Nusse, R. Wnt/B-catenin signaling and disease. Cell 2012, 149, 1192–1205. [Google Scholar] [CrossRef] [PubMed]
- Anastas, J.N.; Moon, R.T. WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer 2012, 13, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Enzo, M.V.; Rastrelli, M.; Rossi, C.R.; Hladnik, U.; Segat, D. The Wnt/β-catenin pathway in human fibrotic-like diseases and its eligibility as a therapeutic target. Mol. Cell. Ther. 2015, 3, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Purro, S.A.; Galli, S.; Salinas, P.C. Dysfunction of Wnt signaling and synaptic disassembly in neurodegenerative diseases. J. Mol. Cell Biol. 2014, 6, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Niehrs, C. The complex world of WNT receptor signalling. Nat. Rev. Mol. Cell Biol. 2012, 13, 767–779. [Google Scholar] [CrossRef]
- Sokol, S.Y. Spatial and temporal aspects of Wnt signaling and planar cell polarity during vertebrate embryonic development. Semin. Cell Dev. Biol. 2015, 42, 78–85. [Google Scholar] [CrossRef]
- Viale-Bouroncle, S.; Klingelhoffer, C.; Ettl, T.; Reichert, T.E.; Morsczeck, C. A protein kinase A (PKA)/beta-catenin pathway sustains the BMP2/DLX3-induced osteogenic differentiation in dental follicle cells (DFCs). Cell Signal 2015, 27, 598–605. [Google Scholar] [CrossRef]
- Kim, S.; Nie, H.; Nesin, V.; Tran, U.; Outeda, P.; Bai, C.-X.; Keeling, J.; Maskey, D.; Watnick, T.; Wessely, O.; et al. The polycystin complex mediates Wnt/Ca2+ signalling. Nat. Cell Biol. 2016, 18, 752–764. [Google Scholar] [CrossRef]
- Mezzacappa, C.; Komiya, Y.; Habas, R. Activation and Function of Small GTPases Rho, Rac, and Cdc42 During Gastrulation. Methods Mol. Biol. 2012, 839, 119–131. [Google Scholar] [CrossRef]
- Puvirajesinghe, T.M.; Bertucci, F.; Jain, A.; Scerbo, P.; Belotti, E.; Restouin, A.; Macara, I.; Birnbaum, D.; Marchetto, S.; Collette, Y.; et al. Identification of p62/SQSTM1 as a component of non-canonical Wnt VANGL2–JNK signalling in breast cancer. Nat. Commun. 2016, 7, 10318. [Google Scholar] [CrossRef] [PubMed]
- Kahn, M. Can we safely target the WNT pathway? Nat. Rev. Drug Discov. 2014, 13, 513–532. [Google Scholar] [CrossRef] [PubMed]
- Nusse, R.; Varmus, H.E. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell 1982, 31, 99–109. [Google Scholar] [CrossRef]
- Korinek, V.; Barker, N.; Morin, P.J.; van Wichen, D.; de Weger, R.; Kinzler, K.W.; Vogelstein, B.; Clevers, H. Constitutive Transcriptional Activation by a β-Catenin-Tcf Complex in APC−/− Colon Carcinoma. Science 1997, 275, 1784–1787. [Google Scholar] [CrossRef] [PubMed]
- Morin, P.J.; Sparks, A.B.; Korinek, V.; Barker, N.; Clevers, H.; Vogelstein, B.; Kinzler, K.W. Activation of β-Catenin-Tcf Signaling in Colon Cancer by Mutations in β-Catenin or APC. Science 1997, 275, 1787–1790. [Google Scholar] [CrossRef]
- Cao, X.; Eu, K.W.; Seow-Choen, F.; Cheah, P.Y. Germline mutations are frequent in the APC gene but absent in the beta-catenin gene in familial adenomatous polyposis patients. Genes Chromosomes Cancer 1999, 25, 396–398. [Google Scholar] [CrossRef]
- Jiang, H.-L.; Jiang, L.-M.; Han, W.-D. Wnt/beta-catenin signaling pathway in lung cancer stem cells is a potential target for the development of novel anticancer drugs. J. BUON 2015, 20, 1094–1100. [Google Scholar]
- Nakashima, N.; Liu, D.; Huang, C.-L.; Ueno, M.; Zhang, X.; Yokomise, H. Wnt3 gene expression promotes tumor progression in non-small cell lung cancer. Lung Cancer 2012, 76, 228–234. [Google Scholar] [CrossRef]
- Gonnissen, A.; Isebaert, S.; Haustermans, K. Targeting the Hedgehog signaling pathway in cancer: Beyond Smoothened. Oncotarget 2015, 6, 13899–13913. [Google Scholar] [CrossRef]
- Gupta, S.; Takebe, N.; Lorusso, P. Targeting the Hedgehog pathway in cancer. Ther. Adv. Med. Oncol. 2010, 2, 237–250. [Google Scholar] [CrossRef]
- Jung, B.; Padula, D.; Burtscher, I.; Landerer, C.; Lutter, D.; Theis, F.; Messias, A.C.; Geerlof, A.; Sattler, M.; Kremmer, E.; et al. Pitchfork and Gprasp2 target smoothened to the primary cilium for hedgehog pathway activation. PLoS ONE 2016, 11, e0149477. [Google Scholar] [CrossRef] [PubMed]
- Hahn, H.; Wicking, C.; Zaphiropoulos, P.G.; Gailani, M.R.; Shanley, S.; Chidambaram, A.; Vorechovsky, I.; Holmberg, E.; Unden, A.B.; Gillies, S.; et al. Mutations of the Human Homolog of Drosophila patched in the Nevoid Basal Cell Carcinoma Syndrome. Cell 1996, 85, 841–851. [Google Scholar] [CrossRef]
- Thalakoti, S.; Geller, T. Basal cell nevus syndrome or Gorlin syndrome. Handb. Clin. Neurol. 2015, 132, 119–128. [Google Scholar] [PubMed]
- Shanley, S.; McCormack, C. Diagnosis and Management of Hereditary Basal Cell Skin Cancer. Rare Hered. Cancers 2016, 205, 191–212. [Google Scholar]
- Cochrane, C.; Szczepny, A.; Watkins, D.; Cain, J. Hedgehog Signaling in the Maintenance of Cancer Stem Cells. Cancers 2015, 7, 1554–1585. [Google Scholar] [CrossRef] [PubMed]
- Cordenonsi, M.; Zanconato, F.; Azzolin, L.; Forcato, M.; Rosato, A.; Frasson, C.; Inui, M.; Montagner, M.; Parenti, A.R.; Poletti, A.; et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 2011, 147, 759–772. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y.; Zhu, Y.; Yuan, C.; Wang, D.; Zhang, W.; Qi, B.; Qiu, J.; Song, X.; Ye, J.; et al. The Hippo transducer TAZ promotes epithelial to mesenchymal transition and cancer stem cell maintenance in oral cancer. Mol. Oncol. 2015, 9, 1091–1105. [Google Scholar] [CrossRef] [PubMed]
- Mo, J.-S.; Park, H.W.; Guan, K.-L. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014, 15, 642–656. [Google Scholar] [CrossRef]
- Moroishi, T.; Hansen, C.G.; Guan, K. The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer 2015, 15, 73–79. [Google Scholar] [CrossRef]
- Meng, Z.; Moroishi, T.; Mottier-Pavie, V.; Plouffe, S.W.; Hansen, C.G.; Hong, A.W.; Park, H.W.; Mo, J.-S.; Lu, W.; Lu, S.; et al. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat. Commun. 2015, 6, 8357. [Google Scholar] [CrossRef]
- Harvey, K.F.; Zhang, X.; Thomas, D.M. The Hippo pathway and human cancer. Nat. Rev. Cancer 2013, 13, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.-X.; Zhao, B.; Guan, K.-L. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 2015, 163, 811–828. [Google Scholar] [CrossRef] [PubMed]
- Steinhardt, A.A.; Gayyed, M.F.; Klein, A.P.; Dong, J.; Maitra, A.; Pan, D.; Montgomery, E.A.; Anders, R.A. Expression of Yes-associated protein in common solid tumors. Hum. Pathol. 2008, 39, 1582–1589. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.-G.; Gumbiner, B.M. Adhesion to fibronectin regulates Hippo signaling via the FAK–Src–PI3K pathway. J. Cell Biol. 2015, 210, 503–515. [Google Scholar] [CrossRef]
- Sorrentino, G.; Ruggeri, N.; Zannini, A.; Ingallina, E.; Bertolio, R.; Marotta, C.; Neri, C.; Cappuzzello, E.; Forcato, M.; Rosato, A.; et al. Glucocorticoid receptor signalling activates YAP in breast cancer. Nat. Commun. 2017, 8, 14073. [Google Scholar] [CrossRef]
- Wang, Y.; Dong, Q.; Zhang, Q.; Li, Z.; Wang, E.; Qiu, X. Overexpression of yes-associated protein contributes to progression and poor prognosis of non-small-cell lung cancer. Cancer Sci. 2010, 101, 1279–1285. [Google Scholar] [CrossRef]
- Lau, A.N.; Curtis, S.J.; Fillmore, C.M.; Rowbotham, S.P.; Mohseni, M.; Wagner, D.E.; Beede, A.M.; Montoro, D.T.; Sinkevicius, K.W.; Walton, Z.E.; et al. Tumor-propagating cells and Yap/Taz activity contribute to lung tumor progression and metastasis. EMBO J. 2014, 33, 468–481. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhong, W.; Ma, G.; Zhang, B.; Tian, H. Yes-associated protein regulates the growth of human non-small cell lung cancer in response to matrix stiffness. Mol. Med. Rep. 2015, 11, 4267–4272. [Google Scholar] [CrossRef]
- Song, S.; Ajani, J.A.; Honjo, S.; Maru, D.M.; Chen, Q.; Scott, A.W.; Heallen, T.R.; Xiao, L.; Hofstetter, W.L.; Weston, B.; et al. Hippo coactivator YAP1 upregulates SOX9 and endows esophageal cancer cells with stem-like properties. Cancer Res. 2014, 74, 4170–4182. [Google Scholar] [CrossRef]
- Basu-Roy, U.; Bayin, N.S.; Rattanakorn, K.; Han, E.; Placantonakis, D.G.; Mansukhani, A.; Basilico, C. Sox2 antagonizes the Hippo pathway to maintain stemness in cancer cells. Nat. Commun. 2015, 6, 6411. [Google Scholar] [CrossRef]
- Bhat, K.P.L.; Salazar, K.L.; Balasubramaniyan, V.; Wani, K.; Heathcock, L.; Hollingsworth, F.; James, J.D.; Gumin, J.; Diefes, K.L.; Kim, S.H.; et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011, 25, 2594–2609. [Google Scholar] [CrossRef] [PubMed]
- Murakami, H.; Mizuno, T.; Taniguchi, T.; Fujii, M.; Ishiguro, F.; Fukui, T.; Akatsuka, S.; Horio, Y.; Hida, T.; Kondo, Y.; et al. LATS2 is a tumor suppressor gene of malignant mesothelioma. Cancer Res. 2011, 71, 873–883. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.M.; Liu, W.W.; Liu, C.J.; Wen, C.; Lu, H.F.; Wan, F.S. Mst1 overexpression inhibited the growth of human non-small cell lung cancer in vitro and in vivo. Cancer Gene Ther. 2013, 20, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Duzagac, F.; Inan, S.; Ela Simsek, F.; Acikgoz, E.; Guven, U.; Khan, S.A.; Rouhrazi, H.; Oltulu, F.; Aktug, H.; Erol, A.; et al. JAK/STAT pathway interacts with intercellular cell adhesion molecule (ICAM) and vascular cell adhesion molecule (VCAM) while prostate cancer stem cells form tumor spheroids. J. BUON 2015, 20, 1250–1257. [Google Scholar]
- Chung, S.S.; Vadgama, J.V. Curcumin and epigallocatechin gallate inhibit the cancer stem cell phenotype via down-regulation of STAT3-NFkappaB signaling. Anticancer Res. 2015, 35, 39–46. [Google Scholar]
- Dutta, P.; Sabri, N.; Li, J.; Li, W.X. Role of STAT3 in lung cancer. Jak-Stat 2014, 3, e999503. [Google Scholar] [CrossRef]
- Patel, N.; Baranwal, S.; Patel, B.B. A Strategic Approach to Identification of Selective Inhibitors of Cancer Stem Cells. Methods Mol. Biol. 2022, 2303, 765–777. [Google Scholar]
- Reynolds, B.A.; Weiss, S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 1992, 255, 1707–1710. [Google Scholar] [CrossRef]
- Bielecka, Z.F.; Maliszewska-Olejniczak, K.; Safir, I.J.; Szczylik, C.; Czarnecka, A.M. Three-dimensional cell culture model utilization in cancer stem cell research. Biol. Rev. Camb. Philos. Soc. 2016, 92, 1505–1520. [Google Scholar] [CrossRef]
- Eramo, A.; Lotti, F.; Sette, G.; Pilozzi, E.; Biffoni, M.; Di, V.A.; Conticello, C.; Ruco, L.; Peschle, C.; De, M.R. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008, 15, 504–514. [Google Scholar] [CrossRef]
- Qiu, X.; Wang, Z.; Li, Y.; Miao, Y.; Ren, Y.; Luan, Y. Characterization of sphere-forming cells with stem-like properties from the small cell lung cancer cell line H446. Cancer Lett. 2012, 323, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Ghani, F.I.; Yamazaki, H.; Iwata, S.; Okamoto, T.; Aoe, K.; Okabe, K.; Mimura, Y.; Fujimoto, N.; Kishimoto, T.; Yamada, T.; et al. Identification of cancer stem cell markers in human malignant mesothelioma cells. Biochem. Biophys. Res. Commun. 2011, 404, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Zakaria, N.; Yusoff, N.M.; Zakaria, Z.; Lim, M.N.; Baharuddin, P.J.N.; Fakiruddin, K.S.; Yahaya, B. Human non-small cell lung cancer expresses putative cancer stem cell markers and exhibits the transcriptomic profile of multipotent cells. BMC Cancer 2015, 15, 84. [Google Scholar] [CrossRef] [PubMed]
- Dou, J.; Pan, M.; Wen, P.; Li, Y.; Tang, Q.; Chu, L.; Zhao, F.; Jiang, C.; Hu, W.; Hu, K.; et al. Isolation and identification of cancer stem-like cells from murine melanoma cell lines. Cell. Mol. Immunol. 2007, 4, 467–472. [Google Scholar] [PubMed]
- Zhang, W.C.; Shyh-Chang, N.; Yang, H.; Rai, A.; Umashankar, S.; Ma, S.; Soh, B.S.; Sun, L.L.; Tai, B.C.; Nga, M.E.; et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell 2012, 148, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-G.; Jiang, A.-G.; Lu, H.-Y.; Zhang, L.-X.; Gao, X.-Y. Isolation, cultivation and identification of human lung adenocarcinoma stem cells. Oncol. Lett. 2015, 9, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Hashida, S.; Yamamoto, H.; Shien, K.; Miyoshi, Y.; Ohtsuka, T.; Suzawa, K.; Watanabe, M.; Maki, Y.; Soh, J.; Asano, H.; et al. Acquisition of cancer stem cell-like properties in non-small cell lung cancer with acquired resistance to afatinib. Cancer Sci. 2015, 106, 1377–1384. [Google Scholar] [CrossRef]
- Chaichana, K.; Zamora-Berridi, G.; Camara-Quintana, J.; Quinones-Hinojosa, A. Neurosphere assays: Growth factors and hormone differences in tumor and nontumor studies. Stem Cells 2006, 24, 2851–2857. [Google Scholar] [CrossRef]
- Pastrana, E.; Silva-Vargas, V.; Doetsch, F. Eyes Wide Open: A Critical Review of Sphere-Formation as an Assay For Stem Cells. Cell Stem Cell 2011, 8, 486–498. [Google Scholar] [CrossRef]
- Yakisich, J.S.; Azad, N.; Venkatadri, R.; Kulkarni, Y.; Wright, C.; Kaushik, V.; Iyer, A.K.V. Formation of Tumorspheres with Increased Stemness without External Mitogens in a Lung Cancer Model. Stem Cells Int. 2016, 2016, 5603135. [Google Scholar] [CrossRef]
- Calvet, C.Y.; Andre, F.M.; Mir, L.M. The culture of cancer cell lines as tumorspheres does not systematically result in cancer stem cell enrichment. PLoS ONE 2014, 9, e89644. [Google Scholar] [CrossRef] [PubMed]
- Weiswald, L.-B.; Bellet, D.; Dangles-Marie, V. Spherical cancer models in tumor biology. Neoplasia 2015, 17, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Singec, I.; Knoth, R.; Meyer, R.P.; Maciaczyk, J.; Volk, B.; Nikkhah, G.; Frotscher, M.; Snyder, E.Y. Defining the actual sensitivity and specificity of the neurosphere assay in stem cell biology. Nat. Methods 2006, 3, 801–806. [Google Scholar] [CrossRef] [PubMed]
- Rota, L.M.; Lazzarino, D.A.; Ziegler, A.N.; LeRoith, D.; Wood, T.L. Determining mammosphere-forming potential: Application of the limiting dilution analysis. J. Mammary Gland. Biol. Neoplasia 2012, 17, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Qureshi-Baig, K.; Ullmann, P.; Rodriguez, F.; Frasquilho, S.; Nazarov, P.V.; Haan, S.; Letellier, E. What Do We Learn from Spheroid Culture Systems? Insights from Tumorspheres Derived from Primary Colon Cancer Tissue. PLoS ONE 2016, 11, e0146052. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, H.; Okudela, K.; Yazawa, T.; Sato, H.; Shimoyamada, H. Cancer stem cell: Implications in cancer biology and therapy with special reference to lung cancer. Lung Cancer 2009, 66, 275–281. [Google Scholar] [CrossRef]
- Malanchi, I.; Santamaria-Martinez, A.; Susanto, E.; Peng, H.; Lehr, H.-A.A.; Delaloye, J.-F.F.; Huelsken, J. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature 2012, 481, 85–89. [Google Scholar] [CrossRef]
- Seo, J.; Park, S.-J.; Kim, J.; Choi, S.-J.; Moon, S.-H.; Chung, H.-M. Effective method for the isolation and proliferation of primary lung cancer cells from patient lung tissues. Biotechnol. Lett. 2013, 35, 1165–1174. [Google Scholar] [CrossRef]
- Kurpios, N.A.; Girgis-Gabardo, A.; Hallett, R.M.; Rogers, S.; Gludish, D.W.; Kockeritz, L.; Woodgett, J.; Cardiff, R.; Hassell, J.A. Single unpurified breast tumor-initiating cells from multiple mouse models efficiently elicit tumors in immune-competent hosts. PLoS ONE 2013, 8, e58151. [Google Scholar] [CrossRef]
- Wicha, M.S.; Liu, S.; Dontu, G. Cancer stem cells: An old idea—A paradigm shift. Cancer Res. 2006, 66, 1883–1890. [Google Scholar] [CrossRef]
- Koch, U.; Krause, M.; Baumann, M. Cancer stem cells at the crossroads of current cancer therapy failures—Radiation oncology perspective. Semin. Cancer Biol. 2010, 20, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Alison, M.R.; Lim, S.M.L.; Nicholson, L.J. Cancer stem cells: Problems for therapy? J. Pathol. 2011, 223, 148–162. [Google Scholar] [CrossRef] [PubMed]
- Herreros-Pomares, A.; Zhou, X.; Calabuig-Fariñas, S.; Lee, S.-J.; Torres, S.; Esworthy, T.; Hann, S.Y.; Jantus-Lewintre, E.; Camps, C.; Lijie Grace Zhang, L.G. 3D printing novel in vitro cancer cell culture model systems for lung cancer stem cell study. Mater. Sci. Eng. C 2021, 122, 111914. [Google Scholar] [CrossRef] [PubMed]
- Clara, J.A.; Monge, C.; Yang, Y.; Takebe, N. Targeting signalling pathways and the immune microenvironment of cancer stem cells—A clinical update. Nat. Rev. Clin. Oncol. 2020, 17, 204–232. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Shi, P.; Zhao, G.; Xu, J.; Peng, W.; Zhang, J.; Zhang, G.; Wang, X.; Dong, Z.; Chen, F.; et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target. Ther. 2020, 5, 8. [Google Scholar] [CrossRef]
- Galassi, C.; Vitale, I.; Galluzzi, L. Using epigenetic modifiers to target cancer stem cell immunoevasion. Cancer Cell 2021, 39, 1573–1575. [Google Scholar] [CrossRef]
- Herreros-Pomares, A.; Aguilar-Gallardo, C.; Calabuig-Fariñas, S.; Sirera, R.; Jantus-Lewintre, E.; Camps, C. EpCAM duality becomes this molecule in a new Dr. Jekyll and Mr. Hyde tale. Crit. Rev. Oncol. Hematol. 2018, 126, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Riethmuller, G.; Schneider-Gadicke, E.; Schlimok, G.; Schmiegel, W.; Raab, R.; Hoffken, K.; Gruber, R.; Pichlmaier, H.; Hirche, H.; Pichlmayr, R. Randomised trial of monoclonal antibody for adjuvant therapy of resected Dukes’ C colorectal carcinoma. German Cancer Aid 17-1A Study Group. Lancet 1994, 343, 1177–1183. [Google Scholar] [CrossRef]
- Riethmuller, G.; Holz, E.; Schlimok, G.; Schmiegel, W.; Raab, R.; Hoffken, K.; Gruber, R.; Funke, I.; Pichlmaier, H.; Hirche, H.; et al. Monoclonal antibody therapy for resected Dukes’ C colorectal cancer: Seven-year outcome of a multicenter randomized trial. J. Clin. Oncol. 1998, 16, 1788–1794. [Google Scholar] [CrossRef]
- Schmoll, H.-J.; Arnold, D. When wishful thinking leads to a misty-eyed appraisal: The story of the adjuvant colon cancer trials with edrecolomab. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009, 27, 1926–1929. [Google Scholar] [CrossRef]
- Niedzwiecki, D.; Bertagnolli, M.M.; Warren, R.S.; Compton, C.C.; Kemeny, N.E.; Benson, A.B., 3rd; Eckhardt, G.S.; Alberts, S.; Porjosh, G.N.; Kerr, D.J.; et al. Documenting the natural history of patients with resected stage II adenocarcinoma of the colon after random assignment to adjuvant treatment with edrecolomab or observation: Results from CALGB 9581. J. Clin. Oncol. 2011, 29, 3146–3152. [Google Scholar] [CrossRef] [PubMed]
- Stoelben, E.; Loibner, H.; Weder, W.; Schmoll, C.; Bijelovic, M.; Hasse, J. Adjuvant active vaccination with IGN101 in patients after radical lung cancer resection in stage Ib-IIIa—A prospective randomized, double-blind, placebo-controlled multicenter phase II/III study. Chir. Forum. 2008, 37, 329–331. [Google Scholar]
- Münz, M.; Murr, A.; Kvesic, M.; Rau, D.; Mangold, S.; Pflanz, S.; Lumsden, J.; Volkland, J.; Fagerberg, J.; Riethmüller, G.; et al. Side-by-side analysis of five clinically tested anti-EpCAM monoclonal antibodies. Cancer Cell Int. 2010, 10, 44. [Google Scholar] [CrossRef] [PubMed]
- Oberneder, R.; Weckermann, D.; Ebner, B.; Quadt, C.; Kirchinger, P.; Raum, T.; Locher, M.; Prang, N.; Baeuerle, P.A.; Leo, E. A phase I study with adecatumumab, a human antibody directed against epithelial cell adhesion molecule, in hormone refractory prostate cancer patients. Eur. J. Cancer 2006, 42, 2530–2538. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Scheulen, M.E.; Dittrich, C.; Obrist, P.; Marschner, N.; Dirix, L.; Schmidt, M.; Ruttinger, D.; Schuler, M.; Reinhardt, C.; et al. An open-label, randomized phase II study of adecatumumab, a fully human anti-EpCAM antibody, as monotherapy in patients with metastatic breast cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2010, 21, 275–282. [Google Scholar] [CrossRef]
- Marschner, N.; Ruttinger, D.; Zugmaier, G.; Nemere, G.; Lehmann, J.; Obrist, P. Phase II study of the human anti-epithelial cell adhesion molecule antibody adecatumumab in prostate cancer patients with increasing serum levels of prostate-specific antigen after radical prostatectomy. Urol. Int. 2010, 85, 386–395. [Google Scholar] [CrossRef]
- Schmidt, M.; Ruttinger, D.; Sebastian, M.; Hanusch, C.A.; Marschner, N.; Baeuerle, P.A.; Wolf, A.; Schmidt, M.; Abrahamsson, P.; Reinhardt, C.; et al. Phase IB study of the EpCAM antibody adecatumumab combined with docetaxel in patients with EpCAM-positive relapsed or refractory advanced-stage breast cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2012, 23, 2306–2313. [Google Scholar] [CrossRef]
- Jager, M.; Schoberth, A.; Ruf, P.; Hess, J.; Hennig, M.; Schmalfeldt, B.; Wimberger, P.; Strohlein, M.; Theissen, B.; Heiss, M.M.; et al. Immunomonitoring results of a phase II/III study of malignant ascites patients treated with the trifunctional antibody catumaxomab (anti-EpCAM x anti-CD3). Cancer Res. 2012, 72, 24–32. [Google Scholar] [CrossRef]
- Baeuerle, P.A.; Gires, O. EpCAM (CD326) finding its role in cancer. Br. J. Cancer 2007, 96, 417–423. [Google Scholar] [CrossRef]
- Kowalski, M.; Guindon, J.; Brazas, L.; Moore, C.; Entwistle, J.; Cizeau, J.; Jewett, M.A.S.; MacDonald, G.C. A phase II study of oportuzumab monatox: An immunotoxin therapy for patients with noninvasive urothelial carcinoma in situ previously treated with bacillus Calmette-Guerin. J. Urol. 2012, 188, 1712–1718. [Google Scholar] [CrossRef]
- Menke-van der Houven van Oordt, C.W.; Gomez-Roca, C.; van Herpen, C.; Coveler, A.L.; Mahalingam, D.; Verheul, H.M.W.; van der Graaf, W.T.A.; Christen, R.; Rüttinger, D.; Weigand, S.; et al. First-in-human phase I clinical trial of RG7356, an anti-CD44 humanized antibody, in patients with advanced, CD44-expressing solid tumors. Oncotarget 2016, 7, 80046–80058. [Google Scholar] [CrossRef] [PubMed]
- Riechelmann, H.; Sauter, A.; Golze, W.; Hanft, G.; Schroen, C.; Hoermann, K.; Erhardt, T.; Gronau, S. Phase I trial with the CD44v6-targeting immunoconjugate bivatuzumab mertansine in head and neck squamous cell carcinoma. Oral Oncol. 2008, 44, 823–829. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, M.; Wu, Z.; Tong, C.; Dai, H.; Guo, Y.; Liu, Y.; Huang, J.; Lv, H.; Luo, C.; et al. CD133-directed CAR T cells for advanced metastasis malignancies: A phase I trial. Oncoimmunology 2018, 7, e1440169. [Google Scholar] [CrossRef] [PubMed]
- Majumder, S.; Crabtree, J.S.; Golde, T.E.; Minter, L.M.; Osborne, B.A.; Miele, L. Targeting Notch in oncology: The path forward. Nat. Rev. Drug Discov. 2021, 20, 125–144. [Google Scholar] [CrossRef] [PubMed]
- Fouladi, M.; Stewart, C.F.; Olson, J.; Wagner, L.M.; Onar-Thomas, A.; Kocak, M.; Packer, R.J.; Goldman, S.; Gururangan, S.; Gajjar, A.; et al. Phase I trial of MK-0752 in children with refractory CNS malignancies: A pediatric brain tumor consortium study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2011, 29, 3529–3534. [Google Scholar] [CrossRef]
- Krop, I.; Demuth, T.; Guthrie, T.; Wen, P.Y.; Mason, W.P.; Chinnaiyan, P.; Butowski, N.; Groves, M.D.; Kesari, S.; Freedman, S.J.; et al. Phase I pharmacologic and pharmacodynamic study of the gamma secretase (Notch) inhibitor MK-0752 in adult patients with advanced solid tumors. J. Clin. Oncol. 2012, 30, 2307–2313. [Google Scholar] [CrossRef]
- Chen, X.; Gong, L.; Ou, R.; Zheng, Z.; Chen, J.; Xie, F.; Huang, X.; Qiu, J.; Zhang, W.; Jiang, Q.; et al. Sequential combination therapy of ovarian cancer with cisplatin and γ-secretase inhibitor MK-0752. Gynecol. Oncol. 2016, 140, 537–544. [Google Scholar] [CrossRef]
- van Groningen, T.; Akogul, N.; Westerhout, E.M.; Chan, A.; Hasselt, N.E.; Zwijnenburg, D.A.; Broekmans, M.; Stroeken, P.; Haneveld, F.; Hooijer, G.K.J.; et al. A NOTCH feed-forward loop drives reprogramming from adrenergic to mesenchymal state in neuroblastoma. Nat. Commun. 2019, 10, 1530. [Google Scholar] [CrossRef]
- Cook, N.; Basu, B.; Smith, D.-M.; Gopinathan, A.; Evans, J.; Steward, W.P.; Palmer, D.; Propper, D.; Venugopal, B.; Hategan, M.; et al. A phase I trial of the γ-secretase inhibitor MK-0752 in combination with gemcitabine in patients with pancreatic ductal adenocarcinoma. Br. J. Cancer 2018, 118, 793–801. [Google Scholar] [CrossRef]
- Zhang, S.; Chung, W.; Miele, L.; Xu, K. Targeting Met and Notch in the Lfng-deficient, Met-amplified triple-negative breast cancer. Cancer Biol. Ther. 2014, 15, 633–642. [Google Scholar] [CrossRef]
- Brana, I.; Berger, R.; Golan, T.; Haluska, P.; Edenfield, J.; Fiorica, J. A parallel-arm phase I trial of the humanised anti-IGF-1R antibody dalotuzumab in combination with the AKT inhibitor MK-2206, the mTOR inhibitor ridaforolimus, or the NOTCH inhibitor MK-0752, in patients with advanced solid tumours. Br. J. Cancer 2014, 111, 1932–1944. [Google Scholar] [CrossRef]
- Luistro, L.; He, W.; Smith, M.; Packman, K.; Vilenchik, M.; Carvajal, D.; Roberts, J.; Cai, J.; Berkofsky-Fessler, W.; Hilton, H.; et al. Preclinical profile of a potent gamma-secretase inhibitor targeting notch signaling with in vivo efficacy and pharmacodynamic properties. Cancer Res. 2009, 69, 7672–7680. [Google Scholar] [CrossRef]
- Tolcher, A.W.; Messersmith, W.A.; Mikulski, S.M.; Papadopoulos, K.P.; Kwak, E.L.; Gibbon, D.G.; Patnaik, A.; Falchook, G.S.; Dasari, A.; Shapiro, G.I.; et al. Phase I study of RO4929097, a gamma secretase inhibitor of notch signaling, in patients with refractory metastatic or locally advanced solid tumors. J. Clin. Oncol. 2012, 30, 2348–2353. [Google Scholar] [CrossRef] [PubMed]
- Richter, S.; Bedard, P.L.; Chen, E.X.; Clarke, B.A.; Tran, B.; Hotte, S.J.; Stathis, A.; Hirte, H.W.; Razak, A.R.A.; Reedijk, M.; et al. A phase I study of the oral gamma secretase inhibitor R04929097 in combination with gemcitabine in patients with advanced solid tumors (PHL-078/CTEP 8575). Investig. New Drugs 2014, 32, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Sahebjam, S.; Bedard, P.L.; Castonguay, V.; Chen, Z.; Reedijk, M.; Liu, G.; Cohen, B.; Zhang, W.J.; Clarke, B.; Zhang, T.; et al. A phase I study of the combination of ro4929097 and cediranib in patients with advanced solid tumours (PJC-004/NCI 8503). Br. J. Cancer 2013, 109, 943–949. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Padilla, I.; Wilson, M.K.; Clarke, B.A.; Hirte, H.W.; Welch, S.A.; Mackay, H.J.; Biagi, J.J.; Reedijk, M.; Weberpals, J.I.; Fleming, G.F.; et al. A phase II study of single-agent RO4929097, a gamma-secretase inhibitor of Notch signaling, in patients with recurrent platinum-resistant epithelial ovarian cancer: A study of the Princess Margaret, Chicago and California phase II consortia. Gynecol. Oncol. 2015, 137, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Strosberg, J.R.; Yeatman, T.; Weber, J.; Coppola, D.; Schell, M.J.; Han, G.; Almhanna, K.; Kim, R.; Valone, T.; Jump, H.; et al. A phase II study of RO4929097 in metastatic colorectal cancer. Eur. J. Cancer 2012, 48, 997–1003. [Google Scholar] [CrossRef]
- De Jesus-Acosta, A.; Laheru, D.; Maitra, A.; Arcaroli, J.; Rudek, M.A.; Dasari, A.; Blatchford, P.J.; Quackenbush, K.; Messersmith, W. A phase II study of the gamma secretaseinhibitor RO4929097 in patients with previously treated metastatic pancreatic adenocarcinoma. Investig. New Drugs 2014, 32, 739–745. [Google Scholar] [CrossRef]
- Messersmith, W.A.; Shapiro, G.I.; Cleary, J.M.; Jimeno, A.; Dasari, A.; Huang, B. A Phase I, dose-finding study in patients with advanced solid malignancies of the oral γ-secretase inhibitor PF-03084014. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2015, 21, 60–67. [Google Scholar] [CrossRef]
- Kummar, S.; O’Sullivan Coyne, G.; Do, K.T.; Turkbey, B.; Meltzer, P.S.; Polley, E.; Choyke, P.L.; Meehan, R.; Vilimas, R.; Horneffer, Y.; et al. Clinical Activity of the γ-Secretase Inhibitor PF-03084014 in Adults With Desmoid Tumors (Aggressive Fibromatosis). J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2017, 35, 1561–1569. [Google Scholar] [CrossRef]
- Papayannidis, C.; DeAngelo, D.J.; Stock, W.; Huang, B.; Shaik, M.N.; Cesari, R.; Zheng, X.; Reynolds, J.M.; English, P.A.; Ozeck, M.; et al. A Phase 1 study of the novel gamma-secretase inhibitor PF-03084014 in patients with T-cell acute lymphoblastic leukemia and T-cell lymphoblastic lymphoma. Blood Cancer J. 2015, 5, e350. [Google Scholar] [CrossRef] [PubMed]
- Zweidler-McKay, P.A.; DeAngelo, D.J.; Douer, D.; Dombret, H.; Ottmann, O.G.; Vey, N.; Thomas, D.A.; Zhu, L.; Huang, F.; Bajaj, G.; et al. The Safety and Activity of BMS-906024, a Gamma Secretase Inhibitor (GSI) with Anti-Notch Activity, in Patients with Relapsed T-Cell Acute Lymphoblastic Leukemia (T-ALL): Initial Results of a Phase 1 Trial. Blood 2014, 124, 968. [Google Scholar] [CrossRef]
- Smith, D.C.; Eisenberg, P.D.; Manikhas, G.; Chugh, R.; Gubens, M.A.; Stagg, R.J.; Ann, M.; Kapoun, A.M.; Xu, L.; Dupont, J.; et al. A phase I dose escalation and expansion study of the anticancer stem cell agent demcizumab (anti-DLL4) in patients with previously treated solid tumors. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 6295–6303. [Google Scholar] [CrossRef] [PubMed]
- Chiorean, E.G.; LoRusso, P.; Strother, R.M.; Diamond, J.R.; Younger, A.; Messersmith, W.A.; Adriaens, L.; Liu, L.; Kao, R.J.; DiCioccio, A.T.; et al. A Phase I First-in-Human Study of Enoticumab (REGN421), a Fully Human Delta-like Ligand 4 (Dll4) Monoclonal Antibody in Patients with Advanced Solid Tumors. Clin. cancer Res. Off. J. Am. Assoc. Cancer Res. 2015, 21, 2695–2703. [Google Scholar] [CrossRef]
- Sekulic, A.; Migden, M.R.; Oro, A.E.; Dirix, L.; Lewis, K.D.; Hainsworth, J.D.; Solomon, J.; Yoo, S.; Arron, S.T.; Friedlander, P.; et al. Efficacy and Safety of Vismodegib in Advanced Basal-Cell Carcinoma. N. Engl. J. Med. 2012, 366, 2171–2179. [Google Scholar] [CrossRef]
- Basset-Séguin, N.; Hauschild, A.; Kunstfeld, R.; Grob, J.; Dréno, B.; Mortier, L.; Ascierto, P.A.; Licitra, L.; Dutriaux, C.; Thomas, L.; et al. Vismodegib in patients with advanced basal cell carcinoma: Primary analysis of STEVIE, an international, open-label trial. Eur. J. Cancer 2017, 86, 334–348. [Google Scholar] [CrossRef]
- Robinson, G.W.; Orr, B.A.; Wu, G.; Gururangan, S.; Lin, T.; Qaddoumi, I.; Roger, J.; Packer, R.J.; Goldman, S.; Prados, M.D.; et al. Vismodegib Exerts Targeted Efficacy Against Recurrent Sonic Hedgehog-Subgroup Medulloblastoma: Results From Phase II Pediatric Brain Tumor Consortium Studies PBTC-025B and PBTC-032. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2015, 33, 2646–2654. [Google Scholar] [CrossRef]
- Gajjar, A.; Stewart, C.F.; Ellison, D.W.; Kaste, S.; Kun, L.E.; Packer, R.J.; Goldman, S.; Chintagumpala, M.; Wallace, D.; Takebe, N.; et al. Phase I study of vismodegib in children with recurrent or refractory medulloblastoma: A pediatric brain tumor consortium study. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 6305–6312. [Google Scholar] [CrossRef]
- Italiano, A.; Le Cesne, A.; Bellera, C.; Piperno-Neumann, S.; Duffaud, F.; Penel, N.; Cassier, P.; Domont, J.; Takebe, N.; Kind, M.; et al. GDC-0449 in patients with advanced chondrosarcomas: A French Sarcoma Group/US and French National Cancer Institute Single-Arm Phase II Collaborative Study. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2013, 24, 2922–2926. [Google Scholar] [CrossRef]
- Berlin, J.; Bendell, J.C.; Hart, L.L.; Firdaus, I.; Gore, I.; Hermann, R.C.; Mulcahy, M.F.; Zalupski, M.M.; Mackey, H.M.; Yauch, R.L.; et al. A randomized phase II trial of vismodegib versus placebo with FOLFOX or FOLFIRI and bevacizumab in patients with previously untreated metastatic colorectal cancer. Clin. Cancer Res. 2013, 19, 258–267. [Google Scholar] [CrossRef]
- Catenacci, D.V.T.; Junttila, M.R.; Karrison, T.; Bahary, N.; Horiba, M.N.; Nattam, S.R.; Marsh, R.; Wallace, J.; Kozloff, M.; Rajdev, L.; et al. Randomized Phase Ib/II Study of Gemcitabine Plus Placebo or Vismodegib, a Hedgehog Pathway Inhibitor, in Patients With Metastatic Pancreatic Cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin Oncol. 2015, 33, 4284–4292. [Google Scholar] [CrossRef] [PubMed]
- Cohen, D.J.; Christos, P.J.; Kindler, H.L.; Catenacci, D.V.T.; Bekaii-Saab, T.B.; Tahiri, S.; Janjigian, Y.Y.; Gibson, M.K.; Chan, E.; Rajdev, L.; et al. Vismodegib (V), a hedgehog (HH) pathway inhibitor, combined with FOLFOX for first-line therapy of patients (pts) with advanced gastric and gastroesophageal junction (GEJ) carcinoma: A New York Cancer Consortium led phase II randomized study. J. Clin. Oncol. 2013, 31, 4011. [Google Scholar] [CrossRef]
- Kaye, S.B.; Fehrenbacher, L.; Holloway, R.; Amit, A.; Karlan, B.; Slomovitz, B.; Sabbatini, P.; Fu, L.; Yauch, R.L.; Chang, I.; et al. A phase II, randomized, placebo-controlled study of vismodegib as maintenance therapy in patients with ovarian cancer in second or third complete remission. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2012, 18, 6509–6518. [Google Scholar] [CrossRef]
- Lear, J.T.; Migden, M.R.; Lewis, K.D.; Chang, A.L.S.; Guminski, A.; Gutzmer, R.; Dirix, L.; Combemale, P.; Stratigos, A.; Plummer, R.; et al. Long-term efficacy and safety of sonidegib in patients with locally advanced and metastatic basal cell carcinoma: 30-month analysis of the randomized phase 2 BOLT study. J. Eur. Acad. Dermatol. Venereol. 2018, 32, 372–381. [Google Scholar] [CrossRef]
- Kieran, M.W.; Chisholm, J.; Casanova, M.; Brandes, A.A.; Aerts, I.; Bouffet, E.; Bailey, S.; Leary, S.; MacDonald, T.J.; Mechinaud, F.; et al. Phase I study of oral sonidegib (LDE225) in pediatric brain and solid tumors and a phase II study in children and adults with relapsed medulloblastoma. Neuro Oncol. 2017, 19, 1542–1552. [Google Scholar] [CrossRef] [PubMed]
- Pietanza, M.C.; Litvak, A.M.; Varghese, A.M.; Krug, L.M.; Fleisher, M.; Teitcher, J.B.; Holodny, A.I.; Sima, C.S.; Woo, K.M.; Ng, K.K.; et al. A phase I trial of the Hedgehog inhibitor, sonidegib (LDE225), in combination with etoposide and cisplatin for the initial treatment of extensive stage small cell lung cancer. Lung Cancer 2016, 99, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.E.; Heidel, F.H.; Hellmann, A.; Fiedler, W.; Smith, B.D.; Robak, T.; Montesinos, P.; Pollyea, D.A.; DesJardins, P.; Ottmann, O.; et al. Randomized comparison of low dose cytarabine with or without glasdegib in patients with newly diagnosed acute myeloid leukemia or high-risk myelodysplastic syndrome. Leukemia 2019, 33, 379–389. [Google Scholar] [CrossRef]
- Cortes, J.E.; Douglas Smith, B.; Wang, E.S.; Merchant, A.; Oehler, V.G.; Arellano, M.; DeAngelo, D.J.; Pollyea, D.A.; Sekeres, M.A.; Robak, T.; et al. Glasdegib in combination with cytarabine and daunorubicin in patients with AML or high-risk MDS: Phase 2 study results. Am. J. Hematol. 2018, 93, 1301–1310. [Google Scholar] [CrossRef]
- Bendell, J.; Andre, V.; Ho, A.; Kudchadkar, R.; Migden, M.; Infante, J.; Tiu, R.V.; Pitou, C.; Tucker, T.; Brail, L.; et al. Phase I Study of LY2940680, a Smo Antagonist, in Patients with Advanced Cancer Including Treatment-Naïve and Previously Treated Basal Cell Carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 2082–2091. [Google Scholar] [CrossRef]
- Bowles, D.W.; Keysar, S.B.; Eagles, J.R.; Wang, G.; Glogowska, M.J.; McDermott, J.D.; Le, P.N.; Gao, D.; Ray, C.E.; Rochon, P.J.; et al. A pilot study of cetuximab and the hedgehog inhibitor IPI-926 in recurrent/metastatic head and neck squamous cell carcinoma. Oral Oncol. 2016, 53, 74–79. [Google Scholar] [CrossRef]
| Molecule | Tumor Type | Function | Refs |
|---|---|---|---|
| ABCG2 | Liver, lung, melanoma, pancreatic | It is an ABC drug transporter that act as efflux pump to protect cells from xenobiotic toxins. | [35,36,37,38] |
| ALCAM (CD166) | Colorectal, head and neck, lung, pancreatic | A highly preserved transmembrane protein that belongs to the immunoglobulin superfamily. It binds to the T cell differentiation antigen CD6 and involves in cell adhesion and migration processes. | [34,39,40,41] |
| ALDH1 | Breast, colorectal, lung, melanoma, ovarian, pancreatic, prostate | A group of NAD(P)+-dependent enzymes that catalyze the oxidization of aldehydes into carboxylic acids, playing a role in drug resistance. | [42,43,44,45,46,47,48] |
| CD133 (PROM1) | Breast, colorectal, glioma, liver, lung, melanoma, ovarian, pancreatic, prostate | A pentaspan transmembrane glycoprotein that maintains lipid composition in cell membranes. Evidence suggest that CD133+ cells display strong resistance to chemo-, radio- and immunotherapy. | [36,42,49,50,51,52,53,54] |
| CD44 | Breast, colorectal, glioma, liver, lung, ovarian, pancreatic, prostate | A cell surface glycoprotein that acts as a receptor for many extracellular matrix components, including acid hyaluronic, collagen, integrins and metalloproteinases, promoting cell migration and self-renewal. | [43,51,55,56,57,58,59,60] |
| CD90 (THY1) | Breast, glioma, liver, lung | A highly conserved glycophosphatidylinositol (GPI)-anchored cell surface glycoprotein that participates in T cell adhesion and signal transduction. | [61] |
| EpCAM (CD326) | Colorectal, liver, lung, ovarian, prostate | A transmembraneglycoprotein expressed on most normal epithelial cells that acts as a homotypic calcium-independent cell adhesion molecule. | [44,62,63] |
| Integrin α6β4 | Breast, colorectal, lung, prostate | A cellular adhesion molecule that binds to laminins in the extracellular matrix and nucleates the formation of hemidesmosomes, enabling cell migration and invasion | [56,64,65,66] |
| Target | Therapeutic Strategy | Class | Ongoing Trial | Identifier | Current Status |
|---|---|---|---|---|---|
| EpCAM | Catumaxomab | Trispecific EpCAM/CD3/Fcc antibody | Phase II in gastric cancer with peritoneal carcinomatosis | NCT01504256 | Completed |
| Vicinium | Immunotoxin | Phase III in bladder cancer | NCT02449239 | Active, not recruting | |
| EpCAM CAR-T | Autologous T cells engineered | Phase I in nasopharyngeal carcinoma and breast cancer | NCT02915445 | Recruiting | |
| EpCAM CAR-T | Autologous T cells engineered | Phase I/II in colon, esophageal, pancreatic, prostate, gastric and hepatic cancer | NCT03013712 | Unknown | |
| EpCAM CAR-T | Autologous T cells engineered | Phase II in liver cancer | NCT02729493 | Unknown | |
| EpCAM CAR-T | Autologous T cells engineered | Phase II in gastric cancer | NCT02725125 | Unknown | |
| CD44 | RO5429083 | Anti-CD44 monoclonal antibody | Phase I in advanced CD44-expressing malignant solid tumors | NCT01358903 | Completed |
| SPL-108 | CD44 targeted agent | Phase I in ovarian epithelial cancer | NCT03078400 | Active, not recruting | |
| AMC303 | CD44v6 inhibitor | Phase I/Ib in solid tumors | NCT03009214 | Completed | |
| CD44 CAR-T | Autologous T cells engineered | Phase I/II in CD44v6 positive multiple myeloma, lymphoma, stomach, breast and prostate cancer | NCT04427449 | Recruiting | |
| CD44 CAR-T | Autologous T cells engineered | Phase I/II in breast cancer | NCT04430595 | Recruiting | |
| CD133 | CD133 CAR-T | Autologous T cells engineered | Phase I in recurrent glioma | NCT03423992 | Recruiting |
| CD166 | CX-2009 | CD166-directed probody drug conjugate | Phase II in advanced breast cancer | NCT04596150 | Recruiting |
| Integrins | PF-04605412 | Anti-α5β1 integrin monoclonal antibody | Phase I in advanced non-Hematologic Malignancies | NCT00915278 | Terminated |
| Cilengitide | Anti-αvβ3, α5β1, and αvβ5 small molecules | Phase III in glioblastoma | NCT00689221 | Completed | |
| ProAgio | Anti-αvβ3 integrin cytotoxin | Phase I in advanced pancreatic cancer | NCT05085548 | Recruiting |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Herreros-Pomares, A. Identification, Culture and Targeting of Cancer Stem Cells. Life 2022, 12, 184. https://doi.org/10.3390/life12020184
Herreros-Pomares A. Identification, Culture and Targeting of Cancer Stem Cells. Life. 2022; 12(2):184. https://doi.org/10.3390/life12020184
Chicago/Turabian StyleHerreros-Pomares, Alejandro. 2022. "Identification, Culture and Targeting of Cancer Stem Cells" Life 12, no. 2: 184. https://doi.org/10.3390/life12020184
APA StyleHerreros-Pomares, A. (2022). Identification, Culture and Targeting of Cancer Stem Cells. Life, 12(2), 184. https://doi.org/10.3390/life12020184






