EVs-miRNA: The New Molecular Markers for Chronic Respiratory Diseases
Abstract
1. Introduction
2. Extracellular Vesicles
3. Micro-RNA
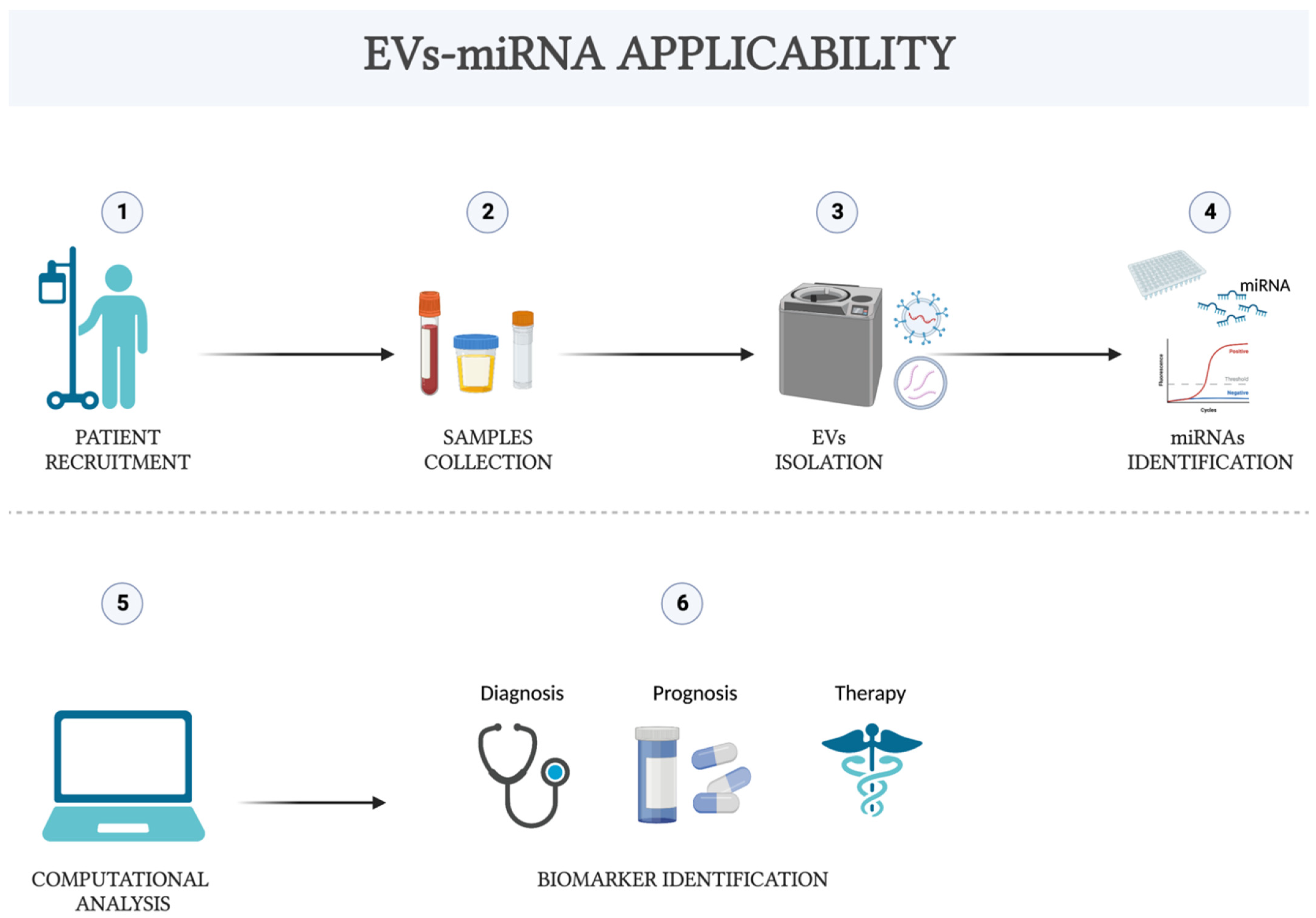
4. EVs-miRNA
- -
- packed inside extracellular vesicles (EVs-miRNA);
- -
- associated with transport proteins (EVs-free).
5. EVs-miRNA as Therapeutic Tools in Chronic Respiratory Diseases
- (1)
- Exhaled breath condensate (EBC): Exhaled breath is saturated with water vapor which can be condensed by cooling and used to sample a wide range of mediators. EBC samples the entire respiratory tract, but newer techniques allow for fractional sampling and provide the ability to collect condensate from different parts of the respiratory tract. Collecting EBCs is a promising sampling method, but several methodological problems hinder its clinical use [61,62,63].
- (2)
- Spontaneous or induced sputum: it is used in clinical practice for microbiological and cell counting studies, while the measurement of inflammatory biomarkers is increasingly implemented in research.
5.1. EVs-miRNAs in Airflow Obstruction
5.1.1. COPD and EVs-miRNAs
5.1.2. Asthma and EVs-miRNAs
5.2. Sleep Disorders and EVs-miRNAs
5.3. Idiopathic Pulmonary Fibrosis and EVs-miRNAs
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Chronic Respiratory Diseases. Available online: https://www.who.int/health-topics/chronic-respiratory-diseases#tab=tab_1 (accessed on 24 October 2021).
- Murray, C.J.; Lopez, A.D. Measuring the global burden of disease. N. Engl. J. Med. 2013, 369, 448–457. [Google Scholar] [CrossRef]
- Vestbo, J.; Hurd, S.S.; Agustií, A.G.; Jones, P.W.; Vogelmeier, C.; Anzueto, A.; Barnes, P.J.; Fabbri, L.M.; Martinez, F.J.; Nishimura, M.; et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am. J. Respir. Crit. Care Med. 2013, 187, 347–365. [Google Scholar] [CrossRef] [PubMed]
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell. Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef] [PubMed]
- Groot, M.; Lee, H. Sorting Mechanisms for MicroRNAs into Extracellular Vesicles and Their Associated Diseases. Cells 2020, 9, 1044. [Google Scholar] [CrossRef]
- Collino, F.; Deregibus, M.C.; Bruno, S.; Sterpone, L.; Aghemo, G.; Viltono, L.; Tetta, C.; Camussi, G. Microvesicles derived from adult human bone marrow and tissue specific mesenchymal stem cells shuttle selected pattern of miRNAs. PLoS ONE 2010, 5, e11803. [Google Scholar] [CrossRef]
- Lacedonia, D.; Scioscia, G.; Soccio, P.; Conese, M.; Catucci, L.; Palladino, G.P.; Simone, F.; Quarato, C.M.I.; Di Gioia, S.; Rana, R.; et al. Downregulation of exosomal let-7d and miR-16 in idiopathic pulmonary fibrosis. BMC Pulm. Med. 2021, 21, 188. [Google Scholar] [CrossRef]
- Lacedonia, D.; Carpagnano, G.E.; Trotta, T.; Palladino, G.P.; Panaro, M.A.; Zoppo, L.D.; Foschino Barbaro, M.P.; Porro, C. Microparticles in sputum of COPD patients: A potential biomarker of the disease? Int. J. Chron. Obstruct. Pulmon. Dis. 2016, 11, 527–533. [Google Scholar] [CrossRef]
- Lin, J.; Li, J.; Huang, B.; Liu, J.; Chen, X.; Chen, X.M.; Xu, Y.M.; Huang, L.F.; Wang, X.Z. Exosomes: Novel biomarkers for clinical diagnosis. Sci. World J. 2015, 2015, 657086. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Bhatnagar, S.; Schorey, J.S. Exosomes released from infected macrophages contain Mycobacterium avium glycopeptidolipids and are proinflammatory. J. Biol. Chem. 2007, 282, 25779–25789. [Google Scholar] [CrossRef] [PubMed]
- Kourembanas, S. Exosomes: Vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu. Rev. Physiol. 2015, 77, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Crescitelli, R.; Lässer, C.; Szabó, T.G.; Kittel, A.; Eldh, M.; Dianzani, I.; Buzás, E.I.; Lötvall, J. Distinct RNA profiles in subpopulations of extracellular vesicles: Apoptotic bodies, microvesicles and exosomes. J. Extracell. Vesicles 2013, 2, 20677. [Google Scholar] [CrossRef] [PubMed]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell. Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Benedikter, B.J.; Wouters, E.F.M.; Savelkoul, P.H.M.; Rohde, G.G.U.; Stassen, F.R.M. Extracellular vesicles released in response to respiratory exposures: Implications for chronic disease. J. Toxicol. Environ. Health B Crit. Rev. 2018, 21, 142–160. [Google Scholar] [CrossRef]
- Gon, Y.; Shimizu, T.; Mizumura, K.; Maruoka, S.; Hikichi, M. Molecular techniques for respiratory diseases: MicroRNA and extracellular vesicles. Respirology 2020, 25, 149–160. [Google Scholar] [CrossRef]
- Ramón-Núñez, L.A.; Martos, L.; Fernández-Pardo, Á.; Oto, J.; Medina, P.; España, F.; Navarro, S. Comparison of protocols and RNA carriers for plasma miRNA isolation. Unraveling RNA carrier influence on miRNA isolation. PLoS ONE 2017, 12, e0187005. [Google Scholar] [CrossRef]
- Livshits, M.A.; Khomyakova, E.; Evtushenko, E.G.; Lazarev, V.N.; Kulemin, N.A.; Semina, S.E.; Generozov, E.V.; Govorun, V.M. Isolation of exosomes by differential centrifugation: Theoretical analysis of a commonly used protocol. Sci. Rep. 2015, 5, 17319. [Google Scholar] [CrossRef]
- Taylor, D.D.; Shah, S. Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods 2015, 87, 3–10. [Google Scholar] [CrossRef]
- Konoshenko, M.Y.; Lekchnov, E.A.; Vlassov, A.V.; Laktionov, P.P. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. Biomed. Res. Int. 2018, 2018, 8545347. [Google Scholar] [CrossRef] [PubMed]
- Dragovic, R.A.; Gardiner, C.; Brooks, A.S.; Tannetta, D.S.; Ferguson, D.J.; Hole, P.; Carr, B.; Redman, C.W.; Harris, A.L.; Dobson, P.J.; et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine 2011, 7, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Coumans, F.A.; van der Pol, E.; Boing, A.N.; Hajji, N.; Sturk, G.; van Leeuwen, T.G.; Nieuwland, R. Reproducible extracellular vesicle size and concentration determination with tunable resistive pulse sensing. J. Extracell. Vesicles 2014, 3, 25922. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Regnault, A.; Garin, J.; Wolfers, J.; Zitvogel, L.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein hsc73. J. Cell Biol. 1999, 147, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Koliha, N.; Wiencek, Y.; Heider, U.; Jungst, C.; Kladt, N.; Krauthauser, S.; Johnston, I.C.; Bosio, A.; Schauss, A.; Wild, S. A novel multiplex bead-based platform highlights the diversity of extracellular vesicles. J. Extracell. Vesicles 2016, 5, 29975. [Google Scholar] [CrossRef]
- Goldie, B.J.; Dun, M.D.; Lin, M.; Smith, N.D.; Verrills, N.M.; Dayas, C.V.; Cairns, M.J. Activity-associated miRNA are packaged in Map1b-enriched exosomes released from depolarized neurons. Nucleic Acids Res. 2014, 42, 9195–9208. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Bhaskaran, M.; Mohan, M. MicroRNAs: History, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 2014, 51, 759–774. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.S.; Hutvágner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef]
- Stolzenburg, L.R.; Harris, A. The role of microRNAs in chronic respiratory disease: Recent insights. Biol. Chem. 2018, 399, 219–234. [Google Scholar] [CrossRef] [PubMed]
- Svitich, O.A.; Sobolev, V.V.; Gankovskaya, L.V.; Zhigalkina, P.V.; Zverev, V.V. The role of regulatory RNAs (miRNAs) in asthma. Allergol. Immunopathol. 2018, 46, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Hanson, E.K.; Lubenow, H.; Ballantyne, J. Identification of forensically relevant body fluids using a panel of differentially expressed microRNAs. Anal. Biochem. 2009, 387, 303–314. [Google Scholar] [CrossRef]
- Chim, S.S.; Shing, T.K.; Hung, E.C.; Leung, T.Y.; Lau, T.K.; Chiu, R.W.; Lo, Y.M. Detection and characterization of placental microRNAs in maternal plasma. Clin. Chem. 2008, 54, 482–490. [Google Scholar] [CrossRef]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Weiz, L.; Burwinkel, B. Extracellular miRNAs: The mystery of their origin and function. Trends Biochem. Sci. 2012, 37, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Ul Hussain, M. Micro-RNAs (miRNAs): Genomic organisation, biogenesis and mode of action. Cell. Tissue Res. 2012, 349, 405–413. [Google Scholar] [CrossRef]
- Hu, G.; Drescher, K.M.; Chen, X.M. Exosomal miRNAs: Biological Properties and Therapeutic Potential. Front. Genet. 2012, 3, 56. [Google Scholar] [CrossRef] [PubMed]
- Ajit, S.K. Circulating microRNAs as biomarkers, therapeutic targets, and signaling molecules. Sensors 2012, 12, 3359–3369. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.X.; Wilfred, B.R.; Hu, Y.; Stromberg, A.J.; Nelson, P.T. Anti-Argonaute RIP-Chip shows that miRNA transfections alter global patterns of mRNA recruitment to microribonucleoprotein complexes. RNA 2010, 16, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell. Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef]
- Tabet, F.; Vickers, K.C.; Cuesta Torres, L.F.; Wiese, C.B.; Shoucri, B.M.; Lambert, G.; Catherinet, C.; Prado-Lourenco, L.; Levin, M.G.; Thacker, S.; et al. HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells. Nat. Commun. 2014, 5, 3292. [Google Scholar] [CrossRef]
- Mitchell, A.J.; Gray, W.D.; Hayek, S.S.; Ko, Y.A.; Thomas, S.; Rooney, K.; Awad, M.; Roback, J.D.; Quyyumi, A.; Searles, C.D. Platelets confound the measurement of extracellular miRNA in archived plasma. Sci. Rep. 2016, 6, 32651. [Google Scholar] [CrossRef]
- Lodes, M.J.; Caraballo, M.; Suciu, D.; Munro, S.; Kumar, A.; Anderson, B. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS ONE 2009, 4, e6229. [Google Scholar] [CrossRef]
- Resnick, K.E.; Alder, H.; Hagan, J.P.; Richardson, D.L.; Croce, C.M.; Cohn, D.E. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol. Oncol. 2009, 112, 55–59. [Google Scholar] [CrossRef]
- Bellingham, S.A.; Coleman, B.M.; Hill, A.F. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012, 40, 10937–10949. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef] [PubMed]
- Mestdagh, P.; Hartmann, N.; Baeriswyl, L.; Andreasen, D.; Bernard, N.; Chen, C.; Cheo, D.; D’Andrade, P.; DeMayo, M.; Dennis, L.; et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat. Methods 2014, 11, 809–815. [Google Scholar] [CrossRef] [PubMed]
- McAlexander, M.A.; Phillips, M.J.; Witwer, K.W. Comparison of Methods for miRNA Extraction from Plasma and Quantitative Recovery of RNA from Cerebrospinal Fluid. Front. Genet. 2013, 4, 83. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Chen, J.; Zhao, X.; Liang, C.; Wang, Y.; Sun, L.; Jiang, Z.; Zhang, Z.; Yang, R.; Chen, J.; et al. MicroRNA expression signatures of bladder cancer revealed by deep sequencing. PLoS ONE 2011, 6, e18286. [Google Scholar] [CrossRef]
- Wach, S.; Nolte, E.; Szczyrba, J.; Stöhr, R.; Hartmann, A.; Ørntoft, T.; Dyrskjøt, L.; Eltze, E.; Wieland, W.; Keck, B.; et al. MicroRNA profiles of prostate carcinoma detected by multiplatform microRNA screening. Int. J. Cancer 2012, 130, 611–621. [Google Scholar] [CrossRef]
- Kosaka, N.; Iguchi, H.; Ochiya, T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010, 101, 2087–2092. [Google Scholar] [CrossRef]
- Ge, Q.; Zhou, Y.; Lu, J.; Bai, Y.; Xie, X.; Lu, Z. miRNA in plasma exosome is stable under different storage conditions. Molecules 2014, 19, 1568–1575. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef]
- Koutsokera, A.; Kostikas, K.; Nicod, L.P.; Fitting, J.W. Pulmonary biomarkers in COPD exacerbations: A systematic review. Respir. Res. 2013, 14, 111. [Google Scholar] [CrossRef]
- Mannino, D.M. Biomarkers in COPD: The search continues! Eur. Respir. J. 2015, 45, 872–874. [Google Scholar] [CrossRef] [PubMed]
- Lacoma, A.; Prat, C.; Andreo, F.; Domínguez, J. Biomarkers in the management of COPD. Eur. Respir. Rev. 2009, 18, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Halushka, M.K. Extracellular vesicle microRNA transfer in cardiovascular disease. Cardiovasc. Pathol. 2015, 24, 199–206. [Google Scholar] [CrossRef]
- Claßen, L.; Tykocinski, L.O.; Wiedmann, F.; Birr, C.; Schiller, P.; Tucher, C.; Krienke, S.; Raab, M.S.; Blank, N.; Lorenz, H.M.; et al. Extracellular vesicles mediate intercellular communication: Transfer of functionally active microRNAs by microvesicles into phagocytes. Eur. J. Immunol. 2017, 47, 1535–1549. [Google Scholar] [CrossRef] [PubMed]
- Aliotta, J.M.; Pereira, M.; Wen, S.; Dooner, M.S.; Del Tatto, M.; Papa, E.; Goldberg, L.R.; Baird, G.L.; Ventetuolo, C.E.; Quesenberry, P.J.; et al. Exosomes induce and reverse monocrotaline-induced pulmonary hypertension in mice. Cardiovasc. Res. 2016, 110, 319–330. [Google Scholar] [CrossRef]
- Serban, K.A.; Rezania, S.; Petrusca, D.N.; Poirier, C.; Cao, D.; Justice, M.J.; Patel, M.; Tsvetkova, I.; Kamocki, K.; Mikosz, A.; et al. Structural and functional characterization of endothelial microparticles released by cigarette smoke. Sci. Rep. 2016, 6, 31596. [Google Scholar] [CrossRef]
- Fujita, Y.; Araya, J.; Ito, S.; Kobayashi, K.; Kosaka, N.; Yoshioka, Y.; Kadota, T.; Hara, H.; Kuwano, K.; Ochiya, T. Suppression of autophagy by extracellular vesicles promotes myofibroblast differentiation in COPD pathogenesis. J. Extracell. Vesicles 2015, 4, 28388. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.; Hu, R.; Runtsch, M.C.; Kagele, D.A.; Mosbruger, T.L.; Tolmachova, T.; Seabra, M.C.; Round, J.L.; Ward, D.M.; O’Connell, R.M. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat. Commun. 2015, 6, 7321. [Google Scholar] [CrossRef]
- Lee, C.; Mitsialis, S.A.; Aslam, M.; Vitali, S.H.; Vergadi, E.; Konstantinou, G.; Sdrimas, K.; Fernandez-Gonzalez, A.; Kourembanas, S. Exosomes mediate the cytoprotective action of mesenchymal stromal cells on hypoxia-induced pulmonary hypertension. Circulation 2012, 126, 2601–2611. [Google Scholar] [CrossRef]
- Huang-Doran, I.; Zhang, C.Y.; Vidal-Puig, A. Extracellular Vesicles: Novel Mediators of Cell Communication In Metabolic Disease. Trends Endocrinol. Metab. 2017, 28, 3–18. [Google Scholar] [CrossRef]
- Nana-Sinkam, S.P.; Acunzo, M.; Croce, C.M.; Wang, K. Extracellular vesicle biology in the pathogenesis of lung disease. Am. J. Respir. Crit. Care Med. 2017, 196, 1510–1518. [Google Scholar] [CrossRef] [PubMed]
- Rogliani, P.; Ora, J.; Puxeddu, E.; Cazzola, M. Airflow obstruction: Is it asthma or is it COPD? Int. J. Chron. Obstruct. Pulmon. Dis. 2016, 11, 3007–3013. [Google Scholar] [CrossRef] [PubMed]
- ISS—Istituto Superiore di Sanità, Centro Nazionale di Epidemiologia, Sorveglianza e Promozione della Salute. Available online: http://www.epicentro.iss.it/ (accessed on 8 April 2022).
- Ishmael, F.T. The inflammatory response in the pathogenesis of asthma. J. Am. Osteopath. Assoc. 2011, 111, 11–17. [Google Scholar] [PubMed]
- Mercado, N.; Ito, K.; Barnes, P.J. Accelerated ageing of the lung in COPD: New concepts. Thorax 2015, 70, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Colarusso, C.; Terlizzi, M.; Molino, A.; Pinto, A.; Sorrentino, R. Role of the inflammasome in chronic obstructive pulmonary disease (COPD). Oncotarget 2017, 8, 81813–81824. [Google Scholar] [CrossRef] [PubMed]
- Barnes, P.J. Immunology of asthma and chronic obstructive pulmonary disease. Nat. Rev. Immunol. 2008, 8, 183–192. [Google Scholar] [CrossRef]
- Savarimuthu Francis, S.M.; Davidson, M.R.; Tan, M.E.; Wright, C.M.; Clarke, B.E.; Duhig, E.E.; Bowman, R.V.; Hayward, N.K.; Fong, K.M.; Yang, I.A. MicroRNA-34c is associated with emphysema severity and modulates SERPINE1 expression. BMC Genomics 2014, 15, 88. [Google Scholar] [CrossRef]
- De Smet, E.G.; Mestdagh, P.; Vandesompele, J.; Brusselle, G.G.; Bracke, K.R. Non-coding RNAs in the pathogenesis of COPD. Thorax 2015, 70, 782–791. [Google Scholar] [CrossRef]
- Salimian, J.; Mirzaei, H.; Moridikia, A.; Harchegani, A.B.; Sahebkar, A.; Salehi, H. Chronic obstructive pulmonary disease: MicroRNAs and exosomes as new diagnostic and therapeutic biomarkers. J. Res. Med. Sci. 2018, 23, 27. [Google Scholar] [CrossRef]
- Xu, H.; Ling, M.; Xue, J.; Dai, X.; Sun, Q.; Chen, C.; Liu, Y.; Zhou, L.; Liu, J.; Luo, F.; et al. Exosomal microRNA-21 derived from bronchial epithelial cells is involved in aberrant epithelium-fibroblast cross-talk in COPD induced by cigarette smoking. Theranostics 2018, 8, 5419–5433. [Google Scholar] [CrossRef]
- O’Farrell, H.E.; Bowman, R.V.; Fong, K.M.; Yang, I.A. Plasma Extracellular Vesicle miRNAs Can Identify Lung Cancer, Current Smoking Status, and Stable COPD. Int. J. Mol. Sci. 2021, 22, 5803. [Google Scholar] [CrossRef] [PubMed]
- Burke, H.; Spalluto, C.M.; Cellura, D.; Staples, K.J.; Wilkinson, T.M.A. Role of exosomal microRNA in driving skeletal muscle wasting in COPD. Eur. Respir. J. 2015, 46, OA2930. [Google Scholar] [CrossRef]
- Wang, J.W.; Li, K.; Hellermann, G.; Lockey, R.F.; Mohapatra, S.; Mohapatra, S. Regulating the Regulators: microRNA and Asthma. World Allergy Organ. J. 2011, 4, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Ariel, D.; Upadhyay, D. The role and regulation of microRNAs in asthma. Curr. Opin. Allergy Clin. Immunol. 2012, 12, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Jardim, M.J.; Dailey, L.; Silbajoris, R.; Diaz-Sanchez, D. Distinct microRNA expression in human airway cells of asthmatic donors identifies a novel asthma-associated gene. Am. J. Respir. Cell Mol. Biol. 2012, 47, 536–542. [Google Scholar] [CrossRef]
- Solberg, O.D.; Ostrin, E.J.; Love, M.I.; Peng, J.C.; Bhakta, N.R.; Hou, L.; Nguyen, C.; Solon, M.; Nguyen, C.; Barczak, A.J.; et al. Airway epithelial miRNA expression is altered in asthma. Am. J. Respir. Crit. Care Med. 2012, 186, 965–974. [Google Scholar] [CrossRef]
- Liu, F.; Qin, H.B.; Xu, B.; Zhou, H.; Zhao, D.Y. Profiling of miRNAs in pediatric asthma: Upregulation of miRNA-221 and miRNA-485-3p. Mol. Med. Rep. 2012, 6, 1178–1182. [Google Scholar] [CrossRef]
- Cañas, J.A.; Sastre, B.; Mazzeo, C.; Fernández-Nieto, M.; Rodrigo-Muñoz, J.M.; González-Guerra, A.; Izquierdo, M.; Barranco, P.; Quirce, S.; Sastre, J.; et al. Exosomes from eosinophils autoregulate and promote eosinophil functions. J. Leukoc. Biol. 2017, 101, 1191–1199. [Google Scholar] [CrossRef]
- Vargas, A.; Roux-Dalvai, F.; Droit, A.; Lavoie, J.P. Neutrophil-Derived Exosomes: A New Mechanism Contributing to Airway Smooth Muscle Remodeling. Am. J. Respir. Cell Mol. Biol. 2016, 55, 450–461. [Google Scholar] [CrossRef]
- Okoye, I.S.; Coomes, S.M.; Pelly, V.S.; Czieso, S.; Papayannopoulos, V.; Tolmachova, T.; Seabra, M.C.; Wilson, M.S. MicroRNA-containing T-regulatory-cell-derived exosomes suppress pathogenic T helper 1 cells. Immunity 2014, 41, 503. [Google Scholar] [CrossRef]
- Fujita, Y.; Yoshioka, Y.; Ito, S.; Araya, J.; Kuwano, K.; Ochiya, T. Intercellular communication by extracellular vesicles and their microRNAs in asthma. Clin. Ther. 2014, 36, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Ekström, K.; Valadi, H.; Sjöstrand, M.; Malmhäll, C.; Bossios, A.; Eldh, M.; Lötvall, J. Characterization of mRNA and microRNA in human mast cell-derived exosomes and their transfer to other mast cells and blood CD34 progenitor cells. J. Extracell. Vesicles 2012, 1, 18389. [Google Scholar] [CrossRef] [PubMed]
- Sinha, A.; Yadav, A.K.; Chakraborty, S.; Kabra, S.K.; Lodha, R.; Kumar, M.; Kulshreshtha, A.; Sethi, T.; Pandey, R.; Malik, G.; et al. Exosome-enclosed microRNAs in exhaled breath hold potential for biomarker discovery in patients with pulmonary diseases. J. Allergy Clin. Immunol. 2013, 132, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Levänen, B.; Bhakta, N.R.; Torregrosa Paredes, P.; Barbeau, R.; Hiltbrunner, S.; Pollack, J.L.; Sköld, C.M.; Svartengren, M.; Grunewald, J.; Gabrielsson, S.; et al. Altered microRNA profiles in bronchoalveolar lavage fluid exosomes in asthmatic patients. J. Allergy Clin. Immunol. 2013, 131, 894–903. [Google Scholar] [CrossRef]
- Gon, Y.; Maruoka, S.; Inoue, T.; Kuroda, K.; Yamagishi, K.; Kozu, Y.; Shikano, S.; Soda, K.; Lötvall, J.; Hashimoto, S. Selective release of miRNAs via extracellular vesicles is associated with house-dust mite allergen-induced airway inflammation. Clin. Exp. Allergy 2017, 47, 1586–1598. [Google Scholar] [CrossRef]
- Qiao, Y.; Liang, X.; Yan, Y.; Lu, Y.; Zhang, D.; Yao, W.; Wu, W.; Yan, Z. Identification of Exosomal miRNAs in Rats With Pulmonary Neutrophilic Inflammation Induced by Zinc Oxide Nanoparticles. Front. Physiol. 2018, 9, 217. [Google Scholar] [CrossRef]
- Gutierrez, M.J.; Gomez, J.L.; Perez, G.F.; Pancham, K.; Val, S.; Pillai, D.K.; Giri, M.; Ferrante, S.; Freishtat, R.; Rose, M.C.; et al. Airway Secretory microRNAome Changes during Rhinovirus Infection in Early Childhood. PLoS ONE 2016, 11, e0162244. [Google Scholar] [CrossRef]
- Tondo, P.; Fanfulla, F.; Sabato, R.; Scioscia, G.; Foschino Barbaro, M.P.; Lacedonia, D. Obstructive Sleep Apnoea-Hypopnoea Syndrome (OSAHS): State of the art. Minerva Med. 2022. [Google Scholar] [CrossRef]
- Jordan, A.S.; McSharry, D.G.; Malhotra, A. Adult obstructive sleep apnoea. Lancet 2014, 383, 736–747. [Google Scholar] [CrossRef]
- Tingting, X.; Danming, Y.; Xin, C. Non-surgical treatment of obstructive sleep apnea syndrome. Eur. Arch. Otorhinolaryngol. 2018, 275, 335–346. [Google Scholar] [CrossRef]
- Lombardi, C.; Mattaliano, P.; Parati, G.; Culebras, A. Sleep and Cardiac Disorders. Medlin. Neurol. 2013. [Google Scholar]
- Kendzerska, T.; Povitz, M.; Leung, R.S.; Boulos, M.I.; McIsaac, D.I.; Murray, B.J.; Bryson, G.L.; Talarico, R.; Hilton, J.F.; Malhotra, A.; et al. Obstructive Sleep Apnea and Incident Cancer: A Large Retrospective Multicenter Clinical Cohort Study. Cancer Epidemiol. Biomark. Prev. 2021, 30, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Parati, G.; Lombardi, C.; Narkiewicz, K. Sleep apnea: Epidemiology, pathophysiology, and relation to cardiovascular risk. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007, 293, R1671–R1683. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, M.P.; Rajapakshe, K.; Hartig, S.M.; Reva, B.; McLellan, M.D.; Kandoth, C.; Ding, L.; Zack, T.I.; Gunaratne, P.H.; Wheeler, D.A.; et al. Identification of a pan-cancer oncogenic microRNA superfamily anchored by a central core seed motif. Nat. Commun. 2013, 4, 2730. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, H.; Du, Y.; Peng, L.; Qin, Y.; Liu, H.; Ma, X.; Wei, Y. Extracellular vesicle microRNA cargoes from intermittent hypoxia-exposed cardiomyocytes and their effect on endothelium. Biochem. Biophys. Res. Commun. 2021, 548, 182–188. [Google Scholar] [CrossRef]
- Khalyfa, A.; Kheirandish-Gozal, L.; Bhattacharjee, R.; Khalyfa, A.A.; Gozal, D. Circulating microRNAs as Potential Biomarkers of Endothelial Dysfunction in Obese Children. Chest 2016, 149, 786–800. [Google Scholar] [CrossRef] [PubMed]
- Khalyfa, A.; Kheirandish-Gozal, L.; Khalyfa, A.A.; Philby, M.F.; Alonso-Álvarez, M.L.; Mohammadi, M.; Bhattacharjee, R.; Terán-Santos, J.; Huang, L.; Andrade, J.; et al. Circulating Plasma Extracellular Microvesicle MicroRNA Cargo and Endothelial Dysfunction in Children with Obstructive Sleep Apnea. Am. J. Respir. Crit. Care Med. 2016, 194, 1116–1126. [Google Scholar] [CrossRef]
- Khalyfa, A.; Gozal, D. Exosomal miRNAs as potential biomarkers of cardiovascular risk in children. J. Transl. Med. 2014, 12, 162. [Google Scholar] [CrossRef]
- Khalyfa, A.; Zhang, C.; Khalyfa, A.A.; Foster, G.E.; Beaudin, A.E.; Andrade, J.; Hanly, P.J.; Poulin, M.J.; Gozal, D. Effect on Intermittent Hypoxia on Plasma Exosomal Micro RNA Signature and Endothelial Function in Healthy Adults. Sleep 2016, 39, 2077–2090. [Google Scholar] [CrossRef]
- Almendros, I.; Wang, Y.; Becker, L.; Lennon, F.E.; Zheng, J.; Coats, B.R.; Schoenfelt, K.S.; Carreras, A.; Hakim, F.; Zhang, S.X.; et al. Intermittent hypoxia-induced changes in tumor-associated macrophages and tumor malignancy in a mouse model of sleep apnea. Am. J. Respir. Crit. Care Med. 2014, 189, 593–601. [Google Scholar] [CrossRef]
- Almendros, I.; Khalyfa, A.; Trzepizur, W.; Gileles-Hillel, A.; Huang, L.; Akbarpour, M.; Andrade, J.; Farré, R.; Gozal, D. Tumor Cell Malignant Properties Are Enhanced by Circulating Exosomes in Sleep Apnea. Chest 2016, 150, 1030–1041. [Google Scholar] [CrossRef] [PubMed]
- Magni, F.; Van Der Burgt, Y.E.; Chinello, C.; Mainini, V.; Gianazza, E.; Squeo, V.; Deelder, A.M.; Kienle, M.G. Biomarkers discovery by peptide and protein profiling in biological fluids based on functionalized magnetic beads purification and mass spectrometry. Blood Transfus. 2010, 8 (Suppl. 3), s92–s97. [Google Scholar] [CrossRef]
- Perera, F.P.; Weinstein, I.B. Molecular epidemiology: Recent advances and future directions. Carcinogenesis 2000, 21, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Mullington, J.M.; Abbott, S.M.; Carroll, J.E.; Davis, C.J.; Dijk, D.J.; Dinges, D.F.; Gehrman, P.R.; Ginsburg, G.S.; Gozal, D.; Haack, M.; et al. Developing Biomarker Arrays Predicting Sleep and Circadian-Coupled Risks to Health. Sleep 2016, 39, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Kalra, H.; Adda, C.G.; Liem, M.; Ang, C.S.; Mechler, A.; Simpson, R.J.; Hulett, M.D.; Mathivanan, S. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics 2013, 13, 3354–3364. [Google Scholar] [CrossRef]
- Pack, A.I. Application of Personalized, Predictive, Preventative, and Participatory (P4) Medicine to Obstructive Sleep Apnea. A Roadmap for Improving Care? Ann. Am. Thorac. Soc. 2016, 13, 1456–1467. [Google Scholar] [CrossRef]
- Properzi, F.; Logozzi, M.; Fais, S. Exosomes: The future of biomarkers in medicine. Biomark. Med. 2013, 7, 769–778. [Google Scholar] [CrossRef]
- Raghu, G.; Remy-Jardin, M.; Myers, J.L.; Richeldi, L.; Ryerson, C.J.; Lederer, D.J.; Behr, J.; Cottin, V.; Danoff, S.K.; Morell, F.; et al. Diagnosis of Idiopathic Pulmonary Fibrosis. An Official ATS/ERS/JRS/ALAT Clinical Practice Guideline. Am. J. Respir. Crit. Care Med. 2018, 198, e44–e68. [Google Scholar] [CrossRef]
- Ley, B.; Collard, H.R.; King, T.E., Jr. Clinical course and prediction of survival in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2011, 183, 431–440. [Google Scholar] [CrossRef]
- Kadota, T.; Yoshioka, Y.; Fujita, Y.; Araya, J.; Minagawa, S.; Hara, H.; Miyamoto, A.; Suzuki, S.; Fujimori, S.; Kohno, T.; et al. Extracellular Vesicles from Fibroblasts Induce Epithelial-Cell Senescence in Pulmonary Fibrosis. Am. J. Respir. Cell Mol. Biol. 2020, 63, 623–636. [Google Scholar] [CrossRef]
- Kadota, T.; Fujita, Y.; Yoshioka, Y.; Araya, J.; Kuwano, K.; Ochiya, T. Emerging role of extracellular vesicles as a senescence-associated secretory phenotype: Insights into the pathophysiology of lung diseases. Mol. Asp. Med. 2018, 60, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Chanda, D.; Otoupalova, E.; Hough, K.P.; Locy, M.L.; Bernard, K.; Deshane, J.S.; Sanderson, R.D.; Mobley, J.A.; Thannickal, V.J. Fibronectin on the Surface of Extracellular Vesicles Mediates Fibroblast Invasion. Am. J. Respir. Cell Mol. Biol. 2019, 60, 279–288. [Google Scholar] [CrossRef]
- Lacy, S.H.; Woeller, C.F.; Thatcher, T.H.; Pollock, S.J.; Small, E.M.; Sime, P.J.; Phipps, R.P. Activated Human Lung Fibroblasts Produce Extracellular Vesicles with Antifibrotic Prostaglandins. Am. J. Respir. Cell Mol. Biol. 2019, 60, 269–278. [Google Scholar] [CrossRef]
- Martin-Medina, A.; Lehmann, M.; Burgy, O.; Hermann, S.; Baarsma, H.A.; Wagner, D.E.; De Santis, M.M.; Ciolek, F.; Hofer, T.P.; Frankenberger, M.; et al. Increased Extracellular Vesicles Mediate WNT5A Signaling in Idiopathic Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2018, 198, 1527–1538. [Google Scholar] [CrossRef]
- Zhu, L.; Chen, Y.; Chen, M.; Wang, W. Mechanism of miR-204-5p in exosomes derived from bronchoalveolar lavage fluid on the progression of pulmonary fibrosis via AP1S2. Ann. Transl. Med. 2021, 9, 1068. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Jiang, T.; Hu, X.; Liu, Z.; Zhao, L.; Liu, H.; Liu, Z.; Ma, L. Downregulation of microRNA-30a in bronchoalveolar lavage fluid from idiopathic pulmonary fibrosis patients. Mol. Med. Rep. 2018, 18, 5799–5806. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, M.; Soccio, P.; Bergantini, L.; Cameli, P.; Scioscia, G.; Foschino Barbaro, M.P.; Lacedonia, D.; Bargagli, E. Extracellular Vesicle Surface Signatures in IPF Patients: A Multiplex Bead-Based Flow Cytometry Approach. Cells 2021, 10, 1045. [Google Scholar] [CrossRef]
- Makiguchi, T.; Yamada, M.; Yoshioka, Y.; Sugiura, H.; Koarai, A.; Chiba, S.; Fujino, N.; Tojo, Y.; Ota, C.; Kubo, H.; et al. Serum extracellular vesicular miR-21-5p is a predictor of the prognosis in idiopathic pulmonary fibrosis. Respir. Res. 2016, 17, 110. [Google Scholar] [CrossRef]
- Njock, M.S.; Guiot, J.; Henket, M.A.; Nivelles, O.; Thiry, M.; Dequiedt, F.; Corhay, J.L.; Louis, R.E.; Struman, I. Sputum exosomes: Promising biomarkers for idiopathic pulmonary fibrosis. Thorax 2019, 74, 309–312. [Google Scholar] [CrossRef]
- Guiot, J.; Cambier, M.; Boeckx, A.; Henket, M.; Nivelles, O.; Gester, F.; Louis, E.; Malaise, M.; Dequiedt, F.; Louis, R.; et al. Macrophage-derived exosomes attenuate fibrosis in airway epithelial cells through delivery of antifibrotic miR-142-3p. Thorax 2020, 75, 870–881. [Google Scholar] [CrossRef]
- Zhou, J.; Lin, Y.; Kang, X.; Liu, Z.; Zhang, W.; Xu, F. microRNA-186 in extracellular vesicles from bone marrow mesenchymal stem cells alleviates idiopathic pulmonary fibrosis via interaction with SOX4 and DKK1. Stem Cell Res. Ther. 2021, 12, 96. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Chen, S.; Fang, Y.; Zuo, W.; Cui, J.; Xie, S. Mesenchymal stem cell-derived extracellular vesicles suppress the fibroblast proliferation by downregulating FZD6 expression in fibroblasts via micrRNA-29b-3p in idiopathic pulmonary fibrosis. J. Cell. Physiol. 2020, 235, 8613–8625. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Sun, J.; Dong, C.; Zhao, M.; Hu, Y.; Jin, F. Extracellular Vesicles Derived from Adipose Mesenchymal Stem Cells Alleviate PM2.5-Induced Lung Injury and Pulmonary Fibrosis. Med. Sci. Monit. 2020, 18, 26. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Ding, Y.; Hou, Y.; Liu, Y.; Mao, K.; Cui, Y.; Nie, H. Exosomal MicroRNA: Diagnostic Marker and Therapeutic Tool for Lung Diseases. Curr. Pharm. Des. 2021, 27, 2934–2942. [Google Scholar] [CrossRef]
- Rabinowits, G.; Gerçel-Taylor, C.; Day, J.M.; Taylor, D.D.; Kloecker, G.H. Exosomal microRNA: A diagnostic marker for lung cancer. Clin. Lung Cancer 2009, 10, 42–46. [Google Scholar] [CrossRef]
| Vesicle | Size | Origin | Markers | Function |
|---|---|---|---|---|
| Exosome | 30–100 nm | Endocytic origin | Alix, Tsg101, Tetraspanins (CD81, CD63, CD9), flotillin | Cell–cell communication. |
| Microvesicle | 100–1000 nm | Plasma membrane budding | Integrins, selectins, CD40 | Cell–cell communication. |
| Apoptotic body | 1000–4000 nm | Blebbing | Annexin V, TSP, C3P, exposed phosphatidylserine | Product of programmed cell death. Facilitate clearance of apoptotic cells. |
| Technique | Advantages | Disadvantages |
|---|---|---|
| RT-PCR |
|
|
| Microarray |
|
|
| Deep sequencing |
|
|
| References | Year of Publication | Study Design | Samples | Altered miRNA | Results |
|---|---|---|---|---|---|
| Fujita et al. [68] | 2015 | Study of EVs derived miRNAs in HBEC from COPD subjects after exposure to cigarette smoke. | HBE cells | miR-210 | miR-210 expression was greater in HBEC cells isolated from lung samples. |
| Xu et al. [82] | 2018 | Analysis of the role of exosomal miR-21 in the dysfunctional epithelium-fibroblast cross-talk caused by cigarette smoke. | SERUM, HBE cells | miR-21 | miR-21 was high in serum of smokers and COPD patients. |
| Serban et al. [67] | 2016 | Investigation of extracellular vesicles following exposure to cigarette smoke. | PLASMA, human lung endothelial cells | Let-7d, miR-191, miR-126, miR-125a | Let-7d, miR-191, miR-126 and miR-125a were upregulated after being exposed to cigarette smoke. |
| O’Farrell et al. [83] | 2021 | Study of extracellular vesicle miRNAs in healthy non-smokers, healthy smokers, lung cancer and COPD patients. | PLASMA | up-regulated: miR-485, miR-27a-3p, miR-106b-3p down-regulated: miR-205-5p, miR-199a-5p, miR-497-5p. | Identification of some dysregulated EV miRNAs which could discriminate between groups of lung cancer cases compared to healthy non-smokers, healthy smokers and stable COPD cases. |
| References | Year of Publication | Study Design | Samples | Altered miRNA | Results |
|---|---|---|---|---|---|
| Sinha et al. [95] | 2013 | Study of approximately 1000 miRNAs from 10 subjects with asthma and 10 healthy subjects. | EBC | 643 miRNA among which: let7, miR181c, miR-1307, miR-574-5p, miR-516a-5p, miR-42, ecc. | A total of 643 miRNAs were found in EBC with a significant difference in expression between the two groups. Furthermore, their presence has been demonstrated in vesicles capable of guaranteeing their stability in body fluids. |
| Levänen et al. [96] | 2013 | Analysis of exosomal miRNAs in a group of asthma patients vs. a group of healthy controls. | BAL | Let-7a, miR-21, miR-658, miR-24, miR-26a, miR-99a, miR-200c, miR-1268 | There are many differences in exosomal miRNA profiles between healthy subjects and patients with unprovoked, mild, stable asthma. |
| Gon et al. [97] | 2017 | Exosomal miRNA analysis in a group of asthmatic mice vs. a group of healthy mice. | BAL | miR-1827, miR-346, miR-574-5p. | The sorting of miRNA into EVs would be involved in the pathogenesis of allergic airway inflammation. |
| Qiao Y et al. [98] | 2018 | Serum exosomal miRNA analysis in rats with airway inflammation caused by zinc oxide nanoparticles and control rats. | SERUM | up-regulated: miR-134-5p, miR-207, miR-465-5p. down-regulated: miR-30b-5p, miR-19a-3p, miR-130a-3p | Serum exosomal miRNAs are involved in lung neutrophilic inflammation induced by zinc oxide. |
| Gutierrez et al. [99] | 2016 | Characterization of nasal exosomal miRNAs in children with or without rhinovirus (RV) infection. | Nasal airway secretions | miR-630, miR-302d-3p, miR-320 e miR-612 | Exosomal miR-155 is overexpressed in the upper airways of RV infected individuals. |
| References | Year of Publication | Study Design | Samples | Altered miRNA | Results |
|---|---|---|---|---|---|
| Khalyfa et al. [109] | 2016 | Circulating exosomal miRNAs analysis in children with OSA. | PLASMA | down-regulated: miRNA-16–5, miRNA-451a, miRNA-5100, miRNA-630. up-regulated: miRNA-4665–3p | Identification of a cluster of plasma-derived exosomal microRNAs to distinguish between normal and abnormal endothelial function in children with obstructive sleep apnea or obesity. |
| Khalyfa et al. [111] | 2016 | Study of microRNA from plasma exosomes in subjects exposed to intermittent hypoxia. | PLASMA | miR-4649-3p, miR-4436b-5p, miR-483-3p, miR-1202, miR-4505 | Intermittent hypoxia alters exosome cargo. |
| Almendros et al. [114] | 2016 | Analysis of exosomal miRNA under intermittent hypoxia conditions. | Human and mice PLASMA | miR-6418-5p, miR-6366, miR-5100, miR-451a, miR-5113, miR-671-5p, miR-709, miR-3082-5p, miR-882, miR-92-3p, miR-2137 | Circulating exosomes released under intermittent hypoxia conditions promote oncogenesis. |
| References | Year of Publication | Study Design | Samples | Altered miRNA | Results |
|---|---|---|---|---|---|
| Lacedonia et al. [8] | 2021 | Characterization of exosomal derived miRNAs in IPF patients and healthy controls. | SERUM | down-regulated: let-7d, miR-16 | Let-7d and miR-16 were found decreased in IPF subjects compared to normal subjects. |
| Zhu et al. [127] | 2021 | Analysis of exosomal miRNAs in rat with pulmonary fibrosis. | BAL | miR-204-5p | miR-204-5p induce the progression of pulmonary fibrosis in rats. |
| Liu et al. [128] | 2018 | Analysis of miRNA from exosomes useful as potential biomarkers for idiopathic pulmonary fibrosis. | BAL | up-pregulated: miR-125b, miR-128, miR-21, miR-100, miR-140-3p, miR-374b. down-regulated: let-7d, miR-103, miR-26 and miR-30a-5p. | Identification of miRNA biomarkers associated with idiopathic pulmonary fibrosis. |
| Makiguchi Y et al. [130] | 2016 | Serum extracellular vesicles miRNA evaluation as putative prognostic biomarkers of idiopathic pulmonary fibrosis. | SERUM | miR-21-5p | Serum extracellular vesicles miR-21-5p seems a potential prognostic biomarker for IPF. |
| Guiot et al. [132] | 2019 | To assess whether the IPF-related exosomal miRNA play a role in the progression of pulmonary fibrosis. | SPUTUM, PLASMA | miR-142-3p | Exosomally derived miR-142 probably plays an antifibrotic role by reducing TGFβ-R1. |
| Zhou et al. [133] | 2021 | Study of exosomal miRNA to improve the therapeutic approach of idiopathic pulmonary fibrosis. | MESENCHYMAL STEM CELLS | miR-186 | miR-186 in extracellular vesicles from bone marrow mesenchymal stem cells relieves idiopathic pulmonary fibrosis, |
| Wan et al. [134] | 2020 | Evaluation of cargo from mesenchymal stem cells-derived EV (extracellular vesicles) in IPF. | MESENCHYMAL STEM CELLS | miRNA-29b-3p | miRNA-29b-3p from mesenchymal stem Cell-derived extracellular vesicles decrease the FZD6 expression repressing the fibroblast proliferation. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soccio, P.; Moriondo, G.; Lacedonia, D.; Tondo, P.; Quarato, C.M.I.; Foschino Barbaro, M.P.; Scioscia, G. EVs-miRNA: The New Molecular Markers for Chronic Respiratory Diseases. Life 2022, 12, 1544. https://doi.org/10.3390/life12101544
Soccio P, Moriondo G, Lacedonia D, Tondo P, Quarato CMI, Foschino Barbaro MP, Scioscia G. EVs-miRNA: The New Molecular Markers for Chronic Respiratory Diseases. Life. 2022; 12(10):1544. https://doi.org/10.3390/life12101544
Chicago/Turabian StyleSoccio, Piera, Giorgia Moriondo, Donato Lacedonia, Pasquale Tondo, Carla Maria Irene Quarato, Maria Pia Foschino Barbaro, and Giulia Scioscia. 2022. "EVs-miRNA: The New Molecular Markers for Chronic Respiratory Diseases" Life 12, no. 10: 1544. https://doi.org/10.3390/life12101544
APA StyleSoccio, P., Moriondo, G., Lacedonia, D., Tondo, P., Quarato, C. M. I., Foschino Barbaro, M. P., & Scioscia, G. (2022). EVs-miRNA: The New Molecular Markers for Chronic Respiratory Diseases. Life, 12(10), 1544. https://doi.org/10.3390/life12101544






