Post-Transcriptional Control in the Regulation of Polyhydroxyalkanoates Synthesis
Abstract
1. Introduction
1.1. The Age of Plastics
1.2. Polyhydroxyalkanoates: Bio-Based Biodegradable Plastics
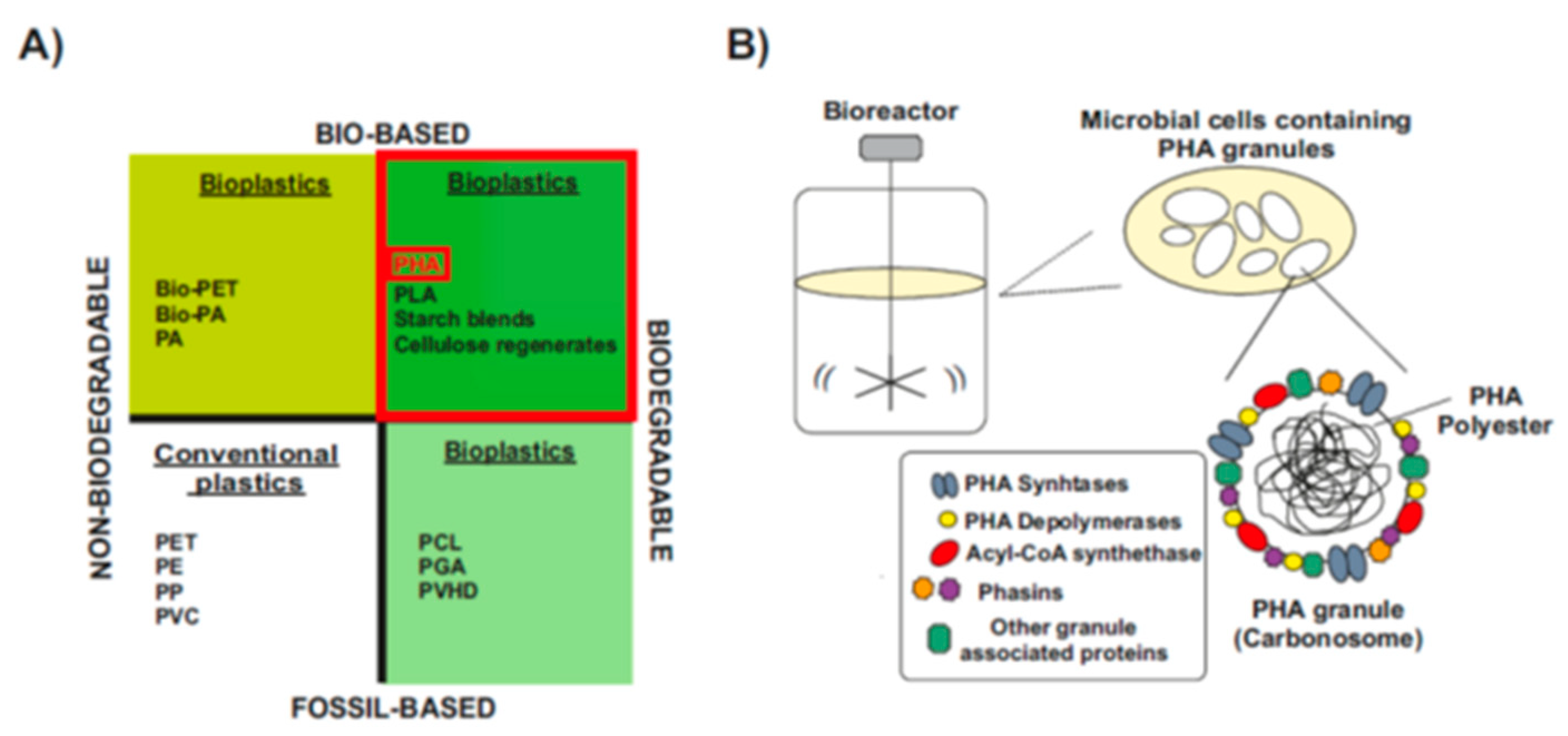
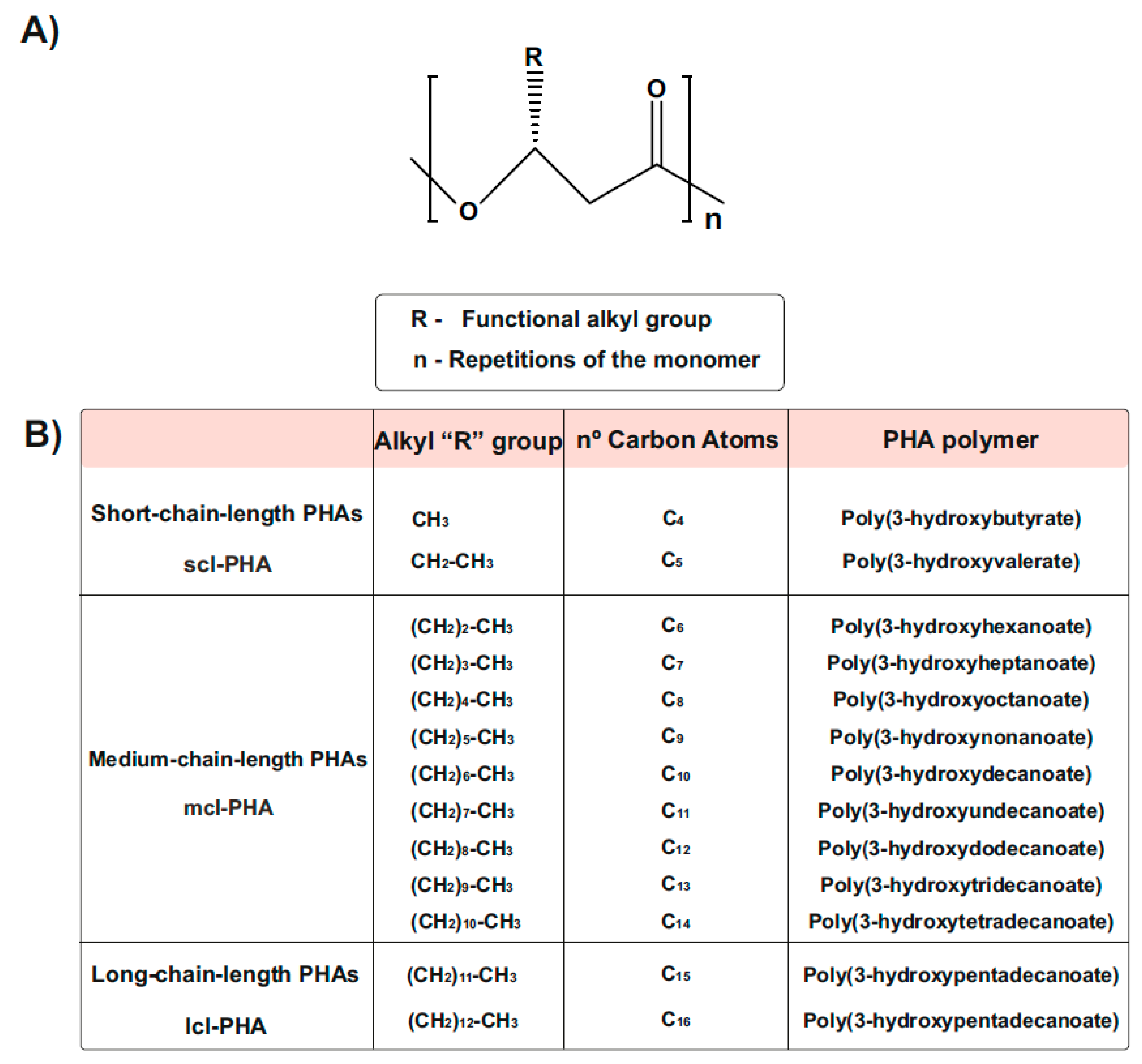
1.3. Types and Chemical Structure of PHAs Polymers
1.4. Natural PHA Producers and Engineering of Non-PHA Producers
1.5. PHA Composition and Preferred Carbon Source
1.6. RNA World
2. RNA-Mediated Control in Native Synthesis of PHAs
2.1. The Expanding RNA World: Non-Coding Bacterial RNome
2.1.1. RNA-Binding Proteins and Regulatory Networks
2.1.2. Post-Transcriptional Regulation by Ribonucleases
2.2. Post-Transcriptional Regulation of sRNAs and Their Implications for Microbial PHAs Synthesis in Different Microorganisms
2.2.1. MmgR sRNA Is a Negative Regulator of PHB Accumulation in Sinorhizobium meliloti
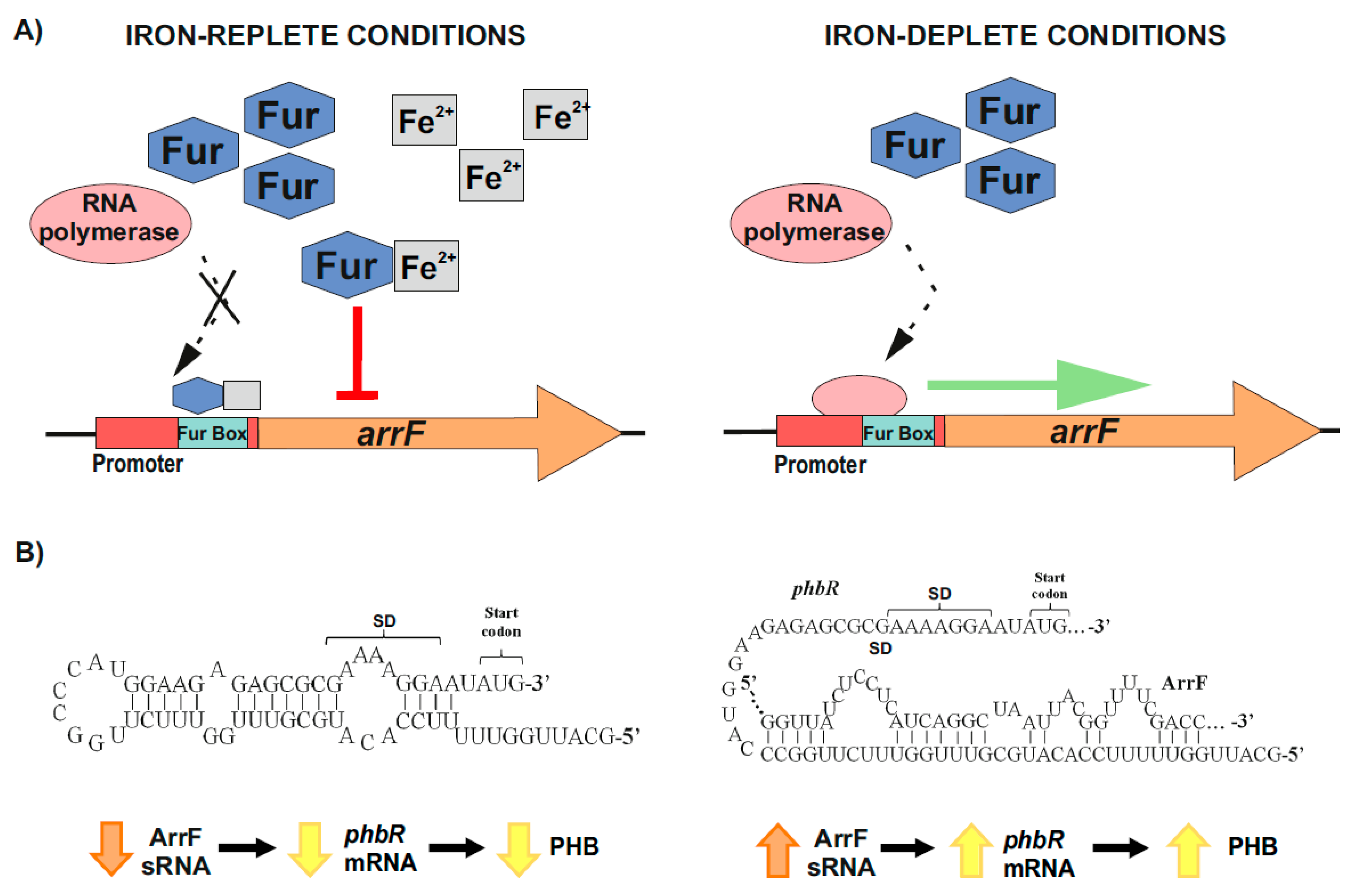
2.2.2. Post-Transcriptional Control of PhbR as Key Step during PHB Production in Azotobacter vinelandii
2.2.3. Global Post-Transcriptional Regulatory Protein Crc as Main Target of sRNAs CrcZ and CrcY in Pseudomonas putida
2.2.4. Post-Transcriptional Control of phaC1 Synthase as a Key Aspect along PHA Synthesis in P. putida CA-3
3. Conclusions and Perspectives
3.1. Role of Post-Transcriptional Regulation during the Native Synthesis of PHAs
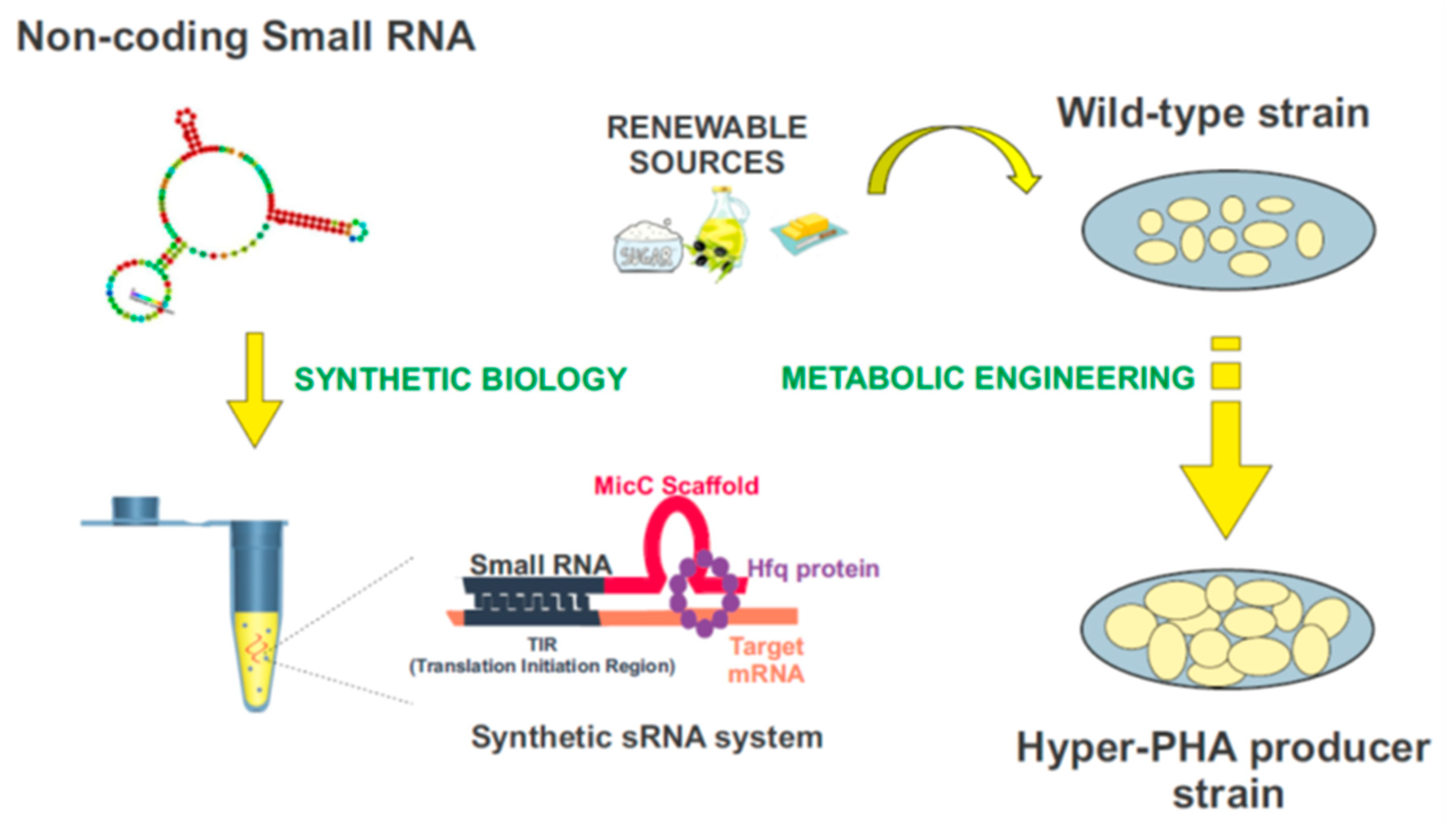
3.2. Controlling PHAs Production in Bacteria via Synthetic Small Non-Coding RNAs
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Kawecki, D.; Scheeder, P.R.W.; Nowack, B. Probabilistic Material Flow Analysis of Seven Commodity Plastics in Europe. Environ. Sci. Technol. 2018, 52, 9874–9888. [Google Scholar] [CrossRef]
- Chae, Y.; An, Y.J.A. Current Research Trends on Plastic Pollution and Ecological Impacts on the Soil Ecosystem: A Review. Environ. Pollut. 2018, 240, 387–395. [Google Scholar] [CrossRef]
- Courtene-Jones, W.; Quinn, B.; Gary, S.F.; Mogg, A.O.M.; Narayanaswamy, B.E. Microplastic Pollution Identified in Deep-Sea Water and Ingested by Benthic Invertebrates in the Rockall Trough, North Atlantic Ocean. Environ. Pollut. 2017, 231, 271–280. [Google Scholar] [CrossRef]
- Wagner, S.; Klöckner, P.; Stier, B.; Römer, M.; Seiwert, B.; Reemtsma, T.; Schmidt, C. Relationship Between Discharge and River Plastic Concentrations in a Rural and an Urban Catchment. Environ. Sci. Technol. 2019, 53, 10082–10091. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, T.; Kang, S.; Sillanpää, M. Importance of Atmospheric Transport for Microplastics Deposited in Remote Areas. Environ. Pollut. 2019, 254, 112953. [Google Scholar] [CrossRef]
- Jepsen, E.M.; de Bruyn, P.J.N. Pinniped Entanglement in Oceanic Plastic Pollution: A Global Review. Mar. Pollut. Bull. 2019, 145, 295–305. [Google Scholar] [CrossRef]
- Calafat, A.M.; Ye, X.; Wong, L.Y.; Reidy, J.A.; Needham, L.L. Exposure of the U.S. population to bisphenol A and 4-tertiary-octylphenol: 2003–2004. Environ. Health Perspect. 2008, 116, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Vandenberg, L.N.; Hauser, R.; Marcus, M.; Olea, N.; Welshons, W.V. Human Exposure to Bisphenol A (BPA). Reprod. Toxicol. 2007, 24, 139–177. [Google Scholar] [CrossRef] [PubMed]
- Luengo, J.M.; Garcia, B.; Sandoval, A.; Naharro, G.; Olivera, E.R. Bioplastics from microorganisms. Curr. Opin. Microbiol. 2003, 6, 251–260. [Google Scholar] [CrossRef]
- Chen, G.Q. A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry. Chem. Soc. Rev. 2009, 38, 2434–2446. [Google Scholar] [CrossRef] [PubMed]
- Patrício, S.A.L.; Prata, J.C.; Walker, T.R.; Campos, D.; Duarte, A.C.; Soares, A.M.V.M.; Barcelò, D.; Rocha-Santos, T. Rethinking and optimising plastic waste management under COVID-19 pandemic: Policy solutions based on redesign and reduction of single-use plastics and personal protective equipment. Sci. Total Environ. 2020, 742, 140565. [Google Scholar] [CrossRef]
- Mezzina, M.P.; Manoli, M.T.; Prieto, M.A.; Nikel, P.I. Engineering Native and Synthetic Pathways in Pseudomonas putida for the Production of Tailored Polyhydroxyalkanoates. Biotechnol. J. 2020, 16, e2000165. [Google Scholar] [CrossRef] [PubMed]
- Elbahloul, Y.; Steinbüchel, A. Large-scale Production of poly(3-hydroxyoctanoic Acid) by Pseudomonas putida GPo1 and a Simplified Downstream Process. Appl. Environ. Microbiol. 2009, 75, 643–651. [Google Scholar] [CrossRef]
- Martínez, V.; García, P.; García, J.L.; Prieto, M.A. Controlled Autolysis Facilitates the Polyhydroxyalkanoate Recovery in Pseudomonas putida KT2440. Microb. Biotechnol. 2011, 4, 533–547. [Google Scholar] [CrossRef]
- Escapa, I.F.; del Cerro, C.; García, J.L.; Prieto, M.A. The Role of GlpR Repressor in Pseudomonas putida KT2440 Growth and PHA Production from Glycerol. Environ. Microbiol. 2013, 15, 93–110. [Google Scholar] [CrossRef]
- Rujnic-Sokele, M.; Pilipovic, A. Challenges and opportunities of biodegradable plastics: A mini review. Waste Manag. Res. 2017, 35, 132–140. [Google Scholar] [CrossRef]
- Serafim, L.S.; Lemos, P.C.; Albuquerque, M.G.; Reis, M.A. Strategies for PHA Production by Mixed Cultures and Renewable Waste Materials. Appl. Microbiol. Biotechnol. 2008, 81, 615–628. [Google Scholar] [CrossRef] [PubMed]
- Castilho, L.R.; Mitchell, D.A.; Freire, D.M. Production of Polyhydroxyalkanoates (PHAs) From Waste Materials and By-Products by Submerged and Solid-State Fermentation. Bioresour. Technol. 2009, 100, 5996–6009. [Google Scholar] [CrossRef]
- European Bioplastics Conference, Berlin. Available online: https://www.european-bioplastics.org/903/ (accessed on 29 November 2016).
- Razza, F.; Innocentii, F. Bioplastics from renewable resources: The benefits of biodegradability. Asia Pac. J. Chem. Eng. 2012, 7, S301–S309. [Google Scholar] [CrossRef]
- Iwata, T. Biodegradable and Bio-Based Polymers: Future Prospects of Eco-Friendly Plastics. Angew. Chem. 2015, 54. [Google Scholar] [CrossRef]
- Rehm, B.H. Polyester synthases: Natural catalysts for plastics. Biochem. J. 2003, 376, 15–33. [Google Scholar] [CrossRef]
- Możejko, J.; Ciesielski, S. Pulsed Feeding Strategy Is More Favorable to Medium-Chain-Length Polyhydroxyalkanoates Production from Waste Rapeseed Oil. Biotechnol. Prog. 2014, 30, 1243–1246. [Google Scholar] [CrossRef]
- Bresan, S.; Sznajder, A.; Hauf, W.; Forchhammer, K.; Pfeiffer, D.; Jendrossek, D. Polyhydroxyalkanoate (PHA) Granules Have No Phospholipids. Sci. Rep. 2016, 6, 26612. [Google Scholar] [CrossRef] [PubMed]
- de Eugenio, L.I.; Escapa, I.F.; Morales, V.; Dinjaski, N.; Galán, B.; García, J.L.; Prieto, M.A. The Turnover of Medium-Chain-Length Polyhydroxyalkanoates in Pseudomonas putida KT2442 and the Fundamental Role of PhaZ Depolymerase for the Metabolic Balance. Environ. Microbiol. 2010, 12, 207–221. [Google Scholar] [CrossRef]
- Lukasiewicz, B.; Basnett, P.; Nigmatullin, R.; Matharu, R.; Knowles, J.C.; Roy, I. Binary Polyhydroxyalkanoate Systems for Soft Tissue Engineering. Acta Biomater. 2018, 71, 225–234. [Google Scholar] [CrossRef]
- Rebocho, A.; Pereira, J.; Freitas, F.; Neves, L.; Alves, V.; Sevrin, C.; Grandfils, C.; Reis, M. Production of Medium-Chain Length Polyhydroxyalkanoates by Pseudomonas citronellolis Grown in Apple Pulp Waste. Appl. Food Biotechnol. 2019, 6, 71–82. [Google Scholar]
- Cruz, M.V.; Freitas, F.; Paiva, A.; Mano, F.; Dionísio, M.; Ramos, A.M.; Reis, M.A. Valorization of fatty acids-containing wastes and byproducts into short- and medium-chain length polyhydroxyalkanoates. New Biotechnol. 2016, 33, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Koller, M. Biodegradable and Biocompatible Polyhydroxy-alkanoates (PHA): Auspicious Microbial Macromolecules for Pharmaceutical and Therapeutic Applications. Molecules 2018, 23, 362. [Google Scholar] [CrossRef]
- Khanna, S.; Srivastava, A.K. Recent advances in microbial polyhydroxyalkanoates. Process Biochem. 2005, 40, 607–619. [Google Scholar] [CrossRef]
- Tan, G.-Y.A.; Chen, C.-L.; Liya, L.L.G.; Wang, L.; Razaad, I.M.N.; Li, Y.; Zhao, L.; Mo, Y.; Wang, J.-Y. Start a Research on Biopolymer Polyhydroxyalkanoate (PHA): A Review. Polymers 2014, 6, 706–754. [Google Scholar] [CrossRef]
- Domínguez-Díaz, M.; Romo-Uribe, A. Viscoelastic behavior of biodegradable polyhydroxyalkanoates. Bioinspired Biomim. Nanobiomaterials 2012, 1, 214–220. [Google Scholar] [CrossRef]
- Mozejko-Ciesielska, J.; Mostek, A. Time-Course Proteomic Analysis of Pseudomonas putida KT2440 during Mcl-Polyhydroxyalkanoate Synthesis under Nitrogen Deficiency. Polymers 2019, 11, 748. [Google Scholar] [CrossRef] [PubMed]
- Kourmentza, C.; Plácido, J.; Venetsaneas, N.; Burniol-Figols, A.; Varrone, C.; Gavala, H.N.; Reis, M.A.M. Recent Advances and Challenges Towards Sustainable Polyhydroxyalkanoate (PHA) Production. Bioengineering 2017, 4, 55. [Google Scholar] [CrossRef] [PubMed]
- Madison, L.L.; Huisman, G.W. Metabolic engineering of poly(3-hydroxyalkanoates): From DNA to plastic. Microbiol. Mol. Biol. Rev. 1999, 63, 21–53. [Google Scholar] [CrossRef]
- Jendrossek, D.; Handrick, R. Microbial Degradation of Polyhydroxyalkanoates. Annu. Rev. Microbiol. 2002, 56, 403–432. [Google Scholar] [CrossRef]
- Williams, S.; Martin, D. Applications of Polyhydroxyalkanoates (PHA) in Medicine and Pharmacy, 4th ed. Biopolymers 2005, 20, 1–38. [Google Scholar] [CrossRef]
- Leong, Y.K.; Show, P.L.; Ooi, C.W.; Ling, T.C.; Lan, J.C. Current trends in polyhydroxyalkanoates (PHAs) biosynthesis: Insights from the recombinant Escherichia coli. J. Biotechnol. 2014, 180, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Chang, H.N. Production of Poly(hydroxyalkanoic Acid). Adv. Biochem. Eng./Biotechnol. 1995, 52, 27–58. [Google Scholar] [CrossRef]
- Cruz, M.V.; Gouveia, A.R.; Dionísio, M.; Freitas, F.; Reis, M.A.M. A Process Engineering Approach to Improve Production of P(3HB) by Cupriavidus necator from Used Cooking Oil. Int. J. Polym. Sci. 2019, 2019, 191650. [Google Scholar] [CrossRef]
- Castro-Mayorga, J.L.; Freitas, F.; Reis, M.A.M.; Prieto, M.A.; Lagaron, J.M. Biosynthesis of Silver Nanoparticles and Polyhydroxybutyrate Nanocomposites of Interest in Antimicrobial Applications. Int. J. Biol. Macromol. 2018, 108, 426–435. [Google Scholar] [CrossRef]
- Escapa, I.; Morales, V.; Martino, V.; Pollet, E.; Avérous, L.; García, J.; Prieto, M. Disruption of β-oxidation pathway in Pseudomonas putida KT2442 to produce new functionalized PHAs with thioester groups. Appl. Microbiol. Biotechnol. 2011, 89, 1583–1598. [Google Scholar] [CrossRef] [PubMed]
- Nikel, P.I.; Pettinari, M.J.; Galvagno, M.A.; Méndez, B.S. Poly(3-hydroxybutyrate) Synthesis from Glycerol by a Recombinant Escherichia coli arcA Mutant in Fed-Batch Microaerobic Cultures. Appl. Microbiol. Biotechnol. 2008, 77, 1337–1343. [Google Scholar] [CrossRef]
- Chen, G.Q.; Jian, X.R. Engineering Bacteria for Enhanced Polyhydroxyalkanoates (PHA) Biosynthesis. Synth. Syst. Biotechnol. 2017, 2, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Khosravi-Darani, K.; Mokhtari, Z.B.; Amai, T.; Tanaka, K. Microbial Production of Poly(hydroxybutyrate) From C₁ Carbon Sources. Appl. Microbiol. Biotechnol. 2013, 97, 1407–1424. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.J.; Dawes, E.A. Occurrence, Metabolism, Metabolic Role, and Industrial Uses of Bacterial Polyhydroxyalkanoates. Microbiol. Rev. 1990, 54, 450–472. [Google Scholar] [CrossRef]
- Steinbüchel, A.; Hein, S. Biochemical and Molecular Basis of Microbial Synthesis of Polyhydroxyalkanoates in Microorganisms. Adv. Biochem. Eng./Biotechnol. 2001, 71, 81–123. [Google Scholar] [CrossRef]
- Jung, Y.M.; Lee, Y.H. Utilization of Oxidative Pressure for Enhanced Production of Poly-Beta-Hydroxybutyrate and poly(3-hydroxybutyrate-3-hydroxyvalerate) in Ralstonia eutropha. J. Biosci. Bioeng. 2000, 90, 266–270. [Google Scholar] [CrossRef]
- Prieto, M.A.; de Eugenio, L.I.; Galán, B.; Luengo, J.M.; Witholt, B.J.M. Synthesis and Degradation of Polyhydroxyalkanoates In Pseudomonas: A Model System in Biology; Springerlink: Berlin, Germany, 2007; Volume V, pp. 397–428. [Google Scholar] [CrossRef]
- Poblete-Castro, I.; Becker, J.; Dohnt, K.; dos Santos, V.M.; Wittmann, C. Industrial biotechnology of Pseudomonas putida and related species. Appl. Microbiol. Biotechnol. 2012, 93, 2279–2290. [Google Scholar] [CrossRef]
- Borrero-de Acuña, J.; Bielecka, A.; Häussler, S.; Schobert, M.; Jahn, M.; Wittmann, C.; Jahn, D.; Poblete-Castro, I. Production of medium chain length polyhydroxyalkanoate in metabolic flux optimized Pseudomonas putida. Microb. Cell Factories 2014, 13, 1–15. [Google Scholar] [CrossRef]
- Mozejko-Ciesielska, J.; Dabrowska, D.; Szalewska-Palasz, A.; Ciesielski, S. Medium-chain-length polyhydroxyalkanoates synthesis by Pseudomonas putida KT2440 relA/spoT mutant: Bioprocess characterization and transcriptome analysis. AMB Express 2017, 7, 92. [Google Scholar] [CrossRef]
- Saramago, M.; Barria, C.; Dos Santos, R.F.; Silva, I.J.; Pobre, V.; Domingues, S.; Andrade, J.M.; Viegas, S.C.; Arraiano, C.M. The role of RNases in the regulation of small RNAs. Curr. Opin. Microbiol. 2014, 18, 105–115. [Google Scholar] [CrossRef]
- Saramago, M.; Peregrina, A.; Robledo, M.; Matos, R.G.; Hilker, R.; Serrania, J.; Becker, A.; Arraiano, C.M.; Jimenez-Zurdo, J.I. Sinorhizobium meliloti YbeY is an endoribonuclease with unprecedented catalytic features, acting as silencing enzyme in riboregulation. Nucleic Acids Res. 2017, 45, 1371–1391. [Google Scholar] [CrossRef] [PubMed]
- Torres-Quesada, O.; Millan, V.; Nisa-Martinez, R.; Bardou, F.; Crespi, M.; Toro, N.; Jimenez-Zurdo, J.I. Independent activity of the homologous small regulatory RNAs AbcR1 and AbcR2 in the legume symbiont Sinorhizobium meliloti. PLoS ONE 2013, 8, e68147. [Google Scholar] [CrossRef]
- Torres-Quesada, O.; Reinkensmeier, J.; Schluter, J.P.; Robledo, M.; Peregrina, A.; Giegerich, R.; Toro, N.; Becker, A.; Jimenez-Zurdo, J.I. Genome-wide profiling of Hfq-binding RNAs uncovers extensive post-transcriptional rewiring of major stress response and symbiotic regulons in Sinorhizobium meliloti. RNA Biol. 2014, 11, 563–579. [Google Scholar] [CrossRef]
- Matos, R.G.; Casinhas, J.; Bárria, C.; Dos Santos, R.F.; Silva, I.J.; Arraiano, C.M. The Role of Ribonucleases and sRNAs in the Virulence of Foodborne Pathogens. Front. Microbiol. 2017, 8, 910. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001, 2, 919–929. [Google Scholar] [CrossRef]
- Gottesman, S.; Storz, G. Bacterial small RNA regulators: Versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 2011, 3, a003798. [Google Scholar] [CrossRef]
- Wagner, E.G.H.; Romby, P. Small RNAs in Bacteria and Archaea: Who They Are, What They Do, and How They Do It. Adv. Genet. 2015, 90, 133–208. [Google Scholar] [CrossRef]
- La Rosa, R.; de la Pena, F.; Prieto, M.A.; Rojo, F. The Crc protein inhibits the production of polyhydroxyalkanoates in Pseudomonas putida under balanced carbon/nitrogen growth conditions. Environ. Microbiol. 2014, 16, 278–290. [Google Scholar] [CrossRef] [PubMed]
- Ryan, W.J.; O’Leary, N.D.; O’Mahony, M.; Dobson, A.D. GacS-dependent Regulation of Polyhydroxyalkanoate Synthesis in Pseudomonas putida CA-3. Appl. Environ. Microbiol. 2013, 79, 1795–1802. [Google Scholar] [CrossRef] [PubMed]
- Muriel-Millán, L.F.; Castellanos, M.; Hernandez-Eligio, J.A.; Moreno, S.; Espín, G. Posttranscriptional Regulation of PhbR, the Transcriptional Activator of Polyhydroxybutyrate Synthesis, by Iron and the sRNA ArrF in Azotobacter Vinelandii. Appl. Microbiol. Biotechnol. 2014, 98, 2173–2182. [Google Scholar] [CrossRef] [PubMed]
- Lagares, A., Jr.; Ceizel Borella, G.; Linne, U.; Becker, A.; Valverde, C. Regulation of polyhydroxybutyrate accumulation in Sinorhizobium meliloti by the trans-encoded small RNA MmgR. J. Bacteriol. 2017, 199, e00776-16. [Google Scholar] [CrossRef]
- Crick, F. Central dogma of molecular biology. Nature 1970, 227, 561–563. [Google Scholar] [CrossRef] [PubMed]
- Temin, H.M. Reverse transcription in the eukaryotic genome: Retroviruses, pararetroviruses, retrotransposons, and retrotranscripts. Mol. Biol. Evol. 1985, 2, 455–468. [Google Scholar] [CrossRef]
- Jimenez-Zurdo, J.I.; Valverde, C.; Becker, A. Insights into the noncoding RNome of nitrogen-fixing endosymbiotic alpha-proteobacteria. Mol. Plant. Microbe Interact. 2013, 26, 160–167. [Google Scholar] [CrossRef]
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by small RNAs in bacteria: Expanding frontiers. Mol. Cell 2011, 43, 880–891. [Google Scholar] [CrossRef]
- Gottesman, S.; McCullen, C.A.; Guillier, M.; Vanderpool, C.K.; Majdalani, N.; Benhammou, J.; Thompson, K.M.; FitzGerald, P.C.; Sowa, N.A.; FitzGerald, D.J. Small RNA Regulators and the Bacterial Response to Stress. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 1–11. [Google Scholar] [CrossRef]
- Updegrove, T.B.; Shabalina, S.A.; Storz, G. How Do Base-Pairing Small RNAs Evolve? FEMS Microbiol. Rev. 2015, 39, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Levine, E.; Zhang, Z.; Kuhlman, T.; Hwa, T. Quantitative Characteristics of Gene Regulation by Small RNA. PLoS Biol. 2007, 5, e229. [Google Scholar] [CrossRef] [PubMed]
- Viegas, S.C.; Arraiano, C.M. Regulating the Regulators: How Ribonucleases Dictate the Rules in the Control of Small Non-Coding RNAs. RNA Biol. 2008, 5, 230–243. [Google Scholar] [CrossRef]
- Storz, G.; Altuvia, S.; Wassarman, K.M. An abundance of RNA regulators. Annu. Rev. Biochem. 2005, 74, 199–217. [Google Scholar] [CrossRef]
- Oliva, G.; Sahr, T.; Buchrieser, C. Small RNAs, 5’ UTR elements and RNA-binding proteins in intracellular bacteria: Impact on metabolism and virulence. FEMS Microbiol. Rev. 2015, 39, 331–349. [Google Scholar] [CrossRef] [PubMed]
- Marzi, S.; Romby, P. RNA Mimicry, a Decoy for Regulatory Proteins. Mol. Microbiol. 2012, 83, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Babitzke, P.; Romeo, T. CsrB sRNA family: Sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 2007, 10, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.; Luisi, B.F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011, 9, 578–589. [Google Scholar] [CrossRef]
- Smirnov, A.; Förstner, K.U.; Holmqvist, E.; Otto, A.; Günster, R.; Becher, D.; Reinhardt, R.; Vogel, J. Grad-seq Guides the Discovery of ProQ as a Major Small RNA-binding Protein. Proc. Natl. Acad. Sci. USA 2016, 113, 11591–11596. [Google Scholar] [CrossRef]
- Melamed, S. New sequencing methodologies reveal interplay between multiple RNA-binding proteins and their RNAs. Curr. Genet. 2020, 66, 713–717. [Google Scholar] [CrossRef]
- Van Assche, E.; Van Puyvelde, S.; Vanderleyden, J.; Steenackers, H.P. RNA-binding proteins involved in post-transcriptional regulation in bacteria. Front. Microbiol. 2015, 6, 141. [Google Scholar] [CrossRef]
- Quendera, A.P.; Seixas, A.F.; Dos Santos, R.F.; Santos, I.; Silva, J.P.N.; Arraiano, C.M.; Andrade, J.M. RNA-Binding Proteins Driving the Regulatory Activity of Small Non-coding RNAs in Bacteria. Front. Mol. Biosci. 2020, 7, 78. [Google Scholar] [CrossRef]
- Herschlag, D. RNA Chaperones and the RNA Folding Problem. J. Biol. Chem. 1995, 270, 20871–20874. [Google Scholar] [CrossRef]
- Torres-Quesada, O.; Oruezabal, R.I.; Peregrina, A.; Jofre, E.; Lloret, J.; Rivilla, R.; Toro, N.; Jimenez-Zurdo, J.I. The Sinorhizobium meliloti RNA chaperone Hfq influences central carbon metabolism and the symbiotic interaction with alfalfa. BMC Microbiol. 2010, 10, 71. [Google Scholar] [CrossRef]
- Sobrero, P.; Valverde, C. The bacterial protein Hfq: Much more than a mere RNA-binding factor. Crit. Rev. Microbiol. 2012, 38, 276–299. [Google Scholar] [CrossRef]
- Franze de Fernandez, M.T.; Eoyang, L.; August, J.T. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature 1968, 219, 588–590. [Google Scholar] [CrossRef]
- Sauer, E.; Weichenrieder, O. Structural basis for RNA 3’-end recognition by Hfq. Proc. Natl. Acad. Sci. USA 2011, 108, 13065–13070. [Google Scholar] [CrossRef] [PubMed]
- De Lay, N.; Schu, D.J.; Gottesman, S. Bacterial Small RNA-based Negative Regulation: Hfq and Its Accomplices. J. Biol. Chem. 2013, 288, 7996–8003. [Google Scholar] [CrossRef]
- Link, T.M.; Valentin-Hansen, P.; Brennan, R.G. Structure of Escherichia Coli Hfq Bound to Polyriboadenylate RNA. Proc. Natl. Acad. Sci. USA 2009, 106, 19292–19297. [Google Scholar] [CrossRef]
- Wang, W.; Wang, L.; Wu, J.; Gong, Q.; Shi, Y. Hfq-bridged ternary complex is important for translation activation of rpoS by DsrA. Nucleic Acids Res. 2013, 41, 5938–5948. [Google Scholar] [CrossRef][Green Version]
- Andrade, J.M.; Dos Santos, R.F.; Chelysheva, I.; Ignatova, Z.; Arraiano, C.M. The RNA-binding protein Hfq is important for ribosome biogenesis and affects translation fidelity. EMBO J. 2018, 37, e97631. [Google Scholar] [CrossRef]
- Moreno, R.; Ruiz-Manzano, A.; Yuste, L.; Rojo, F. The Pseudomonas Putida Crc Global Regulator Is an RNA Binding Protein That Inhibits Translation of the AlkS Transcriptional Regulator. Mol. Microbiol. 2007, 64, 665–675. [Google Scholar] [CrossRef]
- Malecka, E.M.; Bassani, F.; Dendooven, T.; Sonnleitner, E.; Rozner, M.; Albanese, T.G.; Resch, A.; Luisi, B.; Woodson, S.; Bläsi, U. Stabilization of Hfq-mediated translational repression by the co-repressor Crc in Pseudomonas aeruginosa. Nucleic Acids Res. 2021, 49, 7075–7087. [Google Scholar] [CrossRef] [PubMed]
- Moreno, R.; Fonseca, P.; Rojo, F. Two Small RNAs, CrcY and CrcZ, Act in Concert to Sequester the Crc Global Regulator in Pseudomonas putida, Modulating Catabolite Repression. Mol. Microbiol. 2012, 83, 24–40. [Google Scholar] [CrossRef]
- Sonnleitner, E.; Abdou, L.; Haas, D. Small RNA as Global Regulator of Carbon Catabolite Repression in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2009, 106, 21866–21871. [Google Scholar] [CrossRef] [PubMed]
- La Rosa, R.; Nogales, J.; Rojo, F. The Crc/CrcZ-CrcY Global Regulatory System Helps the Integration of Gluconeogenic and Glycolytic Metabolism in Pseudomonas putida. Environ. Microbiol. 2015, 17, 3362–3378. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Arranz, S.; Sanchez-Hevia, D.; Rojo, F.; Moreno, R. Effect of Crc and Hfq proteins on the transcription, processing, and stability of the Pseudomonas putida CrcZ sRNA. RNA 2016, 22, 1902–1917. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Arraiano, C.M.; Maquat, L.E. Post-transcriptional control of gene expression: Effectors of mRNA decay. Mol. Microbiol. 2003, 49, 267–276. [Google Scholar] [CrossRef]
- Ow, M.C.; Perwez, T.; Kushner, S.R. RNase G of Escherichia coli exhibits only limited functional overlap with its essential homologue, RNase E. Mol. Microbiol. 2003, 49, 607–622. [Google Scholar] [CrossRef]
- Morita, T.; Maki, K.; Aiba, H. RNase E-based ribonucleoprotein complexes: Mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes. Dev. 2005, 19, 2176–2186. [Google Scholar] [CrossRef]
- Maki, K.; Uno, K.; Morita, T.; Aiba, H. RNA, but not protein partners, is directly responsible for translational silencing by a bacterial Hfq-binding small RNA. Proc. Natl. Acad. Sci. USA 2008, 105, 10332–10337. [Google Scholar] [CrossRef]
- Moll, I.; Afonyushkin, T.; Vytvytska, O.; Kaberdin, V.R.; Blasi, U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 2003, 9, 1308–1314. [Google Scholar] [CrossRef]
- Dendooven, T.; Sinha, D.; Roeselová, A.; Cameron, T.A.; De Lay, N.R.; Luisi, B.F.; Bandyra, K.J. A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation. Mol. Cell 2021, 81, 2901–2913.e5. [Google Scholar] [CrossRef]
- Santos, J.M.; Drider, D.; Marujo, P.E.; Lopez, P.; Arraiano, C.M. Determinant role of E. coli RNase III in the decay of both specific and heterologous mRNAs. FEMS Microbiol. Lett. 1997, 157, 31–38. [Google Scholar] [CrossRef][Green Version]
- Arraiano, C.M.; Andrade, J.M.; Domingues, S.; Guinote, I.B.; Malecki, M.; Matos, R.G.; Moreira, R.N.; Pobre, V.; Reis, F.P.; Saramago, M.; et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol. Rev. 2010, 34, 883–923. [Google Scholar] [CrossRef] [PubMed]
- Lamontagne, B.; Elela, S.A. Evaluation of the RNA determinants for bacterial and yeast RNase III binding and cleavage. J. Biol. Chem. 2004, 279, 2231–2241. [Google Scholar] [CrossRef] [PubMed]
- Reinecke, F.; Steinbüchel, A. Ralstonia eutropha strain H16 as model organism for PHA metabolism and for biotechnological production of technically interesting biopolymers. J. Mol. Microbiol. Biotechnol. 2009, 16, 91–108. [Google Scholar] [CrossRef]
- del Val, C.; Romero-Zaliz, R.; Torres-Quesada, O.; Peregrina, A.; Toro, N.; Jimenez-Zurdo, J.I. A survey of sRNA families in alpha-proteobacteria. RNA Biol. 2012, 9, 119–129. [Google Scholar] [CrossRef]
- Zevenhuizen, L.P. Cellular glycogen, beta-1,2,-glucan, poly beta-hydroxybutyric acid and extracellular polysaccharides in fast-growing species of Rhizobium. Antonie Leeuwenhoek 1981, 47, 481–497. [Google Scholar] [CrossRef]
- Wang, C.; Saldanha, M.; Sheng, X.; Shelswell, K.J.; Walsh, K.T.; Sobral, B.W.S.; Charles, T.C. Roles of poly-3-hydroxybutyrate (PHB) and glycogen in symbiosis of Sinorhizobium meliloti with Medicago sp. Microbiology 2007, 153, 388–398. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schlüter, J.P.; Reinkensmeier, J.; Barnett, M.J.; Lang, C.; Krol, E.; Giegerich, R.; Long, S.R.; Becker, A. Global mapping of transcription start sites and promoter motifs in the symbiotic alpha-proteobacterium Sinorhizobium meliloti 1021. BMC Genom. 2013, 14, 156. [Google Scholar] [CrossRef]
- Baumgardt, K.; Smidova, K.; Rahn, H.; Lochnit, G.; Robledo, M.; Evguenieva-Hackenberg, E. The stress-related, rhizobial small RNA RcsR1 destabilizes the autoinducer synthase encoding mRNA sinI in Sinorhizobium meliloti. RNA Biol. 2016, 13, 486–499. [Google Scholar] [CrossRef]
- Robledo, M.; Peregrina, A.; Millan, V.; Garcia-Tomsig, N.I.; Torres-Quesada, O.; Mateos, P.F.; Becker, A.; Jimenez-Zurdo, J.I. A conserved alpha-proteobacterial small RNA contributes to osmoadaptation and symbiotic efficiency of rhizobia on legume roots. Environ. Microbiol. 2017. [Google Scholar] [CrossRef]
- Sobrero, P.; Valverde, C. Evidences of autoregulation of hfq expression in Sinorhizobium meliloti strain 2011. Arch. Microbiol. 2011, 193, 629–639. [Google Scholar] [CrossRef]
- Valverde, C.; Livny, J.; Schluter, J.P.; Reinkensmeier, J.; Becker, A.; Parisi, G. Prediction of Sinorhizobium meliloti sRNA genes and experimental detection in strain 2011. BMC Genom. 2008, 9, 416. [Google Scholar] [CrossRef]
- Lagares, A.; Roux, I.; Valverde, C. Phylogenetic distribution and evolutionary pattern of an α-proteobacterial small RNA gene that controls polyhydroxybutyrate accumulation in Sinorhizobium meliloti. Mol. Phylogenetics Evol. 2016, 99, 182–193. [Google Scholar] [CrossRef]
- Ceizel-Borella, G.; Lagares, A., Jr.; Valverde, C. Expression of the small regulatory RNA gene mmgR is regulated negatively by AniA and positively by NtrC in Sinorhizobium meliloti 2011. Microbiology 2018, 164, 88–98. [Google Scholar] [CrossRef]
- Pötter, M.; Steinbüchel, A. Poly(3-hydroxybutyrate) granule-associated proteins: Impacts on poly(3-hydroxybutyrate) synthesis and degradation. Biomacromolecules 2005, 6, 552–560. [Google Scholar] [CrossRef]
- Ceizel-Borella, G.; Lagares, A., Jr.; Valverde, C. Expression of the Sinorhizobium meliloti small RNA gene mmgR is controlled by the nitrogen source. FEMS Microbiol. Lett. 2016, 363, fnw069. [Google Scholar] [CrossRef]
- Rediers, H.; Vanderleyden, J.; De Mot, R. Azotobacter vinelandii: A Pseudomonas in disguise? Microbiology 2004, 150, 1117–1119. [Google Scholar] [CrossRef] [PubMed]
- Özen, A.I.; Ussery, D.W. Defining the Pseudomonas genus: Where do we draw the line with Azotobacter? Microb. Ecol. 2012, 63, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Jensen, H.L. The Azotobacteriaceae. Bacteriol. Rev. 1954, 18, 195–214. [Google Scholar] [CrossRef]
- Gauri, S.S.; Mandal, S.M.; Pati, B.R. Impact of Azotobacter exopolysaccharides on sustainable agriculture. Appl. Microbiol. Biotechnol. 2012, 95, 331–338. [Google Scholar] [CrossRef]
- Page, W.J.; Knosp, O. Hyperproduction of Poly-beta-Hydroxybutyrate during Exponential Growth of Azotobacter vinelandii UWD. Appl. Environ. Microbiol. 1989, 55, 1334–1339. [Google Scholar] [CrossRef]
- Noar, J.D.; Bruno-Bárcena, J.M. Azotobacter vinelandii: The source of 100 years of discoveries and many more to come. Microbiology 2018, 164, 421–436. [Google Scholar] [CrossRef]
- Lee, G.N.; Na, J. Future of microbial polyesters. Microb. Cell Factories 2013, 12, 54. [Google Scholar] [CrossRef]
- Castañeda, M.; Guzmán, J.; Moreno, S.; Espín, G. The GacS Sensor Kinase Regulates Alginate and Poly-β-Hydroxybutyrate Production in Azotobacter vinelandii. J. Bacteriol. 2000, 182, 2624–2628. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Eligio, A.; Castellanos, M.; Moreno, S.; Espín, G. Transcriptional activation of the Azotobacter vinelandii polyhydroxybutyrate biosynthetic genes phbBAC by PhbR and RpoS. Microbiology 2011, 157, 3014–3023. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hernandez-Eligio, A.; Moreno, S.; Castellanos, M.; Castañeda, M.; Nuñez, C.; Muriel-Millan, L.F.; Espín, G. RsmA post-transcriptionally controls PhbR expression and polyhydroxybutyrate biosynthesis in Azotobacter vinelandii. Microbiology 2012, 158, 1953–1963. [Google Scholar] [CrossRef] [PubMed]
- Lapouge, K.; Schubert, M.; Allain, F.H.; Haas, D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: From RNA recognition to regulation of social behaviour. Mol. Microbiol. 2008, 67, 241–253. [Google Scholar] [CrossRef]
- Velázquez-Sánchez, C.; Espín, G.; Peña, C.; Segura, D. The Modification of Regulatory Circuits Involved in the Control of Polyhydroxyalkanoates Metabolism to Improve Their Production. Front. Bioeng. Biotechnol. 2020, 8, 386. [Google Scholar] [CrossRef] [PubMed]
- Manzo, J.; Cocotl-Yañez, M.; Tzontecomani, T.; Martínez, V.M.; Bustillos, R.; Velásquez, C.; Goiz, Y.; Solís, Y.; López, L.; Fuentes, L.E.; et al. Post-transcriptional regulation of the alginate biosynthetic gene algD by the Gac/Rsm system in Azotobacter vinelandii. J. Mol. Microbiol. Biotechnol. 2011, 21, 147–159. [Google Scholar] [CrossRef]
- Bedoya-Pérez, L.P.; Muriel-Millán, L.F.; Moreno, S.; Quiroz-Rocha, E.; Rivera-Gómez, N.; Espín, G. The pyrophosphohydrolase RppH is involved in the control of RsmA/CsrA expression in Azotobacter vinelandii and Escherichia coli. Microbiol. Res. 2018, 214, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Massé, E.; Salvail, H.; Desnoyers, G.; Arguin, M. Small RNAs controlling iron metabolism. Curr. Opin. Microbiol. 2007, 10, 140–145. [Google Scholar] [CrossRef]
- Oglesby-Sherrouse, A.G.; Murphy, E.R. Iron-responsive bacterial small RNAs: Variations on a theme. Met. Integr. Biometal Sci. 2013, 5, 276–286. [Google Scholar] [CrossRef] [PubMed]
- Massé, E.; Gottesman, S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 2002, 99, 4620–4625. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.S.; Kwon, Y.M. Small RNA ArrF regulates the expression of sodB and feSII genes in Azotobacter vinelandii. Curr. Microbiol. 2008, 57, 593–597. [Google Scholar] [CrossRef]
- Pyla, R.; Kim, T.J.; Silva, J.L.; Jung, Y.S. Overproduction of poly-beta-hydroxybutyrate in the Azotobacter vinelandii mutant that does not express small RNA ArrF. Appl. Microbiol. Biotechnol. 2009, 84, 717–724. [Google Scholar] [CrossRef] [PubMed]
- Lugtenberg, B.J.; Dekkers, L.C. What makes Pseudomonas bacteria rhizosphere competent? Environ. Microbiol. 1999, 1, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Nikel, P.I.; de Lorenzo, V. Pseudomonas putida as a functional chassis for industrial biocatalysis: From native biochemistry to trans-metabolism. Metab. Eng. 2018, 50, 142–155. [Google Scholar] [CrossRef]
- Ramos, J.L.; Gallegos, M.T.; Marqués, S.; Ramos-González, M.I.; Espinosa-Urgel, M.; Segura, A. Responses of Gram-negative bacteria to certain environmental stressors. Curr. Opin. Microbiol. 2001, 4, 166–171. [Google Scholar] [CrossRef]
- Dos Santos, V.A.; Heim, S.; Moore, E.R.; Strätz, M.; Timmis, K.N. Insights into the genomic basis of niche specificity of Pseudomonas putida KT2440. Environ. Microbiol. 2004, 6, 1264–1286. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Higdon, R.; Kolker, E. Interplay of heritage and habitat in the distribution of bacterial signal transduction systems. Mol. Biosyst. 2010, 6, 721–728. [Google Scholar] [CrossRef]
- Moreno, R.; Martínez-Gomariz, M.; Yuste, L.; Gil, C.; Rojo, F. The Pseudomonas putida Crc global regulator controls the hierarchical assimilation of amino acids in a complete medium: Evidence from proteomic and genomic analyses. Proteomics 2009, 9, 2910–2928. [Google Scholar] [CrossRef]
- Hernández-Arranz, S.; Moreno, R.; Rojo, F. The translational repressor Crc controls the Pseudomonas putida benzoate and alkane catabolic pathways using a multi-tier regulation strategy. Environ. Microbiol. 2013, 15, 227–241. [Google Scholar] [CrossRef]
- La Rosa, R.; Behrends, V.; Williams, H.D.; Bundy, J.G.; Rojo, F. Influence of the Crc regulator on the hierarchical use of carbon sources from a complete medium in Pseudomonas. Environ. Microbiol. 2016, 18, 807–818. [Google Scholar] [CrossRef]
- Ruiz-Manzano, A.; Yuste, L.; Rojo, F. Levels and activity of the Pseudomonas putida global regulatory protein Crc vary according to growth conditions. J. Bacteriol. 2005, 187, 3678–3686. [Google Scholar] [CrossRef]
- Moreno, R.; Rojo, F. The target for the Pseudomonas putida Crc global regulator in the benzoate degradation pathway is the BenR transcriptional regulator. J. Bacteriol. 2008, 190, 1539–1545. [Google Scholar] [CrossRef] [PubMed][Green Version]
- García-Mauriño, S.M.; Pérez-Martínez, I.; Amador, C.I.; Canosa, I.; Santero, E. Transcriptional activation of the CrcZ and CrcY regulatory RNAs by the CbrB response regulator in Pseudomonas putida. Mol. Microbiol. 2013, 89, 189–205. [Google Scholar] [CrossRef]
- López, N.I.; Pettinari, M.J.; Nikel, P.I.; Méndez, B.S. Polyhydroxyalkanoates: Much More than Biodegradable Plastics. Adv. Appl. Microbiol. 2015, 93, 73–106. [Google Scholar] [CrossRef] [PubMed]
- Sonnleitner, E.; Bläsi, U. Regulation of Hfq by the RNA CrcZ in Pseudomonas aeruginosa carbon catabolite repression. PLoS Genet. 2014, 10, e1004440. [Google Scholar] [CrossRef]
- Moreno, R.; Hernández-Arranz, S.; La Rosa, R.; Yuste, L.; Madhushani, A.; Shingler, V.; Rojo, F. The Crc and Hfq proteins of Pseudomonas putida cooperate in catabolite repression and formation of ribonucleic acid complexes with specific target motifs. Environ. Microbiol. 2015, 17, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Huisman, G.W.; Wonink, E.; Meima, R.; Kazemier, B.; Terpstra, P.; Witholt, B. Metabolism of poly(3-hydroxyalkanoates) (PHAs) by Pseudomonas oleovorans. Identification and sequences of genes and function of the encoded proteins in the synthesis and degradation of PHA. J. Biol. Chem. 1991, 266, 2191–2198. [Google Scholar] [CrossRef]
- Browne, P.; Barret, M.; O’Gara, F.; Morrissey, J.P. Computational prediction of the Crc regulon identifies genus-wide and species-specific targets of catabolite repression control in Pseudomonas bacteria. BMC Microbiol. 2010, 10, 300. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, N.D.; O’Connor, K.E.; Ward, P.; Goff, M.; Dobson, A.D.W. Genetic Characterization of Accumulation of Polyhydroxyalkanoate from Styrene in Pseudomonas putida CA-3. Appl. Environ. Microbiol. 2005, 71, 4380–4387. [Google Scholar] [CrossRef]
- Kalia, V.C.; Lal, S.; Cheema, S. Insight in to the phylogeny of polyhydroxyalkanoate biosynthesis: Horizontal gene transfer. Gene 2007, 389, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Apura, P.; Saramago, M.; Peregrina, A.; Viegas, S.C.; Carvalho, S.M.; Saraiva, L.M.; Arraiano, C.M.; Domingues, S. Tailor-made sRNAs: A plasmid tool to control the expression of target mRNAs in Pseudomonas putida. Plasmid 2020, 109, 102503. [Google Scholar] [CrossRef] [PubMed]
- Freemont, P.S. Synthetic biology industry: Data-driven design is creating new opportunities in biotechnology. Emerg. Top. Life Sci. 2019, 3, 651–657. [Google Scholar] [CrossRef]
- Foley, P.L.; Shuler, M.L. Considerations for the design and construction of a synthetic platform cell for biotechnological applications. Biotechnol. Bioeng. 2010, 105, 26–36. [Google Scholar] [CrossRef]
- Nikel, P.I.; Martínez-García, E.; de Lorenzo, V. Biotechnological domestication of pseudomonads using synthetic biology. Nat. Rev. Microbiol. 2014, 12, 368–379. [Google Scholar] [CrossRef]
- Copeland, M.F.; Politz, M.C.; Pfleger, B.F. Application of TALEs, CRISPR/Cas and sRNAs as trans-acting regulators in prokaryotes. Curr. Opin. Biotechnol. 2014, 29, 46–54. [Google Scholar] [CrossRef][Green Version]
- Chappell, J.; Watters, K.E.; Takahashi, M.K.; Lucks, J.B. A renaissance in RNA synthetic biology: New mechanisms, applications and tools for the future. Curr. Opin. Chem. Biol. 2015, 28, 47–56. [Google Scholar] [CrossRef]
- Isaacs, F.J.; Dwyer, D.J.; Collins, J.J. RNA synthetic biology. Nat. Biotechnol. 2006, 24, 545–554. [Google Scholar] [CrossRef]
- Na, D.; Yoo, S.M.; Chung, H.; Park, H.; Park, J.H.; Lee, S.Y. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat. Biotechnol. 2013, 31, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.M.; Na, D.; Lee, S.Y. Design and use of synthetic regulatory small RNAs to control gene expression in Escherichia coli. Nat. Protoc. 2013, 8, 1694–1707. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.H.; Deng, H.K.; Hou, J.; Wang, L.J. Synthetic small regulatory RNAs in microbial metabolic engineering. Appl. Microbiol. Biotechnol. 2021, 105, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Martínez-García, E.; Nikel, P.I.; Aparicio, T.; de Lorenzo, V. Pseudomonas 2.0: Genetic upgrading of P. putida KT2440 as an enhanced host for heterologous gene expression. Microb. Cell Factories 2014, 13, 159. [Google Scholar] [CrossRef]
- Martínez-García, E.; Goñi-Moreno, A.; Bartley, B.; McLaughlin, J.; Sánchez-Sampedro, L.; Del Pozo, H.P.; Hernández, C.P.; Marletta, A.S.; De Lucrezia, D.; Sánchez-Fernández, G.; et al. SEVA 3.0: An update of the Standard European Vector Architecture for enabling portability of genetic constructs among diverse bacterial hosts. Nucleic Acids Res. 2020, 48, D1164–D1170. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Ma, Y.; Zhang, X.; Zhang, L.; Chen, X.; Ye, J.W.; Chen, G.Q. Tailor-Made Polyhydroxyalkanoates by Reconstructing Pseudomonas entomophila. Adv. Mater. 2021, e2102766. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peregrina, A.; Martins-Lourenço, J.; Freitas, F.; Reis, M.A.M.; Arraiano, C.M. Post-Transcriptional Control in the Regulation of Polyhydroxyalkanoates Synthesis. Life 2021, 11, 853. https://doi.org/10.3390/life11080853
Peregrina A, Martins-Lourenço J, Freitas F, Reis MAM, Arraiano CM. Post-Transcriptional Control in the Regulation of Polyhydroxyalkanoates Synthesis. Life. 2021; 11(8):853. https://doi.org/10.3390/life11080853
Chicago/Turabian StylePeregrina, Alexandra, João Martins-Lourenço, Filomena Freitas, Maria A. M. Reis, and Cecília M. Arraiano. 2021. "Post-Transcriptional Control in the Regulation of Polyhydroxyalkanoates Synthesis" Life 11, no. 8: 853. https://doi.org/10.3390/life11080853
APA StylePeregrina, A., Martins-Lourenço, J., Freitas, F., Reis, M. A. M., & Arraiano, C. M. (2021). Post-Transcriptional Control in the Regulation of Polyhydroxyalkanoates Synthesis. Life, 11(8), 853. https://doi.org/10.3390/life11080853







