Genomic Approaches for Conservation Management in Australia under Climate Change
Abstract
:1. Introduction
2. Climate Change and Conservation Challenges
3. Conservation Genetics in the Genomics Era
4. Application of Conservation Genomics to Climate Change Challenges
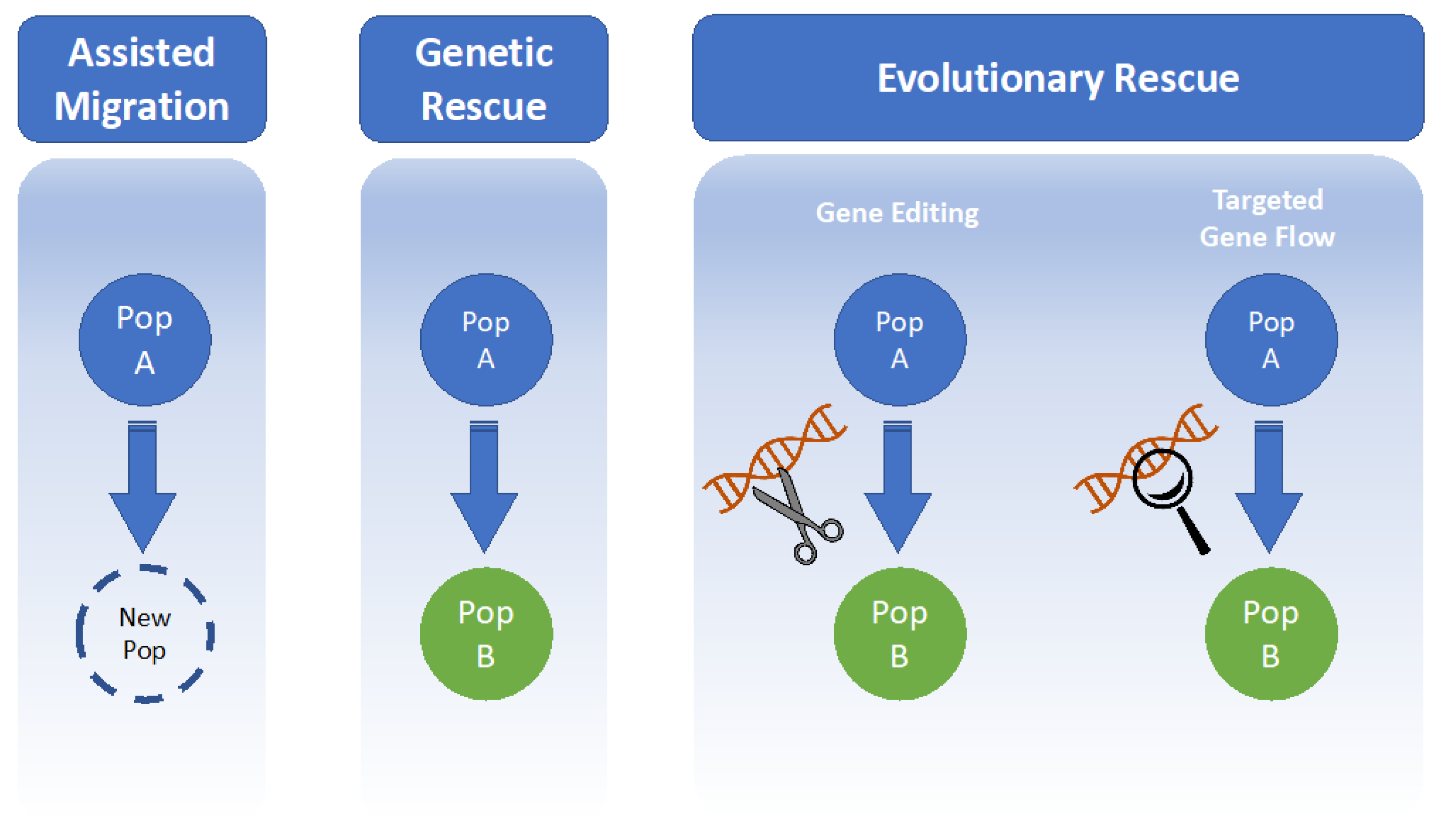
4.1. Assisted Migration
4.2. Genetic Rescue
4.3. Evolutionary Rescue
5. Overcoming Barriers to the Application of Genomics for Conservation Management under Climate Change
6. Future Opportunities and Tools to Harness Conservation Genomics in the Fight against Climate Change
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Woinarski, J.C.Z.; Burbidge, A.A.; Harrison, P. Ongoing unraveling of a continental fauna: Decline and extinction of Australian mammals since European settlement. Proc. Natl. Acad. Sci. USA 2015, 112, 4531–4540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woinarski, J.C.Z.; Braby, M.F.; Burbidge, A.A.; Coates, D.; Garnett, S.T.; Fensham, R.J.; Legge, S.M.; McKenzie, N.L.; Silcock, J.L.; Murphy, B.P. Reading the black book: The number, timing, distribution and causes of listed extinctions in Australia. Biol. Conserv. 2019, 239, 108261. [Google Scholar] [CrossRef]
- Morton, S. The impact of European settlement on the vertebrate animals of arid Australia: A conceptual model. Proc. Ecol. Soc. Aust. 1990, 16, 201–213. [Google Scholar]
- McKenzie, N.L.; Burbidge, A.A.; Baynes, A.; Brereton, R.N.; Dickman, C.; Gordon, G.; Gibson, L.A.; Menkhorst, P.W.; Robinson, A.C.; Williams, M.R.; et al. Analysis of factors implicated in the recent decline of Australia’s mammal fauna. J. Biogeogr. 2007, 34, 597–611. [Google Scholar] [CrossRef]
- McKechnie, A.; Wolf, B. Climate change increases the likelihood of catastrophic avian mortality evens during extreme heat waves. Biol. Lett. 2010, 6, 253–256. [Google Scholar] [CrossRef] [Green Version]
- Lindenmayer, D.B.; Steffen, W.; Burbidge, A.A.; Hughes, L.; Kitching, R.L.; Musgrave, W.; Smith, M.S.; Werner, P.A. Conservation strategies in response to rapid climate change: Australia as a case study. Biol. Conserv. 2010, 143, 1587–1593. [Google Scholar] [CrossRef]
- Ogston, G.; Beatty, S.J.; Morgan, D.L.; Pusey, B.J.; Lymbery, A. Living on burrowed time: Aestivating fishes in south-western Australia face extinction due to climate change. Biol. Conserv. 2016, 195, 235–244. [Google Scholar] [CrossRef]
- Frankham, R. Conservation genetics. Annu. Rev. Genet. 1995, 29, 305–327. [Google Scholar] [CrossRef]
- Godwin, J.; Lumley, A.; Michalczyk, L.; Martin, O.; Gage, M. Mating patterns influence vulnerability to the extinction vortex. Glob. Chang. Biol. 2020, 26, 4226–4239. [Google Scholar] [CrossRef]
- Weeks, A.; Heinze, D.; Perrin, L.; Stoklosa, J.; Hoffmann, A.; van Rooyen, A.; Kelly, T.; Mansergh, I. Genetic rescue increases fitness and aids rapid recovery of an endangered marsupial population. Nat. Commun. 2017, 8, 1071. [Google Scholar] [CrossRef]
- Hedrick, P.W. Conservation genetics: Where are we now? Trends Ecol. Evol. 2001, 16, 629–636. [Google Scholar] [CrossRef]
- Eldridge, M. Marsupial Population and Conservation Genetics. In Marsupial Genetics and Genomics; Waters, J., Marshall, P., Graves, J., Eds.; Springer: Dordrecht, The Netherlands, 2010; pp. 461–497. [Google Scholar]
- Byrne, M. A molecular journey in conservation genetics. Pac. Conserv. Biol. 2018, 24, 235–243. [Google Scholar] [CrossRef]
- White, L.; Moseby, K.; Thomson, V.; Donnellan, S.; Austin, J. Long-term genetic consequences of mammal reintroductions into an Australian conservation reserve. Biol. Conserv. 2018, 219, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Weeks, A.R.; Sgro, C.; Young, A.G.; Frankham, R.; Mitchell, N.J.; Miller, K.A.; Byrne, M.; Coates, D.J.; Eldridge, M.; Sunnucks, P.; et al. Assessing the benefits and risks of translocations in changing environments: A genetic perspective. Evol. Appl. 2011, 4, 709–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouborg, N.; Pertoldi, C.; Loeschcke, V.; Bijlsma, R.; Hedrick, P. Conservation genetics in transition to conservation genomics. Trends Genet. 2010, 26, 177–187. [Google Scholar] [CrossRef]
- Harrisson, K.; Pavlova, A.; Telonis-Scott, M.; Sunnucks, P. Using genomics to characterize evolutionary potential for conservation of wild populations. Evol. Appl. 2014, 9, 1008–1025. [Google Scholar] [CrossRef]
- DeSalle, R.; Amato, G. The expansion of conservation genetics. Nat. Rev. Genet. 2004, 5, 702–712. [Google Scholar] [CrossRef] [PubMed]
- Frankham, R.; Ballou, J.; Ralls, K.; Eldridge, M.; Dudash, M.; Fenster, C.; Lacy, R.; Sunnucks, P. Genetic Management of Fragmented Animal and Plant Populations; Oxford University Press: Oxford, UK, 2017. [Google Scholar]
- Coates, D.J.; Byrne, M.; Moritz, C. Genetic Diversity and Conservation Units: Dealing with the Species-Population Continuum in the Age of Genomics. Front. Ecol. Evol. 2018, 6, 165. [Google Scholar] [CrossRef] [Green Version]
- Milano, I.; Babbucci, M.; Panitz, F.; Ogden, R.; Nielsen, R.; Taylor, M.; Helyar, S.; Carvalho, G.; Espiñeira, M.; Atanassova, M.; et al. Novel tools for conservation genomics: Comparing two high-throughput approaches for SNP discovery in the transcriptome of the European hake. PLoS ONE 2011, 6, e28008. [Google Scholar] [CrossRef]
- Narum, S.; Buerkle, C.; Davey, J.; Miller, M.; Hohenlohe, P. Genotyping-by-sequencing in ecological and conservation genomics. Mol. Ecol. 2013, 22, 2841. [Google Scholar] [CrossRef]
- Meek, M.H.; Larson, W.A. The future is now: Amplicon sequencing and sequence capture usher in the conservation genomics era. Mol. Ecol. Resour. 2019, 19, 795–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bureau of Meteorology. State of the Climate; Bureau of Meteorology: Canberra, Australia, 2019. Available online: http://www.bom.gov.au/state-of-the-climate/ (accessed on 18 March 2019).
- CSIRO. Climate Change in Australia: Australian Climate Futures Tool. Available online: https://www.climatechangeinaustralia.gov.au/ (accessed on 7 November 2020).
- Field, C.; Barros, V.; Stocker, T.; Dahe, Q. Managing the Risks of Extreme Events and Disasters to Advance Climate Change Adaptation: Special Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK, 2012. [Google Scholar]
- Dowdy, A.J. Climatological Variability of Fire Weather in Australia. J. Appl. Meteorol. Clim. 2018, 57, 221–234. [Google Scholar] [CrossRef]
- Walsh, K.J.E.; Ryan, B.F. Tropical Cyclone Intensity Increase near Australia as a Result of Climate Change. J. Clim. 2000, 13, 3029–3036. [Google Scholar] [CrossRef]
- Halgamuge, M.; Nirmalathas, A. Analysis of large flood events: Based on flood data during 1985–2016 in Australian and India. Int. J. Disaster Risk Reduct. 2017, 24, 1–11. [Google Scholar] [CrossRef]
- Ratnayake, H.U.; Kearney, M.R.; Govekar, P.; Karoly, D.; Welbergen, J.A. Forecasting wildlife die-offs from extreme heat events. Anim. Conserv. 2019, 22, 386–395. [Google Scholar] [CrossRef]
- Hannah, L. A Global Conservation System for Climate-Change Adaptation. Conserv. Biol. 2010, 24, 70–77. [Google Scholar] [CrossRef]
- Foden, W.; Mace, G.; Vié, J.; Angulo, A.; Butchart, S.; DeVantier, L.; Dublin, H.; Gutsche, A.; Stuart, S.; Turak, E. Species susceptibility to climate change impacts. In Wildlife in a Changing World—An Analysis of the 2008 IUCN Red List of Threatened Species; International Union for Conservation of Nature (IUCN): Gland, Switzerland, 2009; Volume 77. [Google Scholar]
- Lee, J.R.; Maggini, R.; Taylor, M.F.J.; Fuller, R. Mapping the Drivers of Climate Change Vulnerability for Australia’s Threatened Species. PLoS ONE 2015, 10, e0124766. [Google Scholar] [CrossRef] [Green Version]
- Bond, N.; Thomson, J.; Reich, P.; Stein, J. Using species distribution models to infer potential climate change-induced range shifts of freshwater fish in south-eastern Australia. Mar. Freshw. Res. 2011, 62, 1043–1061. [Google Scholar] [CrossRef] [Green Version]
- Butt, N.; Seabrook, L.; Maron, M.; Law, B.; Dawson, T.; Syktus, J.; McAlpine, C. Cascading effects of climate extremes on vertebrate fauna through changes to low-latitude tree flowering and fruiting phenology. Glob. Chang. Biol. 2015, 21, 3267–3277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urban, M.C. Accelerating extinction risk from climate change. Science 2015, 348, 571–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, A.A.; Rymer, P.D.; Byrne, M.; Ruthrof, K.X.; Whinam, J.; McGeoch, M.; Bergstrom, D.M.; Guerin, G.R.; Sparrow, B.; Joseph, L.; et al. Impacts of recent climate change on terrestrial flora and fauna: Some emerging Australian examples. Austral. Ecol. 2019, 44, 3–27. [Google Scholar] [CrossRef] [Green Version]
- Urban, M.C. Climate-tracking species are not invasive. Nat. Clim. Chang. 2020, 10, 382–384. [Google Scholar] [CrossRef]
- Johnson, C.; Banks, S.; Barrett, N.; Cazassus, F.; Dunstan, P.; Edgar, G.; Frusher, S.; Gardner, C.; Haddon, M.; Helidoniotis, F.; et al. Climate change cascades: Shifts in oceanography, species’ ranges and subtidal marine community dynamics in eastern Tasmania. J. Exp. Mar. Biol. Ecol. 2011, 400, 17–32. [Google Scholar] [CrossRef]
- Davis, J.; Pavlova, A.; Thompson, R.; Sunnucks, P. Evolutionary refugia and ecological refuges: Key concepts for conserving Australian arid zone freshwater biodiversity under climate change. Glob. Chang. Biol. 2013, 19, 1970–1984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wernberg, T.; Russell, B.D.; Moore, P.; Ling, S.; Smale, D.A.; Campbell, A.; Coleman, M.A.; Steinberg, P.D.; Kendrick, G.; Connell, S.D. Impacts of climate change in a global hotspot for temperate marine biodiversity and ocean warming. J. Exp. Mar. Biol. Ecol. 2011, 400, 7–16. [Google Scholar] [CrossRef] [Green Version]
- Williams, S.E.; Bolitho, E.E.; Fox, S. Climate change in Australian tropical rainforests: An impending environmental catastrophe. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2003, 270, 1887–1892. [Google Scholar] [CrossRef]
- Hughes, L. Climate change and Australia: Trends, projections and impacts. Austral. Ecol. 2003, 28, 423–443. [Google Scholar] [CrossRef]
- Pounds, J.A.; Fogden, M.P.L.; Campbell, J.H. Biological response to climate change on a tropical mountain. Nat. Cell Biol. 1999, 398, 611–615. [Google Scholar] [CrossRef]
- Beaumont, L.J.; Hughes, L. Potential changes in the distributions of latitudinally restricted Australian butterfly species in response to climate change. Glob. Chang. Biol. 2002, 8, 954–971. [Google Scholar] [CrossRef]
- Stubbs, J.L.; Marn, N.; Vanderklift, M.A.; Fossette, S.; Mitchell, N.J. Simulated growth and reproduction of green turtles (Chelonia mydas) under climate change and marine heatwave scenarios. Ecol. Model. 2020, 431, 109185. [Google Scholar] [CrossRef]
- Conradie, S.R.; Woodborne, S.M.; Wolf, B.O.; Pessato, A.; Mariette, M.M.; McKechnie, A.E. Avian mortality risk during heat waves will increase greatly in arid Australia during the 21st century. Conserv. Physiol. 2020, 8, coaa048. [Google Scholar] [CrossRef]
- Harada, Y.; Fry, B.; Lee, S.Y.; Maher, D.T.; Sippo, J.Z.; Connolly, R.M. Stable isotopes indicate ecosystem restructuring following climate-driven mangrove dieback. Limnol. Oceanogr. 2020, 65, 1251–1263. [Google Scholar] [CrossRef]
- Arias-Ortiz, A.; Serrano, O.; Masqué, P.; Lavery, P.S.; Mueller, U.; Kendrick, G.A.; Rozaimi, M.; Esteban, A.; Fourqurean, J.W.; Marbà, N.; et al. A marine heatwave drives massive losses from the world’s largest seagrass carbon stocks. Nat. Clim. Chang. 2018, 8, 338–344. [Google Scholar] [CrossRef] [Green Version]
- Miller, A.D.; Nitschke, C.; Weeks, A.R.; Weatherly, W.L.; Heyes, S.D.; Sinclair, S.J.; Holland, O.J.; Stevenson, A.; Broadhurst, L.; Hoebee, S.E.; et al. Genetic data and climate niche suitability models highlight the vulnerability of a functionally important plant species from south-eastern Australia. Evol. Appl. 2020, 13, 2014–2029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butt, N.; Gallagher, R. Using species traits to guide conservation actions under climate change. Clim. Chang. 2018, 151, 317–332. [Google Scholar] [CrossRef]
- Frankham, R. Challenges and opportunities of genetic approaches to biological conservation. Biol. Conserv. 2010, 143, 1919–1927. [Google Scholar] [CrossRef]
- Kohn, M.H.; Murphy, W.J.; Ostrander, E.A.; Wayne, R.K. Genomics and conservation genetics. Trends Ecol. Evol. 2006, 21, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Lacy, R. Should we select genetic alleles in our conservation breeding programs? Zoo Biol. 2000, 19, 279–282. [Google Scholar] [CrossRef]
- Wayne, R.; Morin, P. Conservation genetics in the new molecular age. Front. Ecol. Environ. 2004, 2, 89–97. [Google Scholar] [CrossRef]
- Shafer, A.; Wolf, J.; Alves, P.; Bergström, L.; Bruford, M.; Brännström, I.; Colling, G.; Dalén, L.; De Meester, L.; Ekblom RFawcett, K. Genomics and the challenging translation into conservation practice. Trends Ecol. Evol. 2015, 30, 78–87. [Google Scholar] [CrossRef] [Green Version]
- De Wit, P.; Pespeni, M.; Palumbi, S. SNP genotyping and population genomics from expressed sequences—Current advances and future possibilities. Mol. Ecol. 2015, 24, 2310–2323. [Google Scholar] [CrossRef] [Green Version]
- Grada, A.; Weinbrecht, K. Next-Generation Sequencing: Methodology and Application. J. Investig. Derm. 2013, 133, e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soon, W.W.; Hariharan, M.; Snyder, M.P. High-Throughput sequencing for biology and medicine. Mol. Syst. Biol. 2013, 9, 640. [Google Scholar] [CrossRef]
- Hunter, M.E.; Hoban, S.M.; Bruford, M.W.; Segelbacher, G.; Bernatchez, L. Next-generation conservation genetics and biodiversity monitoring. Evol. Appl. 2018, 11, 1029–1034. [Google Scholar] [CrossRef]
- Gayral, P.; Melo-Ferreira, J.; Glémin, S.; Bierne, N.; Carneiro, M.; Nabholz, B.; Lourenco, J.M.; Alves, P.C.; Ballenghien, M.; Faivre, N.; et al. Reference-Free Population Genomics from Next-Generation Transcriptome Data and the Vertebrate—Invertebrate Gap. PLoS Genet. 2013, 9, e1003457. [Google Scholar] [CrossRef] [Green Version]
- Supple, M.A.; Shapiro, B. Conservation of biodiversity in the genomics era. Genome Biol. 2018, 19, 131. [Google Scholar] [CrossRef]
- Georges, A.; Gruber, B.; Pauly, G.; White, D.; Adams, M.; Young, M.J.; Kilian, A.; Zhang, X.; Shaffer, H.B.; Unmack, P.J. Genomewide SNP markers breathe new life into phylogeography and species delimitation for the problematic short-necked turtles (Chelidae: Emydura) of eastern Australia. Mol. Ecol. 2018, 27, 5195–5213. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, R.M.; Saarman, N.; Ramstad, K.M.; Forester, B.R.; Kelley, J.L.; Hand, B.K.; Malison, R.L.; Ackiss, A.S.; Watsa, M.; Nelson, T.C.; et al. Big Data in Conservation Genomics: Boosting Skills, Hedging Bets, and Staying Current in the Field. J. Hered. 2021. [Google Scholar] [CrossRef] [PubMed]
- Hohenlohe, P.A.; Funk, W.C.; Rajora, O.P. Population genomics for wildlife conservation and management. Mol. Ecol. 2021, 30, 62–82. [Google Scholar] [CrossRef]
- Smith, S.; Bernatchez, L.; Beheregaray, L. RNA-seq analysis reveals extensive transcriptional plasticity to temperature stress in a freshwater fish species. BMC Genom. 2013, 14, 375. [Google Scholar] [CrossRef] [Green Version]
- Tsangaras, K.; Siracusa, M.C.; Nikolaidis, N.; Ishida, Y.; Cui, P.; Vielgrader, H.; Helgen, K.; Roca, A.; Greenwood, A.D. Hybridization Capture Reveals Evolution and Conservation across the Entire Koala Retrovirus Genome. PLoS ONE 2014, 9, e95633. [Google Scholar] [CrossRef]
- Bell, N.; Griffin, P.C.; Hoffmann, A.; Miller, A.D. Spatial patterns of genetic diversity among Australian alpine flora communities revealed by comparative phylogenomics. J. Biogeogr. 2017, 45, 177–189. [Google Scholar] [CrossRef]
- White, L.C.; Thomson, V.A.; West, R.; Ruykys, L.; Ottewell, K.; Kanowski, J.; Moseby, K.E.; Byrne, M.; Donnellan, S.C.; Copley, P.; et al. Genetic monitoring of the greater stick-nest rat meta-population for strategic supplementation planning. Conserv. Genet. 2020, 21, 941–956. [Google Scholar] [CrossRef]
- Wright, B.R.; Farquharson, K.A.; McLennan, E.A.; Belov, K.; Hogg, C.J.; Grueber, C.E. A demonstration of conservation genomics for threatened species management. Mol. Ecol. Resour. 2020, 20, 1526–1541. [Google Scholar] [CrossRef] [PubMed]
- Morin, P.A.; Luikart, G.; Wayne, R.K. The SNP workshop group SNPs in ecology, evolution and conservation. Trends Ecol. Evol. 2004, 19, 208–216. [Google Scholar] [CrossRef]
- Angeloni, F.; Wagemaker, N.; Vergeer, P.; Ouborg, J. Genomic toolboxes for conservation biologists. Evol. Appl. 2011, 5, 130–143. [Google Scholar] [CrossRef]
- Epstein, B.; Jones, M.; Hamede, R.; Hendricks, S.; McCallum, H.; Murchison, E.P.; Schönfeld, B.; Wiench, C.; Hohenlohe, P.; Storfer, A. Rapid evolutionary response to a transmissible cancer in Tasmanian devils. Nat. Commun. 2016, 7, 12684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, A.; Hoffmann, A.; Tan, M.; Young, M.; Ahrens, C.; Cocomazzo, M.; Rattray, A.; Ierodiaconour, D.; Treml, E.; Sherman, C. Local and regional scale habitat heterogeneity contribute to genetic adaptation in a commercially important marine mollusc (Haliotis rubra) from south eastern Australia. Mol. Ecol. 2019, 28, 3053–3072. [Google Scholar] [CrossRef]
- Garner, B.A.; Hand, B.K.; Amish, S.J.; Bernatchez, L.; Foster, J.; Miller, K.M.; Morin, P.; Narum, S.R.; O’Brien, S.J.; Roffler, G.; et al. Genomics in Conservation: Case Studies and Bridging the Gap between Data and Application. Trends Ecol. Evol. 2016, 31, 81–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, G.; Marzinelli, E.; Campbell, A.; Steinberg, P.; Vergés, A.; Coleman, M. Gemomic vulnerability of a dominant seaweed points to future-proofing pathways for Australia’s underwater forests. Glob. Chang. Biol. 2021, 27, 2200–2212. [Google Scholar] [CrossRef] [PubMed]
- Natesh, M.; Taylor, R.W.; Truelove, N.K.; Hadly, E.A.; Palumbi, S.R.; Petrov, D.A.; Ramakrishnan, U. Empowering conservation practice with efficient and economical genotyping from poor quality samples. Methods Ecol. Evol. 2019, 10, 853–859. [Google Scholar] [CrossRef] [PubMed]
- IUCN. Guidelines for Reintroductions and Other Conservation Translocations; IUCN Species Survival Commission: Gland, Switzerland, 2013. [Google Scholar]
- Seddon, P. From reintroduction to assisted colonisation: Moving along the conservation translocation spectrum. Restor. Ecol. 2010, 18, 796–802. [Google Scholar] [CrossRef] [Green Version]
- Moseby, K.; Read, J.; Paton, D.; Copley, P.; Hill, B.; Crisp, H. Predation determines the outcome of 10 reintroduction attempts in arid South Australia. Biol. Conserv. 2011, 144, 2863–2872. [Google Scholar] [CrossRef]
- Sgrò, C.M.; Lowe, A.; Hoffmann, A. Building evolutionary resilience for conserving biodiversity under climate change. Evol. Appl. 2010, 4, 326–337. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, R.V.; Makinson, R.O.; Hogbin, P.M.; Hancock, N. Assisted colonization as a climate change adaptation tool. Austral. Ecol. 2014, 40, 12–20. [Google Scholar] [CrossRef]
- Bell, D.A.; Robinson, Z.L.; Funk, W.C.; Fitzpatrick, S.W.; Allendorf, F.W.; Tallmon, D.A.; Whiteley, A.R. The Exciting Potential and Remaining Uncertainties of Genetic Rescue. Trends Ecol. Evol. 2019, 34, 1070–1079. [Google Scholar] [CrossRef]
- Carlson, S.M.; Cunningham, C.J.; Westley, P.A. Evolutionary rescue in a changing world. Trends Ecol. Evol. 2014, 29, 521–530. [Google Scholar] [CrossRef] [Green Version]
- Bell, G. Evolutionary Rescue. Annu. Rev. Ecol. Evol. Syst. 2017, 48, 605–627. [Google Scholar] [CrossRef]
- Hoffmann, A.; Miller, A.; Weeks, A. Genetic mixing for population management: From genetic rescue to provenancing. Evol. Appl. 2020, 14, 634–652. [Google Scholar] [CrossRef]
- Burbidge, A.; Byrne, M.; Coates, D.; Garnett, S.; Harris, S.; Hatward, M.; Martin, T.; McDonald-Madden, E.; Mitchell, N.; Nally SSetterfield, S. Is Australia ready for assisted colonization? Policy changes required to facilitate translocations under climate change. Pac. Conserv. Biol. 2011, 17, 259–269. [Google Scholar] [CrossRef] [Green Version]
- Breed, M.F.; Ottewell, K.M.; Gardner, M.G.; Lowe, A.J. Clarifying climate change adaptation responses for scattered trees in modified landscapes. J. Appl. Ecol. 2011, 48, 637–641. [Google Scholar] [CrossRef]
- Supple, M.A.; Bragg, J.G.; Broadhurst, L.M.; Nicotra, A.B.; Byrne, M.; Andrew, R.L.; Widdup, A.; Aitken, N.C.; Borevitz, J.O. Landscape genomic prediction for restoration of a Eucalyptus foundation species under climate change. Elife 2018, 7, 31835. [Google Scholar] [CrossRef]
- Hedrick, P.W.; Fredrickson, R. Genetic rescue guidelines with examples from Mexican wolves and Florida panthers. Conserv. Genet. 2009, 11, 615–626. [Google Scholar] [CrossRef]
- Frankham, R. Genetic rescue of small inbred populations: Meta-analysis reveals large and consistent benefits of gene flow. Mol. Ecol. 2015, 24, 2610–2618. [Google Scholar] [CrossRef]
- Whiteley, A.R.; Fitzpatrick, S.W.; Funk, W.C.; Tallmon, D.A. Genetic rescue to the rescue. Trends Ecol. Evol. 2015, 30, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Ralls, K.; Sunnucks, P.; Lacy, R.C.; Frankham, R. Genetic rescue: A critique of the evidence supports maximizing genetic diversity rather than minimizing the introduction of putatively harmful genetic variation. Biol. Conserv. 2020, 251, 108784. [Google Scholar] [CrossRef]
- Pickup, M.; Field, D.; Rowell, D.; Young, A. Source population characteristics affect heterosis following genetic rescue of fragmented plant populations. Proc. R. Soc. B 2013, 280, 20122058. [Google Scholar] [CrossRef] [PubMed]
- Forester, B.; Lasky, J.; Wagner, H.; Urban, D. Comparing methods for detecting multilocus adaptation with multivariate genotype-environment associations. Mol. Ecol. 2018, 27, 2215–2233. [Google Scholar] [CrossRef]
- Fitzpatrick, S.; Funk, W. Genomics for Genetic Rescue. Population Genomics: Wildlife. In Population Genomics; Hohenlohe, P., Rajora, O., Eds.; Springer: Cambridge, UK, 2019. [Google Scholar]
- Fox, C.; Reed, D. Inbreeding depression increases with environmental stress: An experimental study and meta-analysis. Evolution 2011, 65, 246–258. [Google Scholar] [CrossRef]
- Nickolas, H.; Harrison, P.; Tilyard, P.; Vaillancourt, R.; Potts, B. Inbreeding depression and differential maladaptation shape the fitness trajectory of two co-occurring Eucalyptus species. Ann. For. Sci. 2019, 76, 10. [Google Scholar] [CrossRef] [Green Version]
- Kelly, E.; Phillips, B. Targeted gene flow for conservation. Conserv. Biol. 2016, 30, 259–267. [Google Scholar] [CrossRef]
- Kelly, E.; Phillips, B.L. Targeted gene flow and rapid adaptation in an endangered marsupial. Conserv. Biol. 2019, 33, 112–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barua, D.; Heckathorn, S.A.; Coleman, J.S. Variation in Heat-shock Proteins and Photosynthetic Thermotolerance among Natural Populations of Chenopodium album L. from Contrasting Thermal Environments: Implications for Plant Responses to Global Warming. J. Integr. Plant. Biol. 2008, 50, 1440–1451. [Google Scholar] [CrossRef]
- Barah, P.; Jayavelu, N.D.; Mundy, J.; Bones, A.M. Genome scale transcriptional response diversity among ten ecotypes of Arabidopsis thaliana during heat stress. Front. Plant. Sci. 2013, 4, 532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandoval-Castillo, J.; Gates, K.; Brauer, C.; Smith, S.; Bernatchez, L.; Beheregaray, L. Adaptation of plasticity to predicted climates in Australian rainbowfishes (Melanotaenia) across climatically defined bioregions. BioRxiv 2019, 859769. [Google Scholar] [CrossRef] [Green Version]
- Cummins, D.; Kennington, W.J.; Rudin-Bitterli, T.; Mitchell, N.J. A genome-wide search for local adaptation in a terrestrial-breeding frog reveals vulnerability to climate change. Glob. Chang. Biol. 2019, 25, 3151–3162. [Google Scholar] [CrossRef]
- Sandoval-Castillo, J.; Robinson, N.; Hart, A.M.; Strain, L.W.S.; Beheregaray, L.B. Seascape genomics reveals adaptive divergence in a connected and commercially important mollusc, the greenlip abalone (Haliotis laevigata), along a longitudinal environmental gradient. Mol. Ecol. 2018, 27, 1603–1620. [Google Scholar] [CrossRef]
- Bay, R.A.; Palumbi, S.R. Multilocus Adaptation Associated with Heat Resistance in Reef-Building Corals. Curr. Biol. 2014, 24, 2952–2956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Board, O. National Academies of Sciences, Engineering, and Medicine. In A Research Review of Interventions to Increase the Persistence and Resilience of Coral Reefs; National Academies Press: Washington, DC, USA, 2019. [Google Scholar]
- Quigley, K.M.; Bay, L.K.; Van Oppen, M.J.H. The active spread of adaptive variation for reef resilience. Ecol. Evol. 2019, 9, 11122–11135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Oppen, M.J.H.; Oliver, J.K.; Putnam, H.; Gates, R.D. Building coral reef resilience through assisted evolution. Proc. Natl. Acad. Sci. USA 2015, 112, 2307–2313. [Google Scholar] [CrossRef] [Green Version]
- Jordan, R.; Hoffmann, A.; Dillon, S.; Prober, S. Evidence of genomic adaptation to climate in Eucalyptus microcarpa: Implications for adaptive potential to projected climate change. Mol. Ecol. 2017, 21, 6002–6020. [Google Scholar] [CrossRef] [PubMed]
- Steane, D.; Potts, B.; McLean, E.; Prober, S.; Stock, W.; Vaillancourt, R.; Byrne, M. Genome-wide scans detect adaptation to aridity in a widespread forest tree species. Mol. Ecol. 2014, 23, 2500–2513. [Google Scholar] [CrossRef]
- Phelps, M.; Seeb, L.; Seeb, J. Transforming ecology and conservation biology through genome editing. Conserv. Biol. 2020, 34, 54–65. [Google Scholar] [CrossRef]
- Esvelt, K.; Smidler, A.; Catteruccia, F.; Church, G. Emerging technology: Concerning RNA-guided gene drives for the alteration of wild populations. Elife 2014, 3, e03401. [Google Scholar] [CrossRef]
- Champer, J.; Buchman, A.; Akbari, O.S. Cheating evolution: Engineering gene drives to manipulate the fate of wild populations. Nat. Rev. Genet. 2016, 17, 146–159. [Google Scholar] [CrossRef] [Green Version]
- Piaggio, A.J.; Segelbacher, G.; Seddon, P.J.; Alphey, L.; Bennett, E.L.; Carlson, R.H.; Friedman, R.M.; Kanavy, D.; Phelan, R.; Redford, K.H.; et al. Is It Time for Synthetic Biodiversity Conservation? Trends Ecol. Evol. 2017, 32, 97–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, M.; Roemer, G.; Donlan, C.; Dickson, B.; Matocq, M.; Malaney, J. Ecology: Gene tweaking for conservation. Nat. News 2013, 501, 485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Oakes, A.; Newhouse, A.; Baier, K.; Maynard, C.; Powell, W. A threshold level of oxalate oxidase transgene expression reduces Cryphonectria parasitica-induced necrosis in a transgenic American chestnut (Castanea dentate) leaf bioassay. Transgenic Res. 2013, 22, 973–982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohl, P.A.; Brossard, M.; Scheufele, D.A.; Xenos, M. Public views about editing genes in wildlife for conservation. Conserv. Biol. 2019, 33, 1286–1295. [Google Scholar] [CrossRef] [PubMed]
- Westbrook, J.W.; Holliday, J.A.; Newhouse, A.E.; Powell, W.A. A plan to diversify a transgenic blight-tolerant American chestnut population using citizen science. Plants People Planet 2019, 2, 84–95. [Google Scholar] [CrossRef] [Green Version]
- Johnson, J.; Altwegg, R.; Evans, D.; Ewen, J.; Gordon, I. Is there a future for genome-editing technologies in conservation? Anim. Conserv. 2016, 19, 97–101. [Google Scholar] [CrossRef] [Green Version]
- Anthony, K.; Bay, L.K.; Costanza, R.; Firn, J.; Gunn, J.; Harrison, P.; Heyward, A.; Lundgren, P.; Mead, D.; Moore, T.; et al. New interventions are needed to save coral reefs. Nat. Ecol. Evol. 2017, 1, 1420–1422. [Google Scholar] [CrossRef]
- Zafar, S.A.; Zaidi, S.S.-E.-A.; Gaba, Y.; Singla-Pareek, S.L.; Dhankher, O.P.; Li, X.; Mansoor, S.; Pareek, A. Engineering abiotic stress tolerance via CRISPR/ Cas-mediated genome editing. J. Exp. Bot. 2019, 71, 470–479. [Google Scholar] [CrossRef]
- Cleves, P.A.; Strader, M.E.; Bay, L.K.; Pringle, J.R.; Matz, M.V. CRISPR/Cas9-mediated genome editing in a reef-building coral. Proc. Natl. Acad. Sci. USA 2018, 115, 5235–5240. [Google Scholar] [CrossRef] [Green Version]
- Hoban, S.; Kelley, J.L.; Lotterhos, K.E.; Antolin, M.F.; Bradburd, G.; Lowry, D.B.; Poss, M.L.; Reed, L.K.; Storfer, A.; Whitlock, M.C. Finding the Genomic Basis of Local Adaptation: Pitfalls, Practical Solutions, and Future Directions. Am. Nat. 2016, 188, 379–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kardos, M.; Shafer, A. The Peril of Gene-Targeted Conservation. Trends Ecol. Evol. 2018, 33, 827–839. [Google Scholar] [CrossRef] [PubMed]
- Hogg, C.; Grueber, C.; Pemberton, D.; Fox, S.; Lee, A.; Ivy, J.; Belov, K. Devil Tools & Tech: A synergy of conservation research and management practice. Conserv. Lett. 2017, 10, 133–138. [Google Scholar]
- Holderegger, R.; Balkenhol, N.; Bolliger, J.; Engler, J.O.; Gugerli, F.; Hochkirch, A.; Nowak, C.; Segelbacher, G.; Widmer, A.; Zachos, F.E. Conservation genetics: Linking science with practice. Mol. Ecol. 2019, 28, 3848–3856. [Google Scholar] [CrossRef] [Green Version]
- Taylor, H.; Dussex, N.; van Heezik, Y. Bridging the conservation genetics gap by identifying barriers to implementation for conservation practitioners. Glob. Ecol. Conserv. 2017, 10, 231–242. [Google Scholar] [CrossRef]
- Cook, C.N.; Sgrò, C.M. Aligning science and policy to achieve evolutionarily enlightened conservation. Conserv. Biol. 2017, 31, 501–512. [Google Scholar] [CrossRef]
- Kadykalo, A.; Cooke, S.; Young, N. Conservation genomics from a practitioner lens: Evaluating the research-implementation gap in a managed freshwater fishery. Biol. Conserv. 2020, 241, 108350. [Google Scholar] [CrossRef]
- Stowell, S.M.L.; Pinzone, C.A.; Martin, A.P. Overcoming barriers to active interventions for genetic diversity. Biodivers. Conserv. 2017, 26, 1753–1765. [Google Scholar] [CrossRef]
- Krauss, S.; Koch, J.; Vlahos, S. A novel approach for the rapid genetic delineation of provenance for minesite revegetation. Ecol. Manag. Restor. 2005, 6, 153–155. [Google Scholar] [CrossRef]
- Krauss, S.; He, T. Rapid genetic identification of local provenance seed collection zones for ecological restoration and biodiversity conservation. J. Nat. Conserv. 2006, 14, 190–199. [Google Scholar] [CrossRef]
- Robinson, N.; Rhoades, C.; Pierson, J.; Lindenmayer, D.; Banks, S. Prioritising source populations for supplementing genetic diversity of reintroduced southern brown bandicoots Isoodon obesulus obesulus. Conserv. Genet. 2021, 22, 341–353. [Google Scholar] [CrossRef]
- Rodger, Y.S.; Pavlova, A.; Sinclair, S.; Pickup, M.; Sunnucks, P. Evolutionary history and genetic connectivity across highly fragmented populations of an endangered daisy. Heredity 2021, 126, 846–858. [Google Scholar] [CrossRef]
- Britt, M.; Haworth, S.E.; Johnson, J.B.; Martchenko, D.; Shafer, A.B. The importance of non-academic coauthors in bridging the conservation genetics gap. Biol. Conserv. 2018, 218, 118–123. [Google Scholar] [CrossRef]
- Haig, S.M.; Miller, M.P.; Bellinger, M.R.; Draheim, H.M.; Mercer, D.M.; Mullins, T.D. The conservation genetics juggling act: Integrating genetics and ecology, science and policy. Evol. Appl. 2015, 9, 181–195. [Google Scholar] [CrossRef] [Green Version]
- Proft, K.M.; Jones, M.E.; Johnson, C.N.; Burridge, C.P. Making the connection: Expanding the role of restoration genetics in restoring and evaluating connectivity. Restor. Ecol. 2018, 26, 411–418. [Google Scholar] [CrossRef]
- Bucharova, A. Assisted migration within species range ignores biotic interactions and lacks evidence: Missing evidence for assisted migration. Restor. Ecol. 2017, 25, 14–18. [Google Scholar] [CrossRef]
- Ralls, K.; Ballou, J.; Dudash, M.; Eldridge, M.; Fenster, C.; Lacy, R.; Sunnucks, P.; Frankham, R. Call for a paradigm shift in the genetic management of fragmented populations. Conserv. Lett. 2018, 11, e12412. [Google Scholar] [CrossRef]
- Frankham, R. Where are we in conservation genetics and where do we need to go? Conserv. Genet. 2009, 11, 661–663. [Google Scholar] [CrossRef]
- Varshney, R.K.; Bansal, K.C.; Aggarwal, P.K.; Datta, S.K.; Craufurd, P.Q. Agricultural biotechnology for crop improvement in a variable climate: Hope or hype? Trends Plant. Sci. 2011, 16, 363–371. [Google Scholar] [CrossRef] [Green Version]
- Fitzpatrick, S.W.; Bradburd, G.S.; Kremer, C.T.; Salerno, P.E.; Angeloni, L.M.; Funk, W.C. Genomic and Fitness Consequences of Genetic Rescue in Wild Populations. Curr. Biol. 2020, 30, 517–522. [Google Scholar] [CrossRef] [Green Version]
- Browne, L.; Wright, J.; Fitz-Gibbon, S.; Gugger PSork, V. Adaptational lag to temperature in valley oak (Quercus lobate) can be mitigated by genome-informed assisted gene flow. Proc. Natl. Acad. Sci. USA 2019, 116, 25179–25185. [Google Scholar] [CrossRef] [Green Version]
- Reed, T.; Waples, R.; Schindler, D.; Hard, J.; Kinnison, M. Phenotypic plasticity and population viability: The importance of environmental predictability. Proc. R. Soc. B Biol. Sci. 2010, 277, 3391–3400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chevin, L.; Gallet, R.; Gomulkiewicz, R.; Holt, R.; Fellous, S. Phenotypic plasticity in evolutionary rescue experiments. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20120089. [Google Scholar] [CrossRef] [PubMed]
- Liddell, E.; Cook, C.; Sunnucks, P. Evaluating the use of risk assessment frameworks in the identification of population units for biodiversity conservation. Wildl. Res. 2020, 47, 208–216. [Google Scholar] [CrossRef]
- Rossetto, M.; Yap, J.-Y.S.; Lemmon, J.; Bain, D.; Bragg, J.; Hogbin, P.; Gallagher, R.; Rutherford, S.; Summerell, B.; Wilson, T.C. A conservation genomics workflow to guide practical management actions. Glob. Ecol. Conserv. 2021, 26, e01492. [Google Scholar] [CrossRef]
- Ottewell, K.; Bickerton, D.; Byrne, M.; Lowe, A. Bridging the gap: A genetic assessmet framework for population-level threatened plant conservation prioritization and decision-making. Divers. Distrib. 2016, 22, 174–188. [Google Scholar] [CrossRef] [Green Version]
- Rollinson, N.; Keith, D.; Houde, A.; Debes, P.; Mcbride, M.; Hutchings, J. Risk assessment of inbreeding and outbreeding depression in a captive-breeding program. Conserv. Biol. 2014, 28, 529–540. [Google Scholar] [CrossRef]
- McPhee, M. Generations in captivity increases behavioural variance: Considerations for captive breeding and reintroduction programs. Biol. Conserv. 2004, 115, 71–77. [Google Scholar] [CrossRef]
- Wildt, D.E. Genome resource banking for wildlife research, management, and conservation. ILAR J. 2000, 41, 228–234. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, A.; Griffin, P.; Dillon, S.; Catullo, R.; Rane, R.; Byrne, M.; Jordan, R.; Oakeshott, J.; Weeks, A.; Joseph, L.; et al. A framework for incorporating evolutionary genomics into biodiversity conservation and management. Clim. Chang. Responses 2015, 2, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Harley, D.; Mawson, P.; Olds, L.; McFadden, M.; Hogg, C. The contribution of captive breeding in zoos to the conservation of Australia’s threatened fauna. In Recovering Australian Threatened Species: A Book of Hope; Garnett, S., Woinarski, J., Lindenmayer, D., Latch, P., Eds.; CSIRO Publishing: Clayton, Australia, 2018; pp. 281–294. [Google Scholar]
- Kelly, M. Adaptation to climate change through genetic accommodation and assimiliation of plastic phenotypes. Philos. Trans. R. Soc. B 2019, 274, 20180176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bataillon, T.; Galtier, N.; Bernard, A.; Cryer, N.; Faivre, N.; Santoni, S.; Severac, D.; Mikkelsen, T.; Larsen, K.; Beier CSørensen, J. A replicated climate change field experiment reveals rapid evolutionary response in an ecologically important soil invertebrate. Glob. Chang. Biol. 2016, 22, 2370–2379. [Google Scholar] [CrossRef] [Green Version]
- Razgour, O.; Forester, B.; Taggart, J.B.; Bekaert, M.; Juste, J.; Ibanez, C.; Puechmaille, S.J.; Novella-Fernandez, R.; Alberdi, A.; Manel, S. Considering adaptive genetic variation in climate change vulnerability assessment reduces species range loss projections. Proc. Natl. Acad. Sci. USA 2019, 116, 10418–10423. [Google Scholar] [CrossRef] [Green Version]
- Laikre, L.; Allendorf, F.W.; Aroner, L.C.; Baker, C.S.; Gregovich, D.P.; Hansen, M.M.; Jackson, J.A.; Kendall, K.C.; McKelvey, K.; Neel, M.C.; et al. Neglect of Genetic Diversity in Implementation of the Convention on Biological Diversity. Conserv. Biol. 2010, 24, 86–88. [Google Scholar] [CrossRef] [Green Version]
- Pierson, J.C.; Coates, D.J.; Oostermeijer, J.G.B.; Beissinger, S.R.; Bragg, J.; Sunnucks, P.; Schumaker, N.H.; Young, A.G. Genetic factors in threatened species recovery plans on three continents. Front. Ecol. Environ. 2016, 14, 433–440. [Google Scholar] [CrossRef] [Green Version]
- Prober, S.; Byrne, M.; McLean, E.; Steane, D.; Potts, B.; Vaillancourt, R.; Stock, W. Climate-adjusted provenancing: A strategy for climate-resilient ecological restoration. Front. Ecol. Evol. 2015, 3, 65. [Google Scholar] [CrossRef] [Green Version]
- Copley, P. Natural histories of Australia’s stick-nest rats, genus Leporillus (Rodentia: Muridae). Wildl. Res. 1999, 26, 513–539. [Google Scholar] [CrossRef]
- Moseby, K.; Bice, J. A trial re-introduction of the Greater Stick-nest Rat (Leporillus conditor) in arid South Australia. Ecol. Manag. Restor. 2004, 5, 118–124. [Google Scholar] [CrossRef]
- Short, J.; Copley, P.; Ruykys, L.; Morris, K.; Read, J.; Moseby, K. Review of translocations of the greater stick-nest rat (Leporillus conditor): Lessons learnt to facilitate ongoing recovery. Wildl. Res. 2019, 46, 455–475. [Google Scholar] [CrossRef]
- Bolton, J.; Moseby, K. The activity of Sand Goannas Varanus gouldii and their interaction with reintroduced Greater Stick-nest Rats Leporillus conditor. Pac. Conserv. Biol. 2004, 10, 193–201. [Google Scholar] [CrossRef]
- Flanagan, S.; Forester, B.; Latch, E.; Aitken, S.; Hoban, S. Guidelines for planning genomic assessment and monitoring of locally adaptive variation to inform species conservation. Evol. Appl. 2018, 11, 1035–1052. [Google Scholar] [CrossRef] [PubMed]
- Brandies, P.; Peel, E.; Hogg, C.; Belov, K. The value of reference genomes in the conservation of threatened species. Genes 2019, 10, 846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grabowski, P.P.; Morris, G.; Casler, M.D.; Borevitz, J. Population genomic variation reveals roles of history, adaptation and ploidy in switchgrass. Mol. Ecol. 2014, 23, 4059–4073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zalasiewicz, J. The Earth after Us: What Legacy Will Humans Leave in the Rocks? Oxford University Press: Oxford, UK, 2008. [Google Scholar]
- Lewis, S.L.; Maslin, M. Defining the Anthropocene. Nat. Cell Biol. 2015, 519, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.D. Translocation of species, climate change, and the end of trying to recreate past ecological communities. Trends Ecol. Evol. 2011, 26, 216–221. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Eldridge, M.; Lacy, R.C.; Ralls, K.; Dudash, M.R.; Fenster, C.B. Predicting the Probability of Outbreeding Depression. Conserv. Biol. 2011, 25, 465–475. [Google Scholar] [CrossRef]
- Dunlop, J.A.; Birch, N.; Moore, H.; Cowan, M. Pilbara northern quoll research program. In Annual Report; Department of Parks and Wildlife: Perth, Australia, 2017. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Onley, I.R.; Moseby, K.E.; Austin, J.J. Genomic Approaches for Conservation Management in Australia under Climate Change. Life 2021, 11, 653. https://doi.org/10.3390/life11070653
Onley IR, Moseby KE, Austin JJ. Genomic Approaches for Conservation Management in Australia under Climate Change. Life. 2021; 11(7):653. https://doi.org/10.3390/life11070653
Chicago/Turabian StyleOnley, Isabelle R., Katherine E. Moseby, and Jeremy J. Austin. 2021. "Genomic Approaches for Conservation Management in Australia under Climate Change" Life 11, no. 7: 653. https://doi.org/10.3390/life11070653
APA StyleOnley, I. R., Moseby, K. E., & Austin, J. J. (2021). Genomic Approaches for Conservation Management in Australia under Climate Change. Life, 11(7), 653. https://doi.org/10.3390/life11070653





