Impact TMPRSS2–ERG Molecular Subtype on Prostate Cancer Recurrence
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Isolation of RNA and Reverse Transcription
2.2.2. Quantitative PCR (qPCR)
2.2.3. RNA Sequencing
2.2.4. Data Analysis
2.2.5. Statistics
3. Results
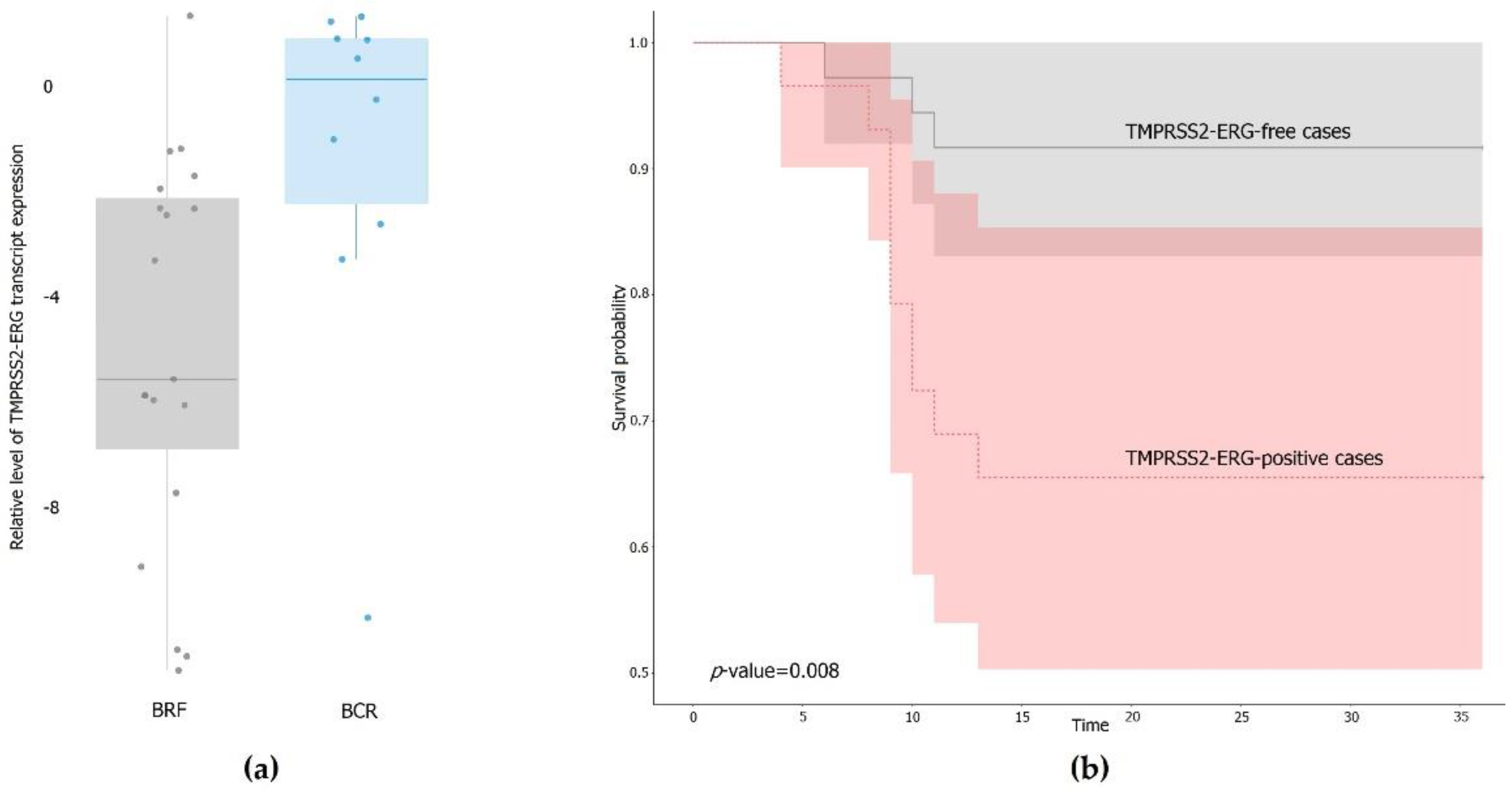
3.1. Expression of the TMPRSS2–ERG Fusion Transcript in PCa Samples
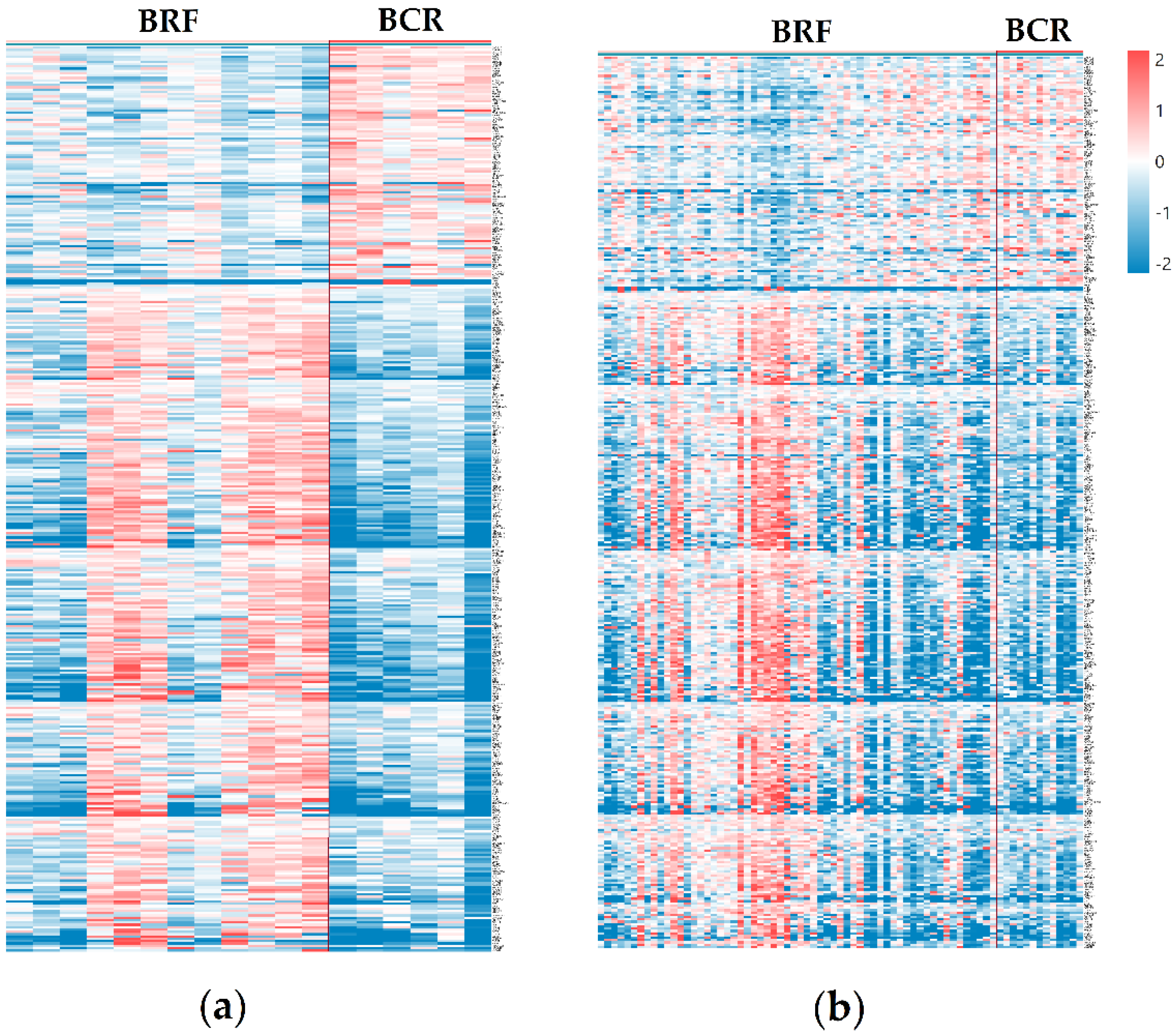
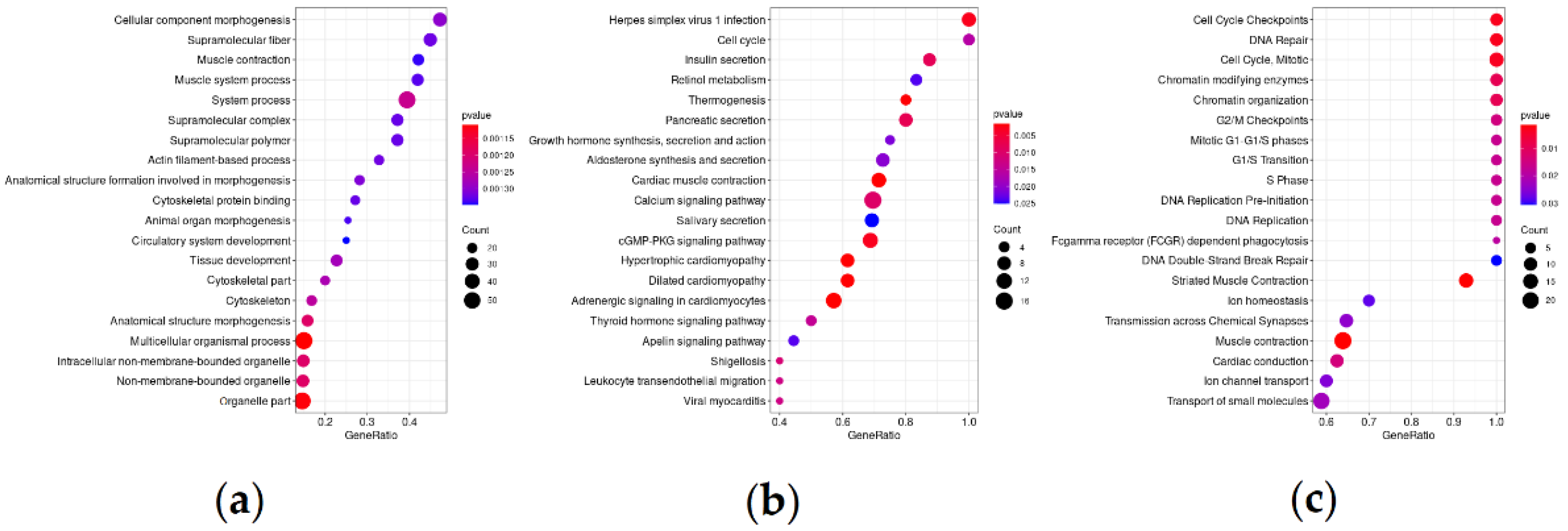
3.2. Differentially Expressed Genes and Significantly Enriched Pathways
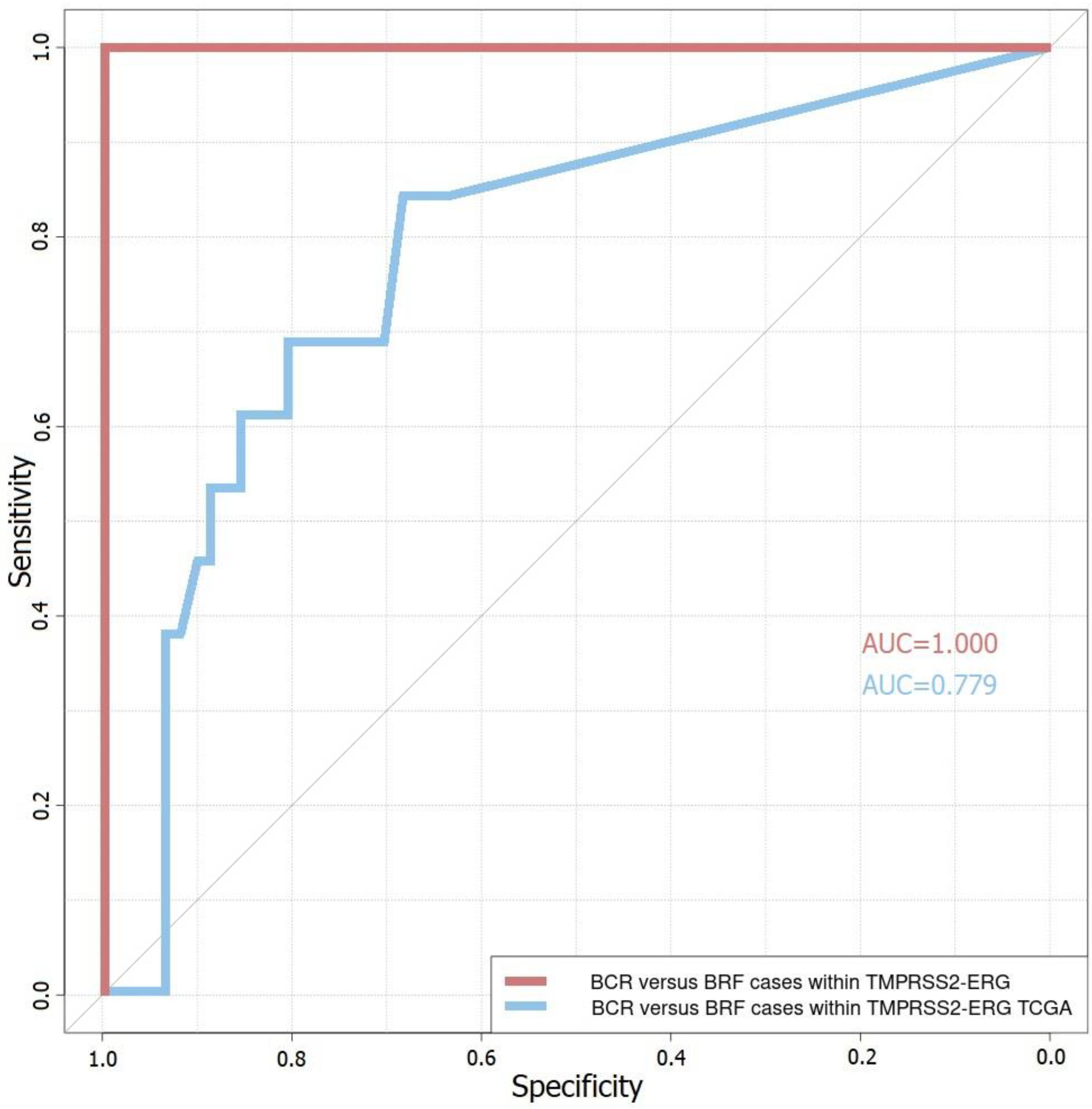
3.3. Four-Gene Prognostic Model
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- The Cancer Genome Atlas Research Network. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015, 163, 1011–1025. [Google Scholar] [CrossRef] [Green Version]
- Arora, K.; Barbieri, C.E. Molecular Subtypes of Prostate Cancer. Curr. Oncol. Rep. 2018, 20, 58. [Google Scholar] [CrossRef]
- The Human Protein Atlas. Available online: https://www.proteinatlas.org/ (accessed on 9 April 2021).
- Tandefelt, D.G.; Boormans, J.; Hermans, K.; Trapman, J. ETS fusion genes in prostate cancer. Endocr. Relat. Cancer 2014, 21, R143–R152. [Google Scholar] [CrossRef]
- Tomlins, S.A.; Rhodes, D.R.; Perner, S.; Dhanasekaran, S.M.; Mehra, R.; Sun, X.-W.; Varambally, S.; Cao, X.; Tchinda, J.; Kuefer, R.; et al. Recurrent Fusion of TMPRSS2 and ETS Transcription Factor Genes in Prostate Cancer. Science 2005, 310, 644–648. [Google Scholar] [CrossRef]
- Esgueva, R.; Perner, S.; LaFargue, C.; Scheble, V.; Stephan, C.; Lein, M.; Fritzsche, F.R.; Dietel, M.; Kristiansen, G.; Rubin, M.A. Prevalence of TMPRSS2–ERG and SLC45A3–ERG gene fusions in a large prostatectomy cohort. Mod. Pathol. 2010, 23, 539–546. [Google Scholar] [CrossRef] [Green Version]
- Mani, R.-S.; Iyer, M.K.; Cao, Q.; Brenner, J.C.; Wang, L.; Ghosh, A.; Cao, X.; Lonigro, R.J.; Tomlins, S.A.; Varambally, S.; et al. TMPRSS2–ERG-Mediated feed-forward regulation of wild-type erg in human prostate cancers. Cancer Res. 2011, 71, 5387–5392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zammarchi, F.; Boutsalis, G.; Cartegni, L. 5′ UTR control of native erg and of Tmprss2:Erg variants activity in prostate cancer. PLoS ONE 2013, 8, e49721. [Google Scholar] [CrossRef] [Green Version]
- Hägglöf, C.; Hammarsten, P.; Strömvall, K.; Egevad, L.; Josefsson, A.; Stattin, P.; Granfors, T.; Bergh, A. TMPRSS2-ERG expression predicts prostate cancer survival and associates with stromal biomarkers. PLoS ONE 2014, 9, e86824. [Google Scholar] [CrossRef] [Green Version]
- Pettersson, A.; Graff, R.E.; Bauer, S.R.; Pitt, M.J.; Lis, R.T.; Stack, E.C.; Martin, N.E.; Kunz, L.; Penney, K.L.; Ligon, A.H.; et al. The TMPRSS2:ERG rearrangement, erg expression, and prostate cancer outcomes: A cohort study and meta-analysis. Cancer Epidemiol. Biomark. Prev. 2012, 21, 1497–1509. [Google Scholar] [CrossRef] [Green Version]
- Sabaliauskaite, R.; Jarmalaite, S.; Petroska, D.; Dasevicius, D.; Laurinavicius, A.; Jankevicius, F.; Lazutka, J.R. Combined analysis ofTMPRSS2-ERGandTERTfor improved prognosis of biochemical recurrence in prostate cancer. Genes Chromosom. Cancer 2012, 51, 781–791. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, A.V.; Whittington, R.; Malkowicz, S.B.; Schultz, D.; Blank, K.; Broderick, G.; Tomaszewski, J.E.; Renshaw, A.A.; Kaplan, I.; Beard, C.J.; et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA 1998, 280, 969–974. [Google Scholar] [CrossRef]
- Cucchiara, V.; Cooperberg, M.R.; Dall’Era, M.; Lin, D.W.; Montorsi, F.; Schalken, J.A.; Evans, C.P. Genomic Markers in Prostate Cancer Decision Making. Eur. Urol. 2018, 73, 572–582. [Google Scholar] [CrossRef]
- Rebello, R.J.; Oing, C.; Knudsen, K.E.; Loeb, S.; Johnson, D.C.; Reiter, R.E.; Gillessen, S.; Van der Kwast, T.; Bristow, R.G. Prostate cancer. Nat. Rev. Dis. Prim. 2021, 7, 1–27. [Google Scholar] [CrossRef]
- Barwick, B.G.; Abramovitz, M.; Kodani, M.; Moreno, C.S.; Nam, R.; Tang, W.; Bouzyk, M.; Seth, A.; Leyland-Jones, B. Prostate cancer genes associated with TMPRSS2–ERG gene fusion and prognostic of biochemical recurrence in multiple cohorts. Br. J. Cancer 2010, 102, 570–576. [Google Scholar] [CrossRef]
- Font-Tello, A.; Juanpere, N.; de Muga, S.; Lorenzo, M.; Lorente, J.A.; Fumado, L.; Serrano, L.; Serrano, S.; Lloreta, J.; Hernández, S. Association of ERG and TMPRSS2-ERG with grade, stage, and prognosis of prostate cancer is dependent on their expression levels. Prostate 2015, 75, 1216–1226. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Kim, S.H.; Joung, J.Y.; Lee, G.K.; Hong, E.K.; Kang, K.M.; Yu, A.; Nam, B.H.; Chung, J.; Seo, H.K.; et al. Overexpression of ERG and Wild-Type PTEN Are Associated with Favorable Clinical Prognosis and Low Biochemical Recurrence in Prostate Cancer. PLoS ONE 2015, 10, e0122498. [Google Scholar] [CrossRef]
- Liu, R.; Zhou, J.; Xia, S.; Li, T. The impact of PTEN deletion and ERG rearrangement on recurrence after treatment for prostate cancer: A systematic review and meta-analysis. Clin. Transl. Oncol. 2020, 22, 694–702. [Google Scholar] [CrossRef] [PubMed]
- Gerke, J.S.; Orth, M.F.; Tolkach, Y.; Romero-Pérez, L.; Wehweck, F.S.; Stein, S.; Musa, J.; Knott, M.M.; Hölting, T.L.; Li, J.; et al. Integrative clinical transcriptome analysis reveals TMPRSS2-ERG dependency of prognostic biomarkers in prostate adenocarcinoma. Int. J. Cancer 2020, 146, 2036–2046. [Google Scholar] [CrossRef] [Green Version]
- GDC. Available online: https://portal.gdc.cancer.gov (accessed on 3 February 2021).
- Krasnov, G.S.; Kudryavtseva, A.V.; Snezhkina, A.V.; Lakunina, V.A.; Beniaminov, A.D.; Melnikova, N.V.; Dmitriev, A.A. Pan-Cancer Analysis of TCGA Data Revealed Promising Reference Genes for qPCR Normalization. Front. Genet. 2019, 10, 97. [Google Scholar] [CrossRef] [Green Version]
- Krasnov, G.; Oparina, N.; Dmitriev, A.A.; Kudryavtseva, A.; Anedchenko, E.A.; Kondrat’Eva, T.T.; Zabarovsky, E.R.; Senchenko, V. RPN1, a new reference gene for quantitative data normalization in lung and kidney cancer. Mol. Biol. 2011, 45, 211–220. [Google Scholar] [CrossRef]
- Melnikova, N.V.; Dmitriev, A.A.; Belenikin, M.S.; Koroban, N.V.; Speranskaya, A.S.; Krinitsina, A.A.; Krasnov, G.S.; Lakunina, V.A.; Snezhkina, A.V.; Sadritdinova, A.F.; et al. Identification, Expression Analysis, and Target Prediction of Flax Genotroph MicroRNAs Under Normal and Nutrient Stress Conditions. Front. Plant Sci. 2016, 7, 399. [Google Scholar] [CrossRef] [Green Version]
- Babraham Bioinformatics—FastQC A Quality Control Tool for High throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 3 March 2021).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- GENCODE—Human Release 19. Available online: https://www.gencodegenes.org/human/release_19.html (accessed on 25 April 2021).
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 3 March 2021).
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- randomForest Package—Rdocumentation. Available online: https://www.rdocumentation.org/packages/randomForest/versions/4.6-14 (accessed on 3 March 2021).
- Robin, X.A.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Muller, M.J. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- survminer: Survival Analysis and Visualization. Available online: https://github.com/kassambara/survminer (accessed on 3 March 2021).
- Pudova, E.A.; Lukyanova, E.N.; Nyushko, K.M.; Mikhaylenko, D.S.; Zaretsky, A.R.; Snezhkina, A.V.; Savvateeva, M.V.; Kobelyatskaya, A.A.; Melnikova, N.V.; Volchenko, N.N.; et al. Differentially Expressed Genes Associated With Prognosis in Locally Advanced Lymph Node-Negative Prostate Cancer. Front. Genet. 2019, 10, 730. [Google Scholar] [CrossRef]
- Pernar, C.H.; Ebot, E.M.; Wilson, K.M.; Mucci, L.A. The Epidemiology of Prostate Cancer. Cold Spring Harb. Perspect. Med. 2018, 8, a030361. [Google Scholar] [CrossRef]
- Kudryavtseva, A.V.; Lukyanova, E.N.; Kharitonov, S.L.; Nyushko, K.M.; Krasheninnikov, A.A.; Pudova, E.A.; Guvatova, Z.G.; Alekseev, B.Y.; Kiseleva, M.V.; Kaprin, A.D.; et al. Bioinformatic identification of differentially expressed genes associated with prognosis of locally advanced lymph node-positive prostate cancer. J. Bioinform. Comput. Biol. 2019, 17, 1950003. [Google Scholar] [CrossRef]
- Lee, M.; Williams, K.A.; Hu, Y.; Andreas, J.; Patel, S.J.; Zhang, S.; Crawford, N.P.S. GNL3 and SKA3 are novel prostate cancer metastasis susceptibility genes. Clin. Exp. Metastasis 2015, 32, 769–782. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Dong, S.; Hu, J.; Duan, B.; Yao, J.; Zhang, R.; Zhou, H.; Sheng, H.; Gao, H.; Li, S.; et al. Guanine nu-cleotide binding protein-like 3 is a potential prognosis indicator of gastric cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 13273–13278. [Google Scholar]
- Tang, X.; Zha, L.; Li, H.; Liao, G.; Huang, Z.; Peng, X.; Wang, Z. Upregulation of GNL3 expression promotes colon cancer cell proliferation, migration, invasion and epithelial-mesenchymal transition via the Wnt/β-catenin signaling pathway. Oncol. Rep. 2017, 38, 2023–2032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sami, M.M.; Hachim, M.Y.; Hachim, I.Y.; Elbarkouky, A.H.; López-Ozuna, V.M. Nucleostemin expression in breast cancer is a marker of more aggressive phenotype and unfavorable patients’ outcome. Medicine 2019, 98, e14744. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Masutomi, K.; Tamura, K.; Moriya, T.; Yamasaki, T.; Fujiwara, Y.; Takahashi, S.; Yamamoto, J.; Tsuda, H. Nucleostemin expression in invasive breast cancer. BMC Cancer 2014, 14, 215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Li, N.; Gao, L.; You, C. Integrated analysis of the roles and prognostic value of RNA binding proteins in lung adenocarcinoma. PeerJ 2020, 8, e8509. [Google Scholar] [CrossRef]
| Criterion | Cohort of Russian Patients with PCa | TCGA–PRAD Cohort | |
|---|---|---|---|
| PCa samples, total Age (years), mean (range) | 72 | 203 | |
| 63 (41–73) | 62 (46–78) | ||
| pT, n | pT3a | 35 | 98 |
| pT3b | 37 | 105 | |
| pN, n | pN0 | 43 | 139 |
| pN1 | 29 | 64 | |
| pM, n | pM0 | 72 | 203 |
| pM1 | 0 | 0 | |
| Gleason score, n | 6 | 7 | 8 |
| 7 | 41 | 77 | |
| 8 | 10 | 30 | |
| 9 | 13 | 8 | |
| 10 | 1 | 2 | |
| Biochemical recurrence (PSA ≥ 0.2 ng/mL), n | 13 | 63 | |
| Russian Patients | TCGA–PRAD | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene | FC | logCPM | QLF p-Value | MW p-Value | FC | logCPM | QLF p-Value | MW p-Value |
| GNL3 | 1.37 | 7.27 | 0.0095 | 0.0032 | 1.29 | 7.27 | 0.0043 | 0.0048 |
| QSOX2 | 1.45 | 5.04 | 0.0086 | 0.0069 | 1.41 | 5.07 | 0.0005 | 0.0002 |
| SSPO | 2.65 | 4.62 | 0.0008 | 0.0083 | 2.08 | 3.32 | 0.0016 | 0.0012 |
| SYS1 | −1.33 | 4.90 | 0.0073 | 0.0001 | −1.23 | 5.40 | 0.0068 | 0.0052 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kobelyatskaya, A.A.; Pudova, E.A.; Snezhkina, A.V.; Fedorova, M.S.; Pavlov, V.S.; Guvatova, Z.G.; Savvateeva, M.V.; Melnikova, N.V.; Dmitriev, A.A.; Trofimov, D.Y.; et al. Impact TMPRSS2–ERG Molecular Subtype on Prostate Cancer Recurrence. Life 2021, 11, 588. https://doi.org/10.3390/life11060588
Kobelyatskaya AA, Pudova EA, Snezhkina AV, Fedorova MS, Pavlov VS, Guvatova ZG, Savvateeva MV, Melnikova NV, Dmitriev AA, Trofimov DY, et al. Impact TMPRSS2–ERG Molecular Subtype on Prostate Cancer Recurrence. Life. 2021; 11(6):588. https://doi.org/10.3390/life11060588
Chicago/Turabian StyleKobelyatskaya, Anastasiya A., Elena A. Pudova, Anastasiya V. Snezhkina, Maria S. Fedorova, Vladislav S. Pavlov, Zulfiya G. Guvatova, Maria V. Savvateeva, Nataliya V. Melnikova, Alexey A. Dmitriev, Dmitry Y. Trofimov, and et al. 2021. "Impact TMPRSS2–ERG Molecular Subtype on Prostate Cancer Recurrence" Life 11, no. 6: 588. https://doi.org/10.3390/life11060588
APA StyleKobelyatskaya, A. A., Pudova, E. A., Snezhkina, A. V., Fedorova, M. S., Pavlov, V. S., Guvatova, Z. G., Savvateeva, M. V., Melnikova, N. V., Dmitriev, A. A., Trofimov, D. Y., Sukhikh, G. T., Nyushko, K. M., Alekseev, B. Y., Razin, S. V., Krasnov, G. S., & Kudryavtseva, A. V. (2021). Impact TMPRSS2–ERG Molecular Subtype on Prostate Cancer Recurrence. Life, 11(6), 588. https://doi.org/10.3390/life11060588










