The Telomeric Protein TRF2 Regulates Replication Origin Activity within Pericentromeric Heterochromatin
Abstract
1. Introduction
2. Materials and Methods
2.1. Transient Transfections
2.2. Real-Time qPCR
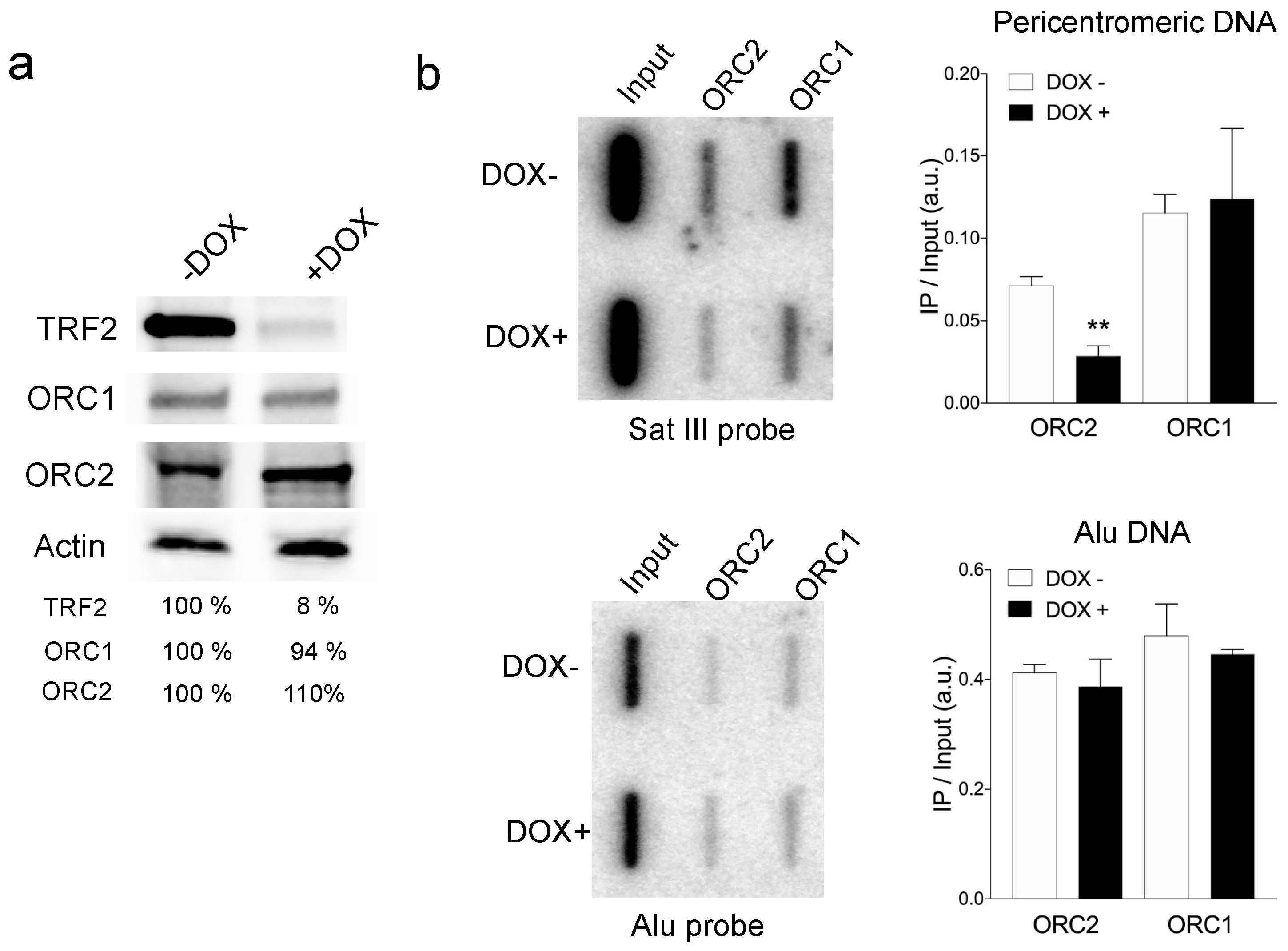
2.3. Western Blotting
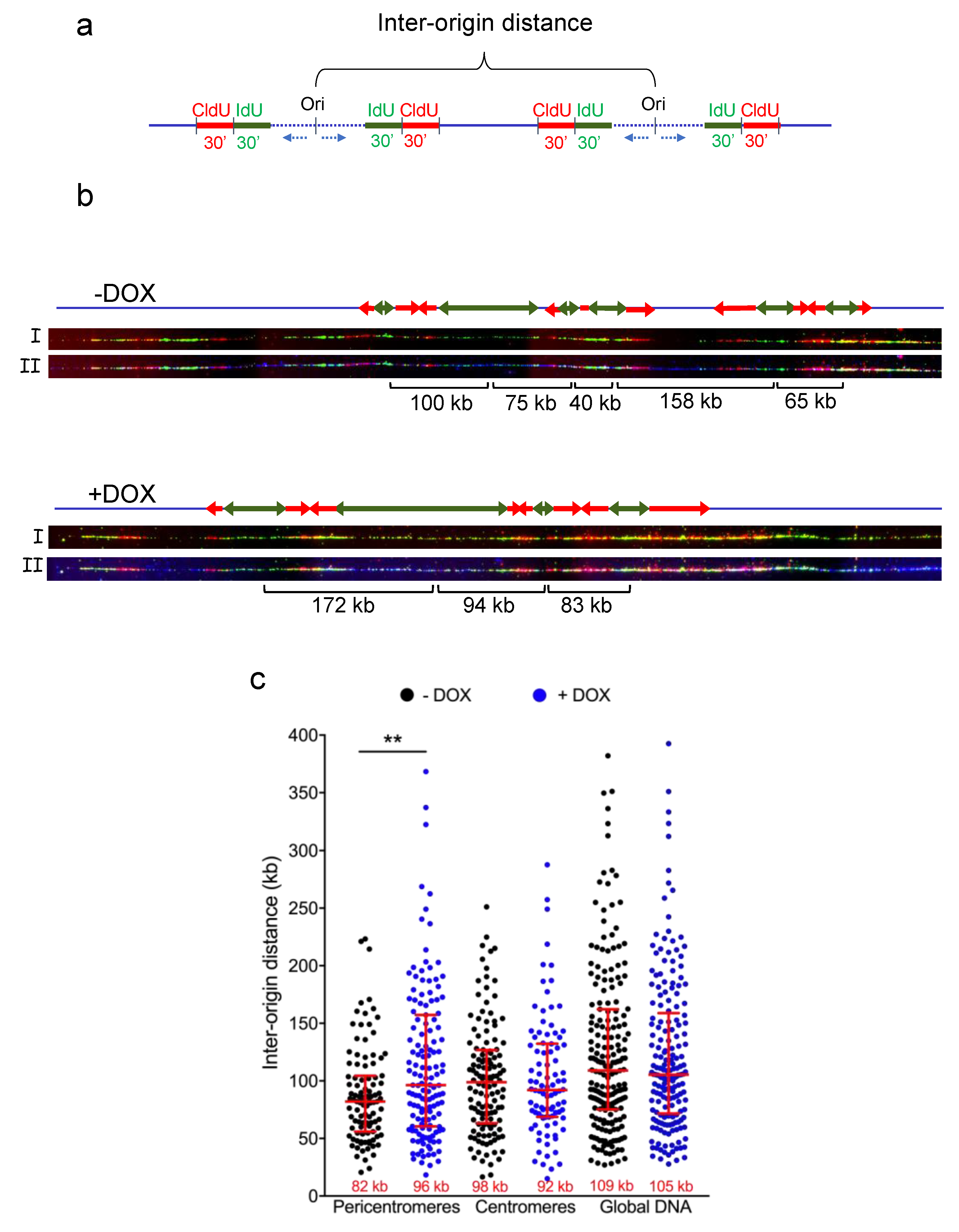
2.4. DNA Combing
2.5. Chromatin Immunoprecipitation (ChIP)
2.6. Statistics
3. Results
3.1. TRF2 Controlled ORC Association to Pericentromeric Satellite DNA
3.2. TRF2 Regulates Origin Density in Pericentromeric Heterochromatin
3.3. The B Domain of TRF2 Is Required for Origin Density Regulation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Janssen, A.; Colmenares, S.U.; Karpen, G.H. Heterochromatin: Guardian of the Genome. Annu. Rev. Cell Dev. Biol. 2018, 34, 265–288. [Google Scholar] [CrossRef]
- Mendez-Bermudez, A.; Giraud-Panis, M.J.; Ye, J.; Gilson, E. Heterochromatin replication goes hand in hand with telomere protection. Nat. Struct. Mol. Biol. 2020, 27, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Tazumi, A.; Fukuura, M.; Nakato, R.; Kishimoto, A.; Takenaka, T.; Ogawa, S.; Song, J.H.; Takahashi, T.S.; Nakagawa, T.; Shirahige, K.; et al. Telomere-binding protein Taz1 controls global replication timing through its localization near late replication origins in fission yeast. Genes Dev. 2012, 26, 2050–2062. [Google Scholar] [CrossRef]
- Zofall, M.; Smith, D.R.; Mizuguchi, T.; Dhakshnamoorthy, J.; Grewal, S.I.S. Taz1-Shelterin Promotes Facultative Heterochromatin Assembly at Chromosome-Internal Sites Containing Late Replication Origins. Mol. Cell 2016, 62, 862–874. [Google Scholar] [CrossRef]
- Zimmermann, M.; Kibe, T.; Kabir, S.; de Lange, T. TRF1 negotiates TTAGGG repeatassociated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes Dev. 2014, 28, 2477–2491. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Lenain, C.; Bauwens, S.; Rizzo, A.; Saint-Léger, A.; Poulet, A.; Benarroch, D.; Magdinier, F.; Morere, J.; Amiard, S.; et al. TRF2 and Apollo Cooperate with Topoisomerase 2α to Protect Human Telomeres from Replicative Damage. Cell 2010, 142, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Kim, H.; Ji, Z.; Zhang, T.; Chen, B.; Ge, Y.; Hu, Y.; Feng, X.; Han, X.; Xu, H.; et al. The BUB3-BUB1 Complex Promotes Telomere DNA Replication. Mol. Cell 2018, 70, 395–407.e4. [Google Scholar] [CrossRef]
- Mendez-Bermudez, A.; Lototska, L.; Bauwens, S.; Giraud-Panis, M.J.; Croce, O.; Jamet, K.; Irizar, A.; Mowinckel, M.; Koundrioukoff, S.; Nottet, N.; et al. Genome-wide Control of Heterochromatin Replication by the Telomere Capping Protein TRF2. Mol. Cell 2018, 70, 449–461.e5. [Google Scholar] [CrossRef]
- Saint-Léger, A.; Koelblen, M.; Civitelli, L.; Bah, A.; Djerbi, N.; Giraud-Panis, M.J.; Londonõ-Vallejo, A.; Ascenzioni, F.; Gilson, E. The basic N-terminal domain of TRF2 limits recombination endonuclease action at human telomeres. Cell Cycle 2014, 13, 2469–2474. [Google Scholar] [CrossRef]
- Poulet, A.; Buisson, R.; Faivre-Moskalenko, C.; Koelblen, M.; Amiard, S.; Montel, F.; Cuesta-Lopez, S.; Bornet, O.; Guerlesquin, F.; Godet, T.; et al. TRF2 promotes, remodels and protects telomeric Holliday junctions. EMBO J. 2009, 28, 641–651. [Google Scholar] [CrossRef]
- Kurth, I.; Gautier, J. Origin-dependent initiation of DNA replication within telomeric sequences. Nucleic Acids Res. 2009, 38, 467–476. [Google Scholar] [CrossRef]
- Deng, Z.; Dheekollu, J.; Broccoli, D.; Dutta, A.; Lieberman, P.M. The Origin Recognition Complex Localizes to Telomere Repeats and Prevents Telomere-Circle Formation. Curr. Biol. 2007, 17, 1989–1995. [Google Scholar] [CrossRef]
- Tatsumi, Y.; Ezura, K.; Yoshida, K.; Yugawa, T.; Narisawa-Saito, M.; Kiyono, T.; Ohta, S.; Obuse, C.; Fujita, M. Involvement of human ORC and TRF2 in pre-replication complex assembly at telomeres. Genes Cells 2008, 13, 1045–1059. [Google Scholar] [CrossRef]
- Zhou, J.; Deng, Z.; Norseen, J.; Lieberman, P.M. Regulation of Epstein-Barr Virus Origin of Plasmid Replication (OriP) by the S-Phase Checkpoint Kinase Chk2. J. Virol. 2010, 84, 4979–4987. [Google Scholar] [CrossRef] [PubMed]
- Atanasiu, C.; Deng, Z.; Wiedmer, A.; Norseen, J.; Lieberman, P.M. ORC binding to TRF2 stimulates OriP replication. EMBO Rep. 2006, 7, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Lezina, L.; Chen, C.J.; Shtivelband, S.; So, W.; Lieberman, P.M. Telomeric proteins regulate episomal maintenance of epstein-barr virus origin of plasmid replication. Mol. Cell 2002, 9, 493–503. [Google Scholar] [CrossRef]
- Grolimund, L.; Aeby, E.; Hamelin, R.; Armand, F.; Chiappe, D.; Moniatte, M.; Lingner, J. A quantitative telomeric chromatin isolation protocol identifies different telomeric states. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef]
- Técher, H.; Koundrioukoff, S.; Azar, D.; Wilhelm, T.; Carignon, S.; Brison, O.; Debatisse, M.; Le Tallec, B. Replication dynamics: Biases and robustness of DNA fiber analysis. J. Mol. Biol. 2013, 425, 4845–4855. [Google Scholar] [CrossRef]
- Anglana, M.; Apiou, F.; Bensimon, A.; Debatisse, M. Dynamics of DNA replication in mammalian somatic cells: Nucleotide pool modulates origin choice and interorigin spacing. Cell 2003, 114, 385–394. [Google Scholar] [CrossRef]
- Miotto, B.; Ji, Z.; Struhl, K. Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers. Proc. Natl. Acad. Sci. USA 2016, 113, E4810–E4819. [Google Scholar] [CrossRef] [PubMed]
- Drosopoulos, W.C.; Deng, Z.; Twayana, S.; Kosiyatrakul, S.T.; Vladimirova, O.; Lieberman, P.M.; Schildkraut, C.L. TRF2 Mediates Replication Initiation within Human Telomeres to Prevent Telomere Dysfunction. Cell Rep. 2020, 33. [Google Scholar] [CrossRef]
- Fox, C.A.; Ehrenhofer-Murray, A.E.; Loo, S.; Rine, J. The origin recognition complex, SIR1, and the S phase requirement for silencing. Science 1997, 276, 1547. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Sathyan, K.M.; Geng, Y.; Zheng, R.; Chakraborty, A.; Freeman, B.; Wang, F.; Prasanth, K.V.; Prasanth, S.G. A WD-repeat protein stabilizes ORC binding to chromatin. Mol. Cell 2010, 40, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Hsu, R.Y.C.; Lin, Y.C.; Redon, C.; Sun, Q.; Singh, D.K.; Wang, Y.; Aggarwal, V.; Mitra, J.; Matur, A.; Moriarity, B.; et al. ORCA/LRWD1 Regulates Homologous Recombination at ALT-Telomeres by Modulating Heterochromatin Organization. iScience 2020, 23. [Google Scholar] [CrossRef] [PubMed]
- Higa, M.; Kushiyama, T.; Kurashige, S.; Kohmon, D.; Enokitani, K.; Iwahori, S.; Sugimoto, N.; Yoshida, K.; Fujita, M. TRF2 recruits ORC through TRFH domain dimerization. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 191–201. [Google Scholar] [CrossRef]
- Akerman, I.; Kasaai, B.; Bazarova, A.; Sang, P.B.; Peiffer, I.; Artufel, M.; Derelle, R.; Smith, G.; Rodriguez-Martinez, M.; Romano, M.; et al. A predictable conserved DNA base composition signature defines human core DNA replication origins. Nat. Commun. 2020, 11. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bauwens, S.; Lototska, L.; Koundrioukoff, S.; Debatisse, M.; Ye, J.; Gilson, E.; Mendez-Bermudez, A. The Telomeric Protein TRF2 Regulates Replication Origin Activity within Pericentromeric Heterochromatin. Life 2021, 11, 267. https://doi.org/10.3390/life11040267
Bauwens S, Lototska L, Koundrioukoff S, Debatisse M, Ye J, Gilson E, Mendez-Bermudez A. The Telomeric Protein TRF2 Regulates Replication Origin Activity within Pericentromeric Heterochromatin. Life. 2021; 11(4):267. https://doi.org/10.3390/life11040267
Chicago/Turabian StyleBauwens, Serge, Liudmyla Lototska, Stephane Koundrioukoff, Michelle Debatisse, Jing Ye, Eric Gilson, and Aaron Mendez-Bermudez. 2021. "The Telomeric Protein TRF2 Regulates Replication Origin Activity within Pericentromeric Heterochromatin" Life 11, no. 4: 267. https://doi.org/10.3390/life11040267
APA StyleBauwens, S., Lototska, L., Koundrioukoff, S., Debatisse, M., Ye, J., Gilson, E., & Mendez-Bermudez, A. (2021). The Telomeric Protein TRF2 Regulates Replication Origin Activity within Pericentromeric Heterochromatin. Life, 11(4), 267. https://doi.org/10.3390/life11040267





