Yuxiensis granularis gen. et sp. nov., a Novel Quellkörper-Bearing Fungal Taxon Added to Scortechiniaceae and Inclusion of Parasympodiellaceae in Coronophorales Based on Phylogenetic Evidence
Abstract
:1. Introduction
2. Materials and Methods
2.1. Specimen Collection and Morphological Studies
2.2. DNA Extraction, PCR Amplification and Sequencing
2.3. Phylogenetic Analyses
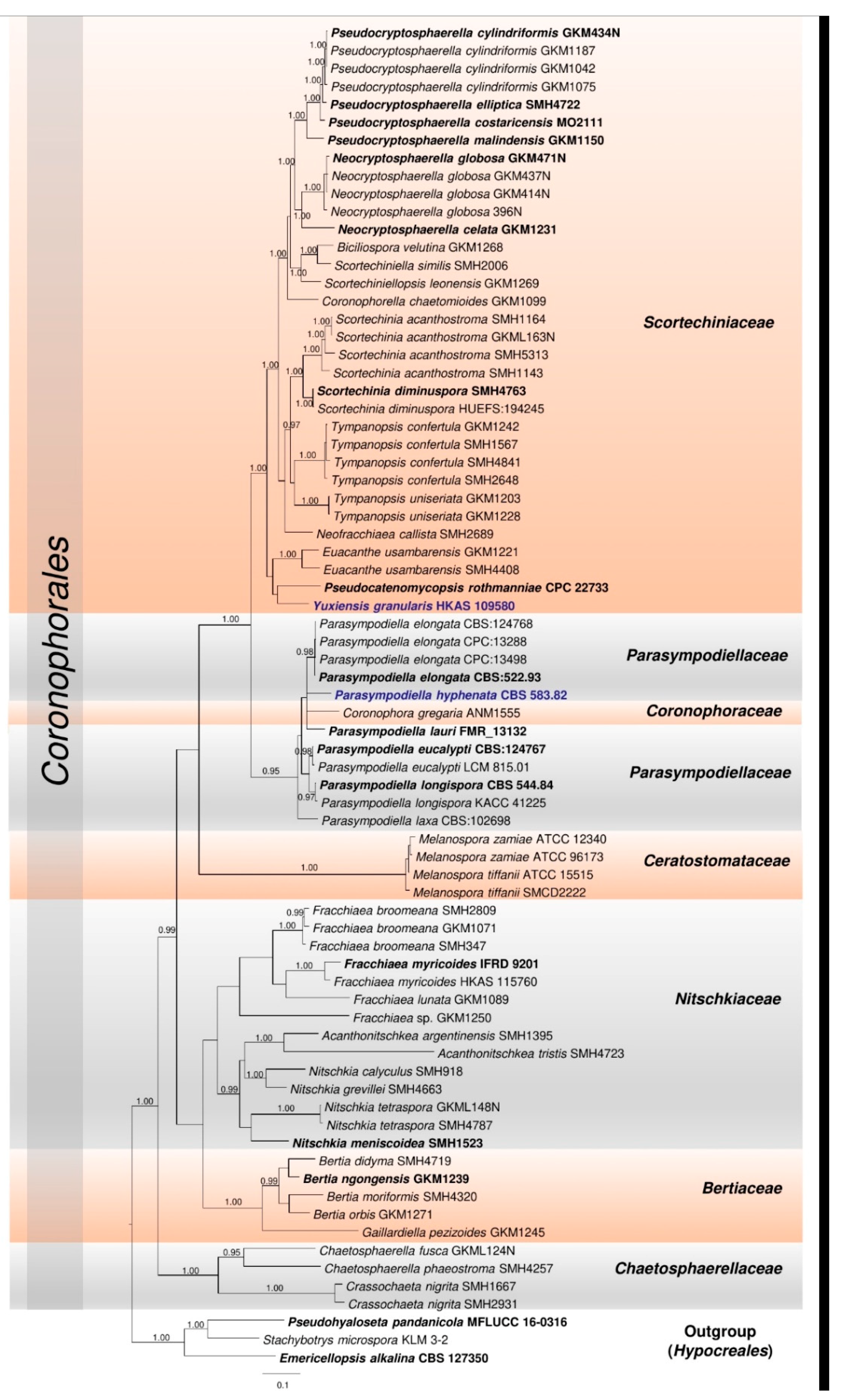
3. Results
3.1. Phylogenetic Analyses
3.2. Taxonomy
3.2.1. Coronophorales Nannf., Nova Acta R. Soc. Scient. Upsal., Ser. 4 8(no. 2): 54 (1932) Amend
3.2.2. Parasympodiellaceae Hern.-Restr., Gené, Guarro & Crous, in Hernández-Restrepo, Gené, Castañeda-Ruiz, Mena-Portales, Crous & Guarro, Stud. Mycol. 86: 87 (2017) Amend
3.2.3. Parasympodiella Ponnappa Trans. Br. Mycol. Soc. 64(2): 344 (1975) Amend
3.2.4. Parasympodiella hyphenata (Sigler, M.T. Dunn & J.W. Carmich.) Bundhun & K.D. Hyde, comb. nov.
3.2.5. Yuxiensis Bundhun & K.D. Hyde, gen. nov.
3.2.6. Yuxiensis granularis Bundhun, Wanas. & K.D. Hyde, sp. nov.
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Nannfeldt, J.A. Studien über die Morphologie und Systematik der nichtlichenisierten inoperculaten Discomyceten. Nova Acta Regiae Soc. Sci. Upsal. 1932, 8, 1–368. [Google Scholar]
- Mugambi, G.K.; Huhndorf, S.M. Multigene phylogeny of the Coronophorales: morphology and new species in the order. Mycologia 2010, 102, 185–210. [Google Scholar] [CrossRef] [Green Version]
- Carneiro de Almeida, D.A.; Gusmão, L.F.P.; Miller, A.N. Brazilian Semi-Arid Ascomycetes II: New and interesting records of Bertiaceae, Nitschkiaceae and Scortechiniaceae (Coronophorales, Sordariomycetes). Nova Hedwigia 2016, 102, 513–522. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Hyde, K.D.; Jones, E.B.G.; McKenzie, E.H.C.; Bhat, J.D.; Dayarathne, M.D.; Huang, S.K.; Norphanphoun, C.; Senanayake, I.C.; Perera, R.H.; et al. Families of Sordariomycetes. Fungal Divers. 2016, 79, 1–317. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Hyde, K.D.; Jones, E.B.G.; McKenzie, E.H.C.; Huang, S.K.; Abdel-Wahab, M.A.; Daranagama, D.A.; Dayarathne, M.D.; D’souza, M.J.; Goonasekara, I.D.; et al. Towards a natural classification and backbone tree for Sordariomycetes. Fungal Divers. 2015, 72, 199–301. [Google Scholar] [CrossRef]
- Hongsanan, S.; Maharachchikumbura, S.S.N.; Hyde, K.D.; Samarakoon, M.C.; Jeewon, R.; Zhao, Q.; Al-Sadi, A.M.; Bahkali, A.H. An updated phylogeny of Sordariomycetes based on phylogenetic and molecular clock evidence. Fungal Divers. 2017, 84, 25–41. [Google Scholar] [CrossRef]
- Hyde, K.D.; Maharachchikumbura, S.S.N.; Hongsanan, S.; Samarakoon, M.C.; Lücking., R.; Pem, D.; Harishchandra, D.; Jeewon, R.; Zhao, R.L.; Xu, J.C.; et al. The ranking of fungi: A tribute to David L. Hawksworth on his 70th birthday. Fungal Divers. 2017, 84, 1–23. [Google Scholar] [CrossRef]
- Hyde, K.D.; Norphanphoun, C.; Maharachchikumbura, S.S.N.; Bhat, D.J.; Jones, E.B.G.; Bundhun, D.; Chen, Y.J.; Bao, D.F.; Boonmee, S.; Calabon, M.S.; et al. Refined families of Sordariomycetes. Mycosphere 2020, 11, 305–1059. [Google Scholar] [CrossRef]
- Huhndorf, S.M.; Miller, A.N.; Fernandez, F.A. Molecular systematics of the Coronophorales and new species of Bertia, Lasiobertia and Nitschkia. Mycol. Res. 2004, 108, 1384–1398. [Google Scholar] [CrossRef]
- Nannfeldt, J.A. Stray studies in the Coronophorales (Pyrenomycetes) 1–3. Svensk Botanisk Tidskrift 1975, 69, 49–66. [Google Scholar]
- Crous, P.W.; Wingfield, M.J.; Guarro, J.; Cheewangkoon, R.; van der Bank, M.; Swart, W.J.; Stchigel, A.M.; Cano-Lira, J.F.; Roux, J.; Madrid, H.; et al. Fungal Planet description sheets: 154–213. Persoonia 2013, 31, 188–296. [Google Scholar] [CrossRef]
- Huang, S.K.; Hyde, K.D.; Maharachchikumbura, S.S.N.; McKenzie, E.H.C.; Wen, T.C. Taxonomic studies of Coronophorales and Niessliaceae (Hypocreomycetidae). Mycosphere 2021, 12, 875–992. [Google Scholar] [CrossRef]
- Hernández-Restrepo, M.; Gené, J.; Castañeda-Ruiz, R.F.; Mena-Portales, J. Phylogeny of saprobic microfungi from Southern Europe. Stud. Mycol. 2017, 86, 53–97. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, C.V.; Vittal, B.P. Three new Hyphomycetes from litter. Can J Bot 1973, 51, 1127–1132. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Kendrick, W.B. Foliicolous dematiaceous hyphomycetes from Syzygium cordatum. Can J Bot 1995, 73, 224–234. [Google Scholar] [CrossRef]
- Cheewangkoon, R.; Groenewald, J.Z.; Summerell, B.A.; Hyde, K.D.; To-anun, C.; Crous, P.W. Myrtaceae, a cache of fungal biodiversity. Persoonia 2009, 23, 55–85. [Google Scholar] [CrossRef] [Green Version]
- Sigler, L.; Dunn, M.T.; Carmichael, J.W. Arthrocristula and Arthropsis, two new Hyphomycetes with dematiaceous arthroconidia. Mycotaxon 1982, 15, 409–419. [Google Scholar]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.K.T.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; Zhao, R.L.; Aptroot, A.; Leontyev, D.V.; Saxena, R.K.; et al. Outline of Fungi and fungus-like taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; De Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef]
- Jayasiri, S.C.; Hyde, K.D.; Ariyawansa, H.A.; Bhat, J.; Buyck, B.; Cai, L.; Dai, Y.C.; Abd-Elsalam, K.A.; Ertz, D.; Hidayat, I.; et al. The Faces of Fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015, 74, 3–18. [Google Scholar] [CrossRef]
- Index Fungorum. 2021. Available online: http://www.indexfungorum.org/names/Names.asp (accessed on 30 May 2021).
- Wanasinghe, D.N.; Phukhamsakda, C.; Hyde, K.D.; Jeewon, R.; Lee, H.B.; Jones, E.B.G.; Tibpromma, S.; Tennakoon, D.S.; Dissanayake, A.J.; Jayasiri, S.C.; et al. Fungal diversity notes 709–839: Taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 2018, 89, 1–236. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. Res. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR—Protoc. Appl. 1990, 18, 315–322. [Google Scholar]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Sung, G.-H.; Sung, J.-M.; Hywel-Jones, N.L.; Spatafora, J.W. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenet. Evol. 2007, 44, 1204–1223. [Google Scholar] [CrossRef] [PubMed]
- Wanasinghe, D.N.; Wijayawardene, N.N.; Xu, J.; Cheewangkoon, R.; Mortimer, P.E. Taxonomic novelties in Magnolia-associated pleosporalean fungi in the Kunming Botanical Gardens (Yunnan, China). PLoS One 2020, 15, e0235855. [Google Scholar] [CrossRef]
- Dissanayake, A.J.; Bhunjun, C.S.; Maharachchikumbura, S.S.N.; Liu, J.K. Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere 2020, 11, 2652–2676. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar] [CrossRef]
- Glez-Pea, D.; Gmez-Blanco, D.; Reboiro-Jato, M.; Fdez-Riverola, F.; Posada, D. ALTER: Program-oriented conversion of DNA and protein alignments. Nucleic Acids Res 2010, 38, 14–18. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [Green Version]
- Zhaxybayeva, O.; Gogarten, J.P. Bootstrap, Bayesian probability and maximum likelihood mapping: Exploring new tools for comparative genome analyses. BMC Genomics 2002, 3, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nylander, J.A.A. MrModeltest, Version 2; Program Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004; Available online: https://github.com/Nylander (accessed on 30 May 2021).
- Swofford, D.L. PAUP* Version 4.0 b10. Phylogenetic Analysis Using Parsimony (*and Other Methods); Sinauer Associates: Sunderland, Massachusetts, 2002. [Google Scholar]
- Rambaut, A. FigTree. v. 1.4.0. 2012. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 2 June 2021).
- Wijayawardene, N.N.; Hyde, K.D.; Lumbsch, H.T.; Liu, J.K.; Maharachchikumbura, S.S.N.; Ekanayaka, A.H.; Tian, Q.; Phookamsak, R. Outline of Ascomycota: 2017. Fungal Divers. 2018, 88, 167–263. [Google Scholar] [CrossRef]
- Holubová-Jechová, V. Studies on Hyphomycetes from Cuba VII. Seven new taxa of dematiaceous Hyphomycetes. Česká Mykologie 1988, 42, 23–30. [Google Scholar]
- Liu, M.; Rombach, M.C.; Humber, R.A.; Hodge, K.T. What’s in a Name? Aschersonia insperata: A new pleoanamorphic fungus with characteristics of Archersonia and Hirsutella. Mycologia 2005, 97, 246–253. [Google Scholar] [CrossRef] [Green Version]
- Ulloa, M.; Hanlin, R.T. Nuevo Diccionario Ilustrado de Micología; American Phytopathological Society: Saint Paul, MN, USA, 2006; p. 674. [Google Scholar]
- Seifert, K.A.; Morgan-Jones, G.; Gams, W.; Kendrick, B. The Genera of Hyphomycetes; CBS Biodiversity Series; CBS-KNAW Fungal Biodiversity Centre: Utrecht, The Netherlands, 2011; Volume 9, pp. 1–997. [Google Scholar]
- Subramanian, C.V.; Sekar, G. Coronophorales from India – a monograph. Kavaka 1990, 18, 19–91. [Google Scholar]
- Huang, S.K.; Hyde, K.D.; Mapook, A.; Maharachchikumbura, S.S.N.; Bhat, D.J.; McKenzie, E.H.C.; Jeewon, R.; Wen, T.C. Taxonomic studies of some often over-looked Diaporthomycetidae and Sordariomycetidae. Fungal Divers 2021, in press. [Google Scholar]
- Sivanesan, A. Two new genera of Coronophorales with descriptions and key. Trans Brit Mycol Soc. 1974, 62, 35–43. [Google Scholar] [CrossRef]
- Hyde, K.D.; Hongsanan, S.; Jeewon, R.; Bhat, D.J.; McKenzie, E.H.C.; Jones, E.B.G.; Phookamsak, R.; Ariyawansa, H.A.; Boonmee, S.; Zhao, Q.; et al. Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016, 80, 1–270. [Google Scholar] [CrossRef]
- Tokumasu, S.; Tubaki, K. Bahusakala longispora sp. nov., and its geographical distribution in the pine forests of Japan. Trans. Mycol. Soc. Japan 1983, 24, 425–431. [Google Scholar]
- Tokumasu, S. Parasympodiella longispora, comb. nov., and its distribution in pine forests. Trans. Mycol. Soc. Japan 1987, 28, 19–26. [Google Scholar]
- Guarro, J.; Calvo, M.A.; Vicente, E. Contribución al estudio de los Hyphomycetes de España. IV. Acta Bot. Malac. 1980, 6, 43–52. [Google Scholar] [CrossRef]
- Pem, D.; Jeewon, R.; Bhat, D.J.; Doilom, M.; Boonmee, S.; Hongsanan, S.; Promputtha, I.; Xu, J.C.; Hyde, K.D. Mycosphere notes 275-324: A morpho-taxonomic revision and typification of obscure Dothideomycetes genera (incertae sedis). Mycosphere 2019, 10, 1115–1246. [Google Scholar] [CrossRef]
- Hyde, K.D.; Norphanphoun, C.; Chen, J.; Dissanayake, A.J.; Doilom, M.; Hongsanan, S.; Jayawardena, R.S.; Jeewon, R.; Perera, R.H.; Thongbai, B.; et al. Thailand’s amazing diversity – up to 96% of fungi in northern Thailand are novel. Fungal Divers. 2018, 93, 215–239. [Google Scholar] [CrossRef]
- Hyde, K.D.; Tennakoon, D.S.; Jeewon, R.; Bhat, D.J.; Maharachchikumbura, S.S.N.; Rossi, W.; Leonardi, M.; Lee, H.B.; Mun, H.Y.; Houbraken, J.; et al. Fungal diversity notes 1036–1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2019, 96, 1–242. [Google Scholar] [CrossRef]
- Hyde, K.D.; Dong, Y.; Phookamsak, R.; Jeewon, R.; Bhat, D.J.; Jones, E.B.G.; Liu, N.-G.; Abeywickrama, P.D.; Mapook, A.; Wei, D.; et al. Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020, 100, 5–277. [Google Scholar] [CrossRef] [Green Version]
- Hyde, K.D.; Jeewon, R.; Chen, Y.J.; Bhunjun, C.S.; Calabon, M.S.; Jiang, H.B.; Lin, C.G.; Norphanphoun, C.; Sysouphanthong, P.; Pem, D.; et al. The numbers of fungi: Is the descriptive curve flattening? Fungal Divers. 2020, 103, 219–271. [Google Scholar] [CrossRef]




| Taxa | Strains | GenBank Accession Numbers | |||
|---|---|---|---|---|---|
| LSU | ITS | tef1 | rpb2 | ||
| Acanthonitschkea argentinensis | SMH1395 | AY695259 | - | FJ969042 | FJ968943 |
| Acanthonitschkea tristis | SMH4723 | FJ968949 | - | FJ969043 | - |
| Bertia didyma | SMH4719 | FJ968958 | - | - | - |
| Bertia ngongensis | GKM1239 * | FJ968954 | - | - | - |
| Bertia moriformis | SMH4320 | AY695260 | - | - | AY780151 |
| Bertia orbis | GKM1271 | FJ968955 | - | FJ969009 | - |
| Biciliospora velutina | GKM1268 | FJ968964 | - | FJ969018 | FJ968932 |
| Chaetosphaerella fusca | GKML124N | FJ968967 | - | FJ969002 | - |
| Chaetosphaerella phaeostroma | SMH4257 | AY695264 | - | FJ969004 | FJ968940 |
| Coronophora gregaria | ANM1555 | - | - | FJ969007 | FJ968938 |
| Coronophorella chaetomioides | GKM1099 | FJ968969 | - | FJ969034 | FJ968922 |
| Crassochaeta nigrita | SMH1667 | AY695265 | - | - | - |
| SMH2931 | AY695266 | - | - | - | |
| Emericellopsis alkalina | CBS 127350 * | MH875970 | MH864534 | KC998993 | KC999029 |
| Euacanthe usambarensis | GKM1221 | FJ968978 | - | FJ969026 | FJ968927 |
| SMH4408 | AY695267 | - | - | - | |
| Fracchiaea broomeana | SMH347 | FJ968979 | - | FJ969041 | FJ968947 |
| SMH2809 | AY695268 | - | FJ969039 | FJ968942 | |
| GKM1071 | - | - | FJ969040 | FJ968919 | |
| Fracchiaea myricoides | IFRD 9201 * | KX856174 | KX856173 | - | - |
| HKAS 115760 | MZ713199 | MZ713184 | MZ712579 | MZ712580 | |
| Fracchiaea lunata | GKM1089 | - | - | - | FJ968921 |
| Fracchiaea sp. | GKM1250 | - | - | FJ969005 | - |
| Gaillardiella pezizoides | GKM1245 | FJ968981 | - | FJ969006 | - |
| Melanospora tiffanii | ATCC 15515 | AY015630 | - | - | AY015637 |
| SMCD2222 | FJ748915 | FJ748921 | - | - | |
| Melanospora zamiae | ATCC 12340 | AY046579 | - | - | AY046580 |
| ATCC 96173 | AY057906 | - | - | - | |
| Neocryptosphaerella celata | GKM1231 * | FJ968975 | - | FJ969035 | FJ968929 |
| Neocryptosphaerella globosa | GKM471N * | FJ968977 | - | FJ969036 | FJ968935 |
| GKM437N | - | - | FJ969038 | - | |
| GKM414N | - | - | FJ969037 | - | |
| 396N | FJ968976 | - | - | - | |
| Neofracchiaea callista | SMH2689 | AY695269 | - | FJ969020 | FJ968941 |
| Nitschkia calyculus | SMH918 | FJ968983 | - | - | - |
| Nitschkia grevillei | SMH4663 | AY346294 | - | - | - |
| Nitschkia meniscoidea | SMH1523 * | AY695270 | - | - | - |
| Nitschkia tetraspora | GKML148N | FJ968987 | - | FJ969011 | FJ968936 |
| SMH4787 | FJ968984 | - | FJ969010 | - | |
| Parasympodiella elongata | CBS:522.93 * | GQ303314 | GQ303283 | - | - |
| CBS:124768 | GQ303311 | GQ303280 | - | - | |
| CPC:13288 | GQ303312 | GQ303281 | - | - | |
| CPC:13498 | GQ303313 | GQ303282 | - | - | |
| Parasympodiella eucalypti | CBS:124767 * | GQ303315 | GQ303284 | - | - |
| LCM 815.01 | - | MF495381 | - | - | |
| Parasympodiella hyphenata | CBS 583.82 * | MH873274 | MH861530 | - | - |
| Parasympodiella lauri | FMR_13132 * | KY853518 | KY853457 | - | - |
| Parasympodiella laxa | CBS 102698 | GQ303316 | GQ303285 | - | - |
| Parasympodiella longispora | CBS 544.84 * | MH873476 | MH861778 | - | - |
| KACC 41225 | - | GQ272636 | - | - | |
| Pseudocatenomycopsis rothmanniae | CPC 22733 * | KF777237 | KF777185 | - | - |
| Pseudocryptosphaerella costaricensis | MO2111 * | FJ968971 | - | FJ969028 | - |
| Pseudocryptosphaerella cylindriformis | GKM434N * | FJ968972 | - | FJ969031 | FJ968934 |
| GKM1187 | GQ217531 | - | FJ969033 | FJ968925 | |
| GKM1042 | FJ968973 | - | FJ969032 | FJ968918 | |
| GKM1075 | - | - | FJ969030 | FJ968920 | |
| Pseudocryptosphaerella elliptica | SMH4722 * | FJ968974 | - | FJ969029 | FJ968944 |
| Pseudocryptosphaerella malindensis | GKM1150 * | FJ968970 | - | FJ969027 | FJ968923 |
| Pseudohyaloseta pandanicola | MFLUCC 16-0316 * | MH376737 | MH388363 | MH388398 | MH412733 |
| Scortechinia acanthostroma | SMH1164 | FJ968989 | - | FJ969014 | FJ968924 |
| SMH1143 | FJ968988 | - | FJ969012 | FJ968948 | |
| GKML163N | FJ968991 | - | FJ969015 | - | |
| SMH5313 | FJ968990 | - | FJ969013 | - | |
| Scortechinia diminuspora | SMH4763 * | FJ968992 | - | - | - |
| HUEFS:194245 | KT003703 | - | - | - | |
| Scortechiniella similis | SMH2006 | FJ968994 | - | FJ969019 | FJ968945 |
| Scortechiniellopsis leonensis | GKM1269 | FJ968993 | - | FJ969021 | FJ968933 |
| Stachybotrys microspora | KLM 3-2 | KU760387 | KU760377 | KU760392 | KU760397 |
| Tympanopsis confertula | GKM1242 | FJ968997 | - | FJ969023 | FJ968930 |
| SMH1567 | FJ969001 | - | - | FJ968939 | |
| SMH4841 | FJ968998 | - | FJ969024 | FJ968946 | |
| SMH2648 | AY695272 | - | - | - | |
| Tympanopsis uniseriata | GKM1203 | FJ968999 | - | FJ969016 | FJ968926 |
| GKM1228 | FJ969000 | - | FJ969017 | - | |
| Yuxiensis granularis | HKAS 109580 * | MZ713198 | MZ713183 | MZ712577 | MZ712578 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bundhun, D.; Wanasinghe, D.N.; Maharachchikumbura, S.S.N.; Bhat, D.J.; Huang, S.-K.; Lumyong, S.; Mortimer, P.E.; Hyde, K.D. Yuxiensis granularis gen. et sp. nov., a Novel Quellkörper-Bearing Fungal Taxon Added to Scortechiniaceae and Inclusion of Parasympodiellaceae in Coronophorales Based on Phylogenetic Evidence. Life 2021, 11, 1011. https://doi.org/10.3390/life11101011
Bundhun D, Wanasinghe DN, Maharachchikumbura SSN, Bhat DJ, Huang S-K, Lumyong S, Mortimer PE, Hyde KD. Yuxiensis granularis gen. et sp. nov., a Novel Quellkörper-Bearing Fungal Taxon Added to Scortechiniaceae and Inclusion of Parasympodiellaceae in Coronophorales Based on Phylogenetic Evidence. Life. 2021; 11(10):1011. https://doi.org/10.3390/life11101011
Chicago/Turabian StyleBundhun, Digvijayini, Dhanushka N. Wanasinghe, Sajeewa S. N. Maharachchikumbura, Darbhe J. Bhat, Shi-Ke Huang, Saisamorn Lumyong, Peter E. Mortimer, and Kevin D. Hyde. 2021. "Yuxiensis granularis gen. et sp. nov., a Novel Quellkörper-Bearing Fungal Taxon Added to Scortechiniaceae and Inclusion of Parasympodiellaceae in Coronophorales Based on Phylogenetic Evidence" Life 11, no. 10: 1011. https://doi.org/10.3390/life11101011
APA StyleBundhun, D., Wanasinghe, D. N., Maharachchikumbura, S. S. N., Bhat, D. J., Huang, S.-K., Lumyong, S., Mortimer, P. E., & Hyde, K. D. (2021). Yuxiensis granularis gen. et sp. nov., a Novel Quellkörper-Bearing Fungal Taxon Added to Scortechiniaceae and Inclusion of Parasympodiellaceae in Coronophorales Based on Phylogenetic Evidence. Life, 11(10), 1011. https://doi.org/10.3390/life11101011








