Mechanotransduction in Prokaryotes: A Possible Mechanism of Spaceflight Adaptation
Abstract
1. Introduction
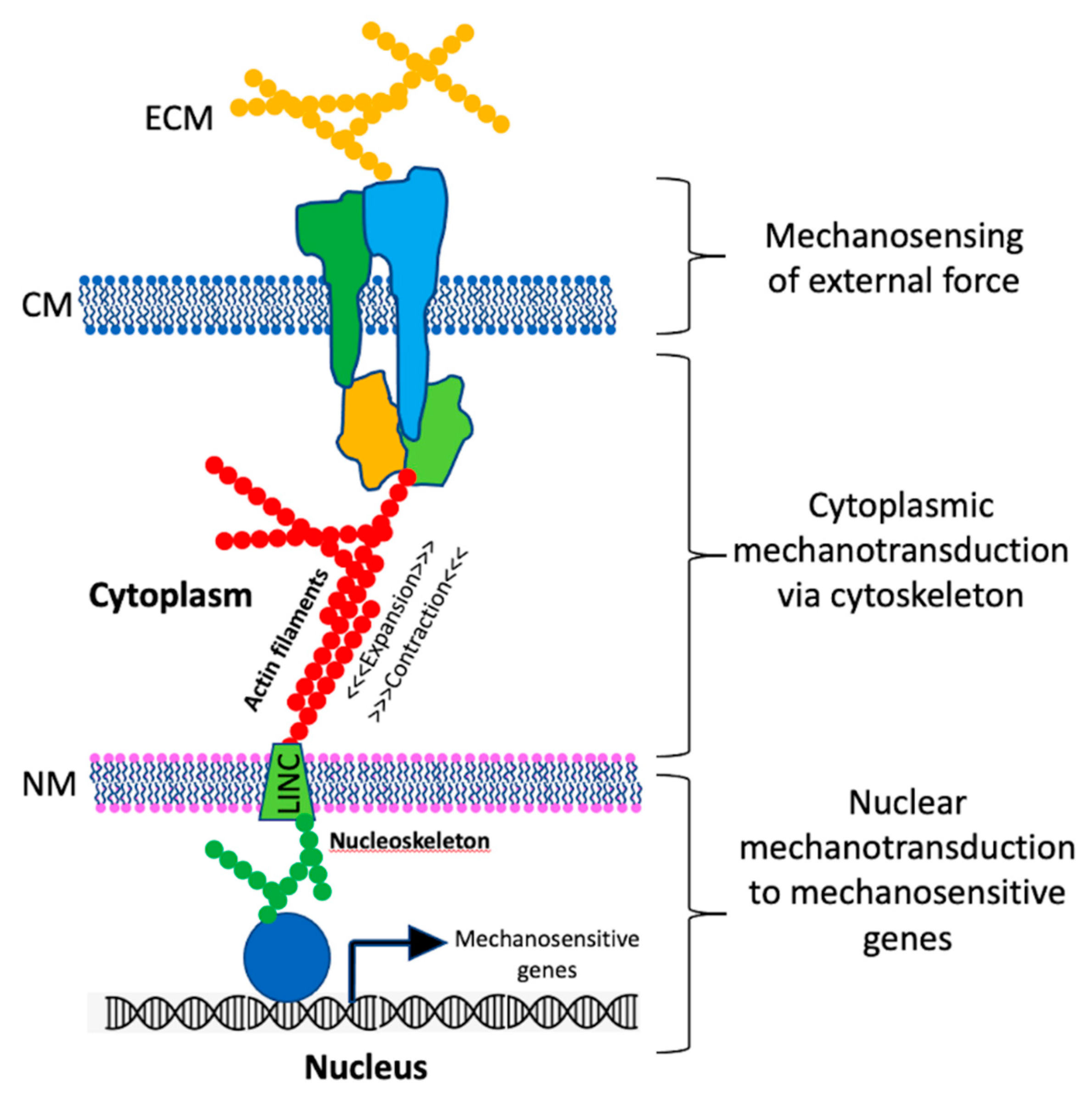
2. Mechanosensing in Eukaryotes
3. Does a Microbial Analog of Eukaryotic Mechanotransduction Exist?
3.1. Microbial Internal Structure
3.2. Mechanosensing in Prokaryotes
3.3. Mechanotransduction in Prokaryotes
3.3.1. Cell Growth and Division
3.3.2. DNA Division, Repair, and Gene Expression
3.4. Prokaryotic Nucleoid Architecture
3.5. DNA Supercoiling
3.6. Epigenetic Modification of DNA
4. Does Mechanotransduction Play a Role in the Prokaryotic Microgravity Response?
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Demets, R. Biology on sounding rockets: History, requirements, results, and scientific interpretation. In Proceedings of the 20th Symposium on European Rocket and Balloon Programmes and Related Research, Hyère, France, 22–26 May 2011; Ouwehand, L., Ed.; ESA Communications: Hyère, France, 2011; pp. 63–66. [Google Scholar]
- Pietsch, J.; Bauer, J.; Egli, M.; Infanger, M.; Wise, P.; Ulbrich, C.; Grimm, D. The effects of weightlessness on the human organism and mammalian cells. Curr. Mol. Med. 2011, 11, 350–364. [Google Scholar] [CrossRef] [PubMed]
- Lychakov, D.V. Behavioural and functional vestibular disorders after space flight: 2. Fish, amphibians and birds. J. Evol. Biochem. Physiol. 2016, 52, 1–16. [Google Scholar] [CrossRef]
- Vandenbrink, J.P.; Kiss, J.Z. Space, the final frontier: A critical review of recent experiments performed in microgravity. Plant. Sci. 2016, 243, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Hader, D.P.; Braun, M.; Grimm, D.; Hemmersbach, R. Gravireceptors in eukaryotes-a comparison of case studies on the cellular level. NPJ Microgravity 2017, 3, 8. [Google Scholar] [CrossRef]
- Rosenzweig, J.A.; Abogunde, O.; Thomas, K.; Lawal, A.; Nguyen, Y.U.; Sodipe, A.; Jejelowo, O. Spaceflight and modeled microgravity effects on microbial growth and virulence. Appl. Microbiol. Biotechnol. 2010, 85, 885–891. [Google Scholar] [CrossRef]
- Rosenzweig, J.A.; Ahmed, S.; Eunson, J.; Chopra, A.K. Low-shear force associated with modeled microgravity and spaceflight does not similarly impact the virulence of notable bacterial pathogens. Appl. Microbiol. Biotechnol. 2014. [Google Scholar] [CrossRef]
- Taylor, P.W. Impact of space flight on bacterial virulence and antibiotic susceptibility. Infect. Drug Resist. 2015, 8, 249–262. [Google Scholar] [CrossRef]
- Nickerson, C.A.; Ott, C.M.; Wilson, J.W.; Ramamurthy, R.; Pierson, D.L. Microbial responses to microgravity and other low-shear environments. Microbiol. Mol. Biol. Rev. 2004, 68, 345–361. [Google Scholar] [CrossRef]
- Najrana, T.; Sanchez-Esteban, J. Mechanotransduction as an adaptation to gravity. Front. Pediatr 2016, 4, 140. [Google Scholar] [CrossRef]
- DuFort, C.C.; Paszek, M.J.; Weaver, V.M. Balancing forces: Architectural control of mechanotransduction. Nat. Rev. Mol. Cell Biology 2011, 12, 308–319. [Google Scholar] [CrossRef]
- Luo, T.; Mohan, K.; Iglesias, P.A.; Robinson, D.N. Molecular mechanisms of cellular mechanosensing. Nat. Mater. 2013, 12, 1064–1071. [Google Scholar] [CrossRef] [PubMed]
- Moulia, B.; Coutand, C.; Julien, J.L. Mechanosensitive control of plant growth: Bearing the load, sensing, transducing, and responding. Front. Plant. Sci. 2015, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Lindstrom, J.B.; Pierce, N.T.; Latz, M.I. Role of trp channels in dinoflagellate mechanotransduction. Biol. Bull. 2017, 233, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Martino, F.; Perestrelo, A.R.; Vinarský, V.; Pagliari, S.; Forte, G. Cellular mechanotransduction: From tension to function. Front. Physiol. 2018, 9, 824. [Google Scholar] [CrossRef] [PubMed]
- Sankaran, J.; Uzer, G.; van Wijnen, A.J.; Rubin, J. Gene regulation through dynamic actin control of nuclear structure. Exp. Biol. Med. 2019, 244, 1345–1353. [Google Scholar] [CrossRef] [PubMed]
- Piccus, R.; Brayson, D. The nuclear envelope: Lincing tissue mechanics to genome regulation in cardiac and skeletal muscle. Biol. Lett. 2020, 16, 9. [Google Scholar] [CrossRef] [PubMed]
- Haswell, E.S.; Phillips, R.; Rees, D.C. Mechanosensitive channels: What can they do and how do they do it? Structure 2011, 19, 1356–1369. [Google Scholar] [CrossRef]
- Aventaggiato, M.; Barreca, F.; Vernucci, E.; Bizzarri, M.; Ferretti, E.; Russo, M.A.; Tafani, M. Putative receptors for gravity sensing in mammalian cells: The effects of microgravity. Appl. Sci. 2020, 10, 28. [Google Scholar] [CrossRef]
- Bradbury, P.; Wu, H.; Choi, J.U.; Rowan, A.E.; Zhang, H.; Poole, K.; Lauko, J.; Chou, J. Modeling the impact of microgravity at the cellular level: Implications for human disease. Front. Cell Dev. Biol. 2020, 8, 96. [Google Scholar] [CrossRef]
- Uhler, C.; Shivashankar, G.V. Regulation of genome organization and gene expression by nuclear mechanotransduction. Nat. Rev. Molec. Cell Biol. 2017, 18, 717–727. [Google Scholar] [CrossRef]
- Paul, A.L.; Zupanska, A.K.; Schultz, E.R.; Ferl, R.J. Organ-specific remodeling of the Arabidopsis transcriptome in response to spaceflight. BMC Plant Biol. 2013, 13, 10. [Google Scholar] [CrossRef] [PubMed]
- Vorselen, D.; Roos, W.H.; MacKintosh, F.C.; Wuite, G.J.L.; van Loon, J. The role of the cytoskeleton in sensing changes in gravity by nonspecialized cells. FASEB J. 2014, 28, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Thiel, C.S.; Tauber, S.; Christoffel, S.; Huge, A.; Lauber, B.A.; Polzer, J.; Paulsen, K.; Lier, H.; Engelmann, F.; Schmitz, B.; et al. Rapid coupling between gravitational forces and the transcriptome in human myelomonocytic U937 cells. Sci. Rep. 2018, 8, 13267. [Google Scholar] [CrossRef] [PubMed]
- Pollard, E.C. Theoretical studies on living systems in the absence of mechanical stress. J. Theor. Biol. 1965, 8, 113–123. [Google Scholar] [CrossRef]
- Horneck, G.; Klaus, D.M.; Mancinelli, R.L. Space microbiology. Microbiol. Mol. Biol. Rev. 2010, 74, 121–156. [Google Scholar] [CrossRef]
- Anken, R. Simulation of microgravity for studies in gravitational biology: Principles, devices, and applications. Curr. Biotechnol. 2013, 2, 192–200. [Google Scholar] [CrossRef]
- Herranz, R.; Anken, R.; Boonstra, J.; Braun, M.; Christianen, P.C.; de Geest, M.; Hauslage, J.; Hilbig, R.; Hill, R.J.; Lebert, M.; et al. Ground-based facilities for simulation of microgravity: Organism-specific recommendations for their use, and recommended terminology. Astrobiology 2013, 13, 1–17. [Google Scholar] [CrossRef]
- Fang, A.; Pierson, D.L.; Koenig, D.W.; Mishra, S.K.; Demain, A.L. Effect of simulated microgravity and shear stress on microcin B17 production by Escherichia coli and on its excretion into the medium. Appl. Environ. Microbiol. 1997, 63, 4090–4092. [Google Scholar] [CrossRef]
- Nickerson, C.A.; Ott, C.M.; Mister, S.J.; Morrow, B.J.; Burns-Keliher, L.; Pierson, D.L. Microgravity as a novel environmental signal affecting Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 2000, 68, 3147–3152. [Google Scholar] [CrossRef]
- Crabbé, A.; Nielsen-Preiss, S.M.; Woolley, C.M.; Barrila, J.; Buchanan, K.; McCracken, J.; Inglis, D.O.; Searles, S.C.; Nelman-Gonzalez, M.A.; Ott, C.M.; et al. Spaceflight enhances cell aggregation and random budding in Candida albicans. PLoS ONE 2013, 8, e80677. [Google Scholar] [CrossRef]
- Aunins, T.R.; Erickson, K.E.; Prasad, N.; Levy, S.E.; Jones, A.; Shrestha, S.; Mastracchio, R.; Stodieck, L.; Klaus, D.; Zea, L.; et al. Spaceflight modifies Escherichia coli gene expression in response to antibioticc exposure and reveals role of oxidative stress response. Front. Microbiol. 2018, 9, 310. [Google Scholar] [CrossRef] [PubMed]
- Crabbé, A.; Schurr, M.J.; Monsieurs, P.; Morici, L.; Schurr, J.; Wilson, J.W.; Ott, C.M.; Tsaprailis, G.; Pierson, D.L.; Stefanyshyn-Piper, H.; et al. Transcriptional and proteomic responses of Pseudomonas aeruginosa PAO1 to spaceflight conditions involve Hfq regulation and reveal a role for oxygen. Appl. Environ. Microbiol. 2011, 77, 1221–1230. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.W.; Ott, C.M.; Bentrup, K.H.Z.; Ramamurthy, R.; Quick, L.; Porwollik, S.; Cheng, P.; McClelland, M.; Tsaprailis, G.; Radabaugh, T.; et al. Space flight alters bacterial gene expression and virulence and reveals a role for global regulator Hfq. Proc. Natl. Acad. Sci. USA 2007, 104, 16299–16304. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, F.; Wang, Q.; Ge, P.; Woo, P.C.; Yan, J.; Zhao, Y.; Gao, G.F.; Liu, C.H.; Liu, C. Genomic and transcriptomic analysis of NDM-1 Klebsiella pneumoniae in spaceflight reveal mechanisms underlying environmental adaptability. Sci. Rep. 2014, 4, 6216. [Google Scholar] [CrossRef] [PubMed]
- Crabbé, A.; Pycke, B.; Van Houdt, R.; Monsieurs, P.; Nickerson, C.; Leys, N.; Cornelis, P. Response of Pseudomonas aeruginosa PAO1 to low shear modelled microgravity involves AlgU regulation. Environ. Microbiol. 2010, 12, 1545–1564. [Google Scholar] [PubMed]
- Mastroleo, F.; Van Houdt, R.; Leroy, B.; Benotmane, M.A.; Janssen, A.; Mergeay, M.; Vanhavere, F.; Hendrickx, L.; Wattiez, R.; Leys, N. Experimental design and environmental parameters affect Rhodospirillum rubrum S1H response to space flight. ISME J. 2009, 3, 1402–1419. [Google Scholar] [CrossRef] [PubMed]
- Morrison, M.D.; Fajardo-Cavazos, P.; Nicholson, W.L. Comparison of Bacillus subtilis transcriptome profiles from two separate missions to the International Space Station. NPJ Microgravity 2019, 5, 1. [Google Scholar] [CrossRef]
- Wilson, J.W.; Ott, C.M.; Quick, L.; Davis, R.; Bentrup, K.H.Z.; Crabbé, A.; Richter, E.; Sarker, S.; Barrila, J.; Porwollik, S.; et al. Media ion composition controls regulatory and virulence response of Salmonella in spaceflight. PLoS ONE 2008, 3, e3923. [Google Scholar] [CrossRef] [PubMed]
- Morrison, M.D.; Nicholson, W.L. Meta-analysis of data from spaceflight transcriptome experiments does not support the idea of a common bacterial “spaceflight response”. Sci. Rep. 2018, 8, 14403. [Google Scholar] [CrossRef]
- Morrison, M.D.; Nicholson, W.L. Comparisons of transcriptome profiles from Bacillus subtilis cells grown in space versus High Aspect Ratio Vessel (HARV) clinostats reveal a low degree of concordance. Astrobiology 2020, 20, 1498–1509. [Google Scholar] [CrossRef]
- Kentner, D.; Sourjik, V. Use of fluorescence microscopy to study intracellular signaling in bacteria. In Annual Review of Microbiology; Gottesman, S., Harwood, C.S., Eds.; Palo Alto: Santa Clara, CA, USA, 2010; Volume 64, pp. 373–390. [Google Scholar]
- Guerrero, R.; Berlanga, M. The hidden side of the prokaryotic cell: Rediscovering the microbial world. Int. Microbiol. 2007, 10, 157–168. [Google Scholar] [PubMed]
- Saier, M.H. Microcompartments and protein machines in prokaryotes. J. Mol. Microbiol. Biotechnol. 2013, 23, 243–269. [Google Scholar] [CrossRef]
- Dufrene, Y.F.; Persat, A. Mechanomicrobiology: How bacteria sense and respond to forces. Nat. Rev. Microbiol. 2020, 14, 227–240. [Google Scholar] [CrossRef] [PubMed]
- Persat, A. Bacterial mechanotransduction. Curr. Opin. Microbiol. 2017, 36, 1–6. [Google Scholar] [CrossRef]
- Rudner, D.Z.; Losick, R. Protein subcellular localization in bacteria. Cold Spring Harbor Perspect. Biol. 2010, 2, 14. [Google Scholar] [CrossRef] [PubMed]
- Wickstead, B.; Gull, K. The evolution of the cytoskeleton. J. Cell Biol. 2011, 194, 513–525. [Google Scholar] [CrossRef]
- Ghosal, D.; Löwe, J. Collaborative protein filaments. EMBO J. 2015, 34, 2312–2320. [Google Scholar] [CrossRef]
- Wagstaff, J.; Löwe, J. Prokaryotic cytoskeletons: Protein filaments organizing small cells. Nat. Rev. Microbiol. 2018, 16, 187–201. [Google Scholar] [CrossRef]
- Celler, K.; Koning, R.I.; Koster, A.J.; van Wezel, G.P. Multidimensional view of the bacterial cytoskeleton. J. Bacteriol. 2013, 195, 1627–1636. [Google Scholar] [CrossRef]
- Schaechter, M. A brief history of bacterial growth physiology. Front. Microbiol. 2015, 6, 5. [Google Scholar] [CrossRef]
- Booth, I.R. Bacterial mechanosensitive channels: Progress towards an understanding of their roles in cell physiology. Curr. Opin. Microbiol. 2014, 18, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.M.; Guttenplan, S.B.; Kearns, D.B. Defects in the flagellar motor increase synthesis of poly-gamma-glutamate in Bacillus subtilis. J. Bacteriol. 2014, 196, 740–753. [Google Scholar] [CrossRef] [PubMed]
- Cairns, L.S.; Marlow, V.L.; Bissett, E.; Ostrowski, A.; Stanley-Wall, N.R. A mechanical signal transmitted by the flagellum controls signalling in Bacillus subtilis. Mol. Microbiol. 2013, 90, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Egan, A.J.F.; Errington, J.; Vollmer, W. Regulation of peptidoglycan synthesis and remodelling. Nat. Rev. Microbiol. 2020, 18, 446–460. [Google Scholar] [CrossRef] [PubMed]
- Thanbichler, M.; Shapiro, L. Chromosome organization and segregation in bacteria. J. Struct. Biol. 2006, 156, 292–303. [Google Scholar] [CrossRef]
- Wang, X.D.; Rudner, D.Z. Spatial organization of bacterial chromosomes. Curr. Opin. Microbiol. 2014, 22, 66–72. [Google Scholar] [CrossRef]
- Le, T.B.K.; Imakaev, M.V.; Mirny, L.A.; Laub, M.T. High-resolution mapping of the spatial organization of a bacterial chromosome. Science 2013, 342, 731–734. [Google Scholar] [CrossRef]
- Lioy, V.S.; Cournac, A.; Marbouty, M.; Duigou, S.; Mozziconacci, J.; Espéli, O.; Boccard, F.; Koszul, R. Multiscale structuring of the E. coli chromosome by nucleoid-associated and condensin proteins. Cell 2018, 172, 771–783.e718. [Google Scholar] [CrossRef]
- Marbouty, M.; Le Gall, A.; Cattoni, D.I.; Cournac, A.; Koh, A.; Fiche, J.B.; Mozziconacci, J.; Murray, H.; Koszul, R.; Nollmann, M. Condensin- and replication-mediated bacterial chromosome folding and origin condensation revealed by Hi-C and super-resolution imaging. Mol. Cell 2015, 59, 588–602. [Google Scholar] [CrossRef]
- Wang, X.D.; Le, T.B.K.; Lajoie, B.R.; Dekker, J.; Laub, M.T.; Rudner, D.Z. Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes Dev. 2015, 29, 1661–1675. [Google Scholar] [CrossRef]
- Wang, X.D.; Brandao, H.B.; Le, T.B.K.; Laub, M.T.; Rudner, D.Z. Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science 2017, 355, 524–527. [Google Scholar] [CrossRef] [PubMed]
- Benza, V.G.; Bassetti, B.; Dorfman, K.D.; Scolari, V.F.; Bromek, K.; Cicuta, P.; Lagomarsino, M.C. Physical descriptions of the bacterial nucleoid at large scales, and their biological implications. Rep. Prog. Phys. 2012, 75, 20. [Google Scholar] [CrossRef] [PubMed]
- Dorman, C.J. DNA supercoiling and transcription in bacteria: A two-way street. BMC Mol. Cell Biol. 2019, 20, 9. [Google Scholar] [CrossRef]
- Dorman, C.J.; Dorman, M.J. DNA supercoiling is a fundamental regulatory principle in the control of bacterial gene expression. Biophys. Rev. 2016, 8, 209–220. [Google Scholar] [CrossRef] [PubMed]
- de la Campa, A.G.; Ferrándiz, M.J.; Martín-Galiano, A.J.; García, M.T.; Tirado-Vélez, J.M. The transcriptome of Streptococcus pneumoniae induced by local and global changes in supercoiling. Front. Microbiol. 2017, 8, 1447. [Google Scholar] [CrossRef] [PubMed]
- Le, T.B.K.; Laub, M.T. Transcription rate and transcript length drive formation of chromosomal interaction domain boundaries. EMBO J. 2016, 35, 1582–1595. [Google Scholar] [CrossRef]
- Atack, J.M.; Tan, A.; Bakaletz, L.O.; Jennings, M.P.; Seib, K.L. Phasevarions of bacterial pathogens: Methylomics sheds new light on old enemies. Trends Microbiol. 2018, 26, 715–726. [Google Scholar] [CrossRef]
- Sanchez-Romero, M.A.; Cota, I.; Casadesus, J. DNA methylation in bacteria: From the methyl group to the methylome. Curr. Opin. Microbiol. 2015, 25, 9–16. [Google Scholar] [CrossRef]
- Sanchez-Romero, M.A.; Casadesús, J. The bacterial epigenome. Nat. Rev. Microbiol. 2020, 18, 7–20. [Google Scholar] [CrossRef]
- Gonzalez, D.; Kozdon, J.B.; McAdams, H.H.; Shapiro, L.; Collier, J. The functions of DNA methylation by CcrM in Caulobacter crescentus: A global approach. Nucleic Acids Res. 2014, 42, 3720–3735. [Google Scholar] [CrossRef]
- Cota, I.; Bunk, B.; Spröer, C.; Overmann, J.; König, C.; Casadesús, J. OxyR-dependent formation of DNA methylation patterns in OpvABOFF and OpvABON cell lineages of Salmonella enterica. Nucleic Acids Res. 2016, 44, 3595–3609. [Google Scholar] [CrossRef] [PubMed]
- Camacho, E.M.; Casadesús, J. Regulation of traJ transcription in the Salmonella virulence plasmid by strand-specific DNA adenine hemimethylation. Mol. Microbiol. 2005, 57, 1700–1718. [Google Scholar] [CrossRef] [PubMed]
- Fioravanti, A.; Fumeaux, C.; Mohapatra, S.S.; Bompard, C.; Brilli, M.; Frandi, A.; Castric, V.; Villeret, V.; Viollier, P.H.; Biondi, E.G. DNA binding of the cell cycle transcriptional regulator gcrA depends on N6-adenosine methylation in Caulobacter crescentus and other Alphaproteobacteria. PLoS Genet. 2013, 9, e1003541. [Google Scholar] [CrossRef] [PubMed]
- Broadbent, S.E.; Davies, M.R.; van der Woude, M.W. Phase variation controls expression of Salmonella lipopolysaccharide modification genes by a DNA methylation-dependent mechanism. Mol. Microbiol. 2010, 77, 337–353. [Google Scholar] [CrossRef]
- Nou, X.; Skinner, B.; Braaten, B.; Blyn, L.; Hirsch, D.; Low, D. Regulation of pyelonephritis-associated pili phase-variation in Escherichia coli: Binding of the PapI and the Lrp regulatory proteins is controlled by DNA methylation. Mol. Microbiol. 1993, 7, 545–553. [Google Scholar] [CrossRef]
- Sánchez-Romero, M.A.; Busby, S.J.; Dyer, N.P.; Ott, S.; Millard, A.D.; Grainger, D.C. Dynamic distribution of SeqA protein across the chromosome of Escherichia coli K-12. MBio 2010, 1. [Google Scholar] [CrossRef]
- Waldminghaus, T.; Weigel, C.; Skarstad, K. Replication fork movement and methylation govern SeqA binding to the Escherichia coli chromosome. Nucleic Acids Res. 2012, 40, 5465–5476. [Google Scholar] [CrossRef]
| Eukaryotic Cytoskeletal Structure | Eukaryotic Subunit Analogs | Prokaryotic Subunit Analogs | Function in Prokaryotes |
|---|---|---|---|
| Microtubules | α-Tubulin, β-Tubulin | FtsZ | Cytokinesis |
| TubZ | DNA positioning | ||
| RepX | Plasmid replication | ||
| Microfilaments | Actin Superfamily | MreB | Cell shape, chromosome segregation |
| FtsA | Cytokinesis | ||
| ParM | DNA segregation | ||
| Intermediate Filaments | Keratins, Vimentin, Desmin, Neurofilament Proteins, Lamins | CreS (crescentin) | Cell shape |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fajardo-Cavazos, P.; Nicholson, W.L. Mechanotransduction in Prokaryotes: A Possible Mechanism of Spaceflight Adaptation. Life 2021, 11, 33. https://doi.org/10.3390/life11010033
Fajardo-Cavazos P, Nicholson WL. Mechanotransduction in Prokaryotes: A Possible Mechanism of Spaceflight Adaptation. Life. 2021; 11(1):33. https://doi.org/10.3390/life11010033
Chicago/Turabian StyleFajardo-Cavazos, Patricia, and Wayne L. Nicholson. 2021. "Mechanotransduction in Prokaryotes: A Possible Mechanism of Spaceflight Adaptation" Life 11, no. 1: 33. https://doi.org/10.3390/life11010033
APA StyleFajardo-Cavazos, P., & Nicholson, W. L. (2021). Mechanotransduction in Prokaryotes: A Possible Mechanism of Spaceflight Adaptation. Life, 11(1), 33. https://doi.org/10.3390/life11010033




