Optimization of Molecular Dynamics Simulations of c-MYC1-88—An Intrinsically Disordered System
Abstract
1. Introduction
1.1. Optimized Force Fields for IDP Simulations
1.2. Optimizing Explicit and Implicit Water Models
1.3. Assessing Simulation Convergence
2. Materials and Methods
2.1. MD Set-up
2.2. Trajectory Analysis
2.3. Experimental Data
2.4. Markov Chain Monte Carlo
3. Results
3.1. Histatin 5
3.2. c-MYC1-88
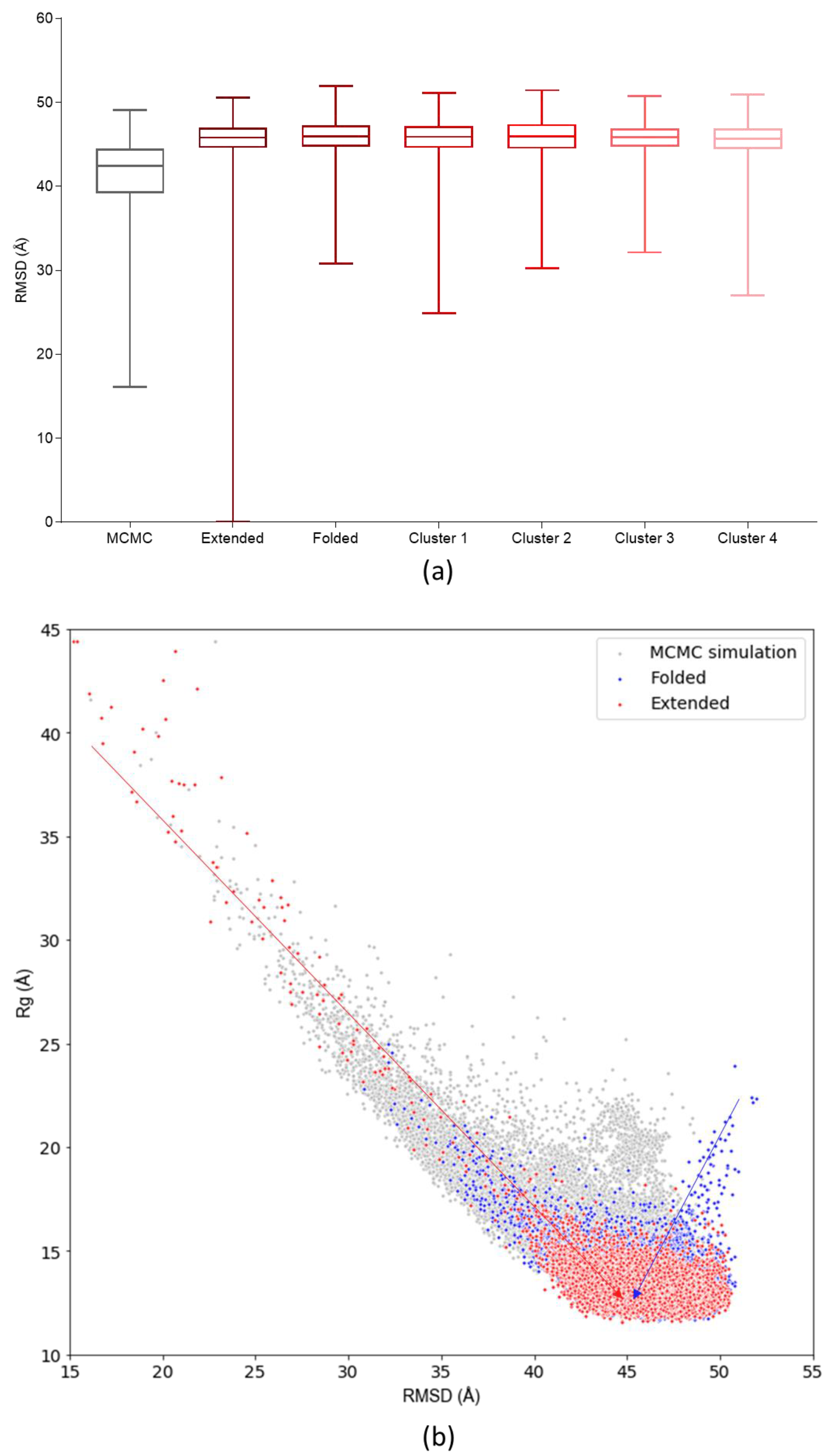
3.3. Assessing Convergence
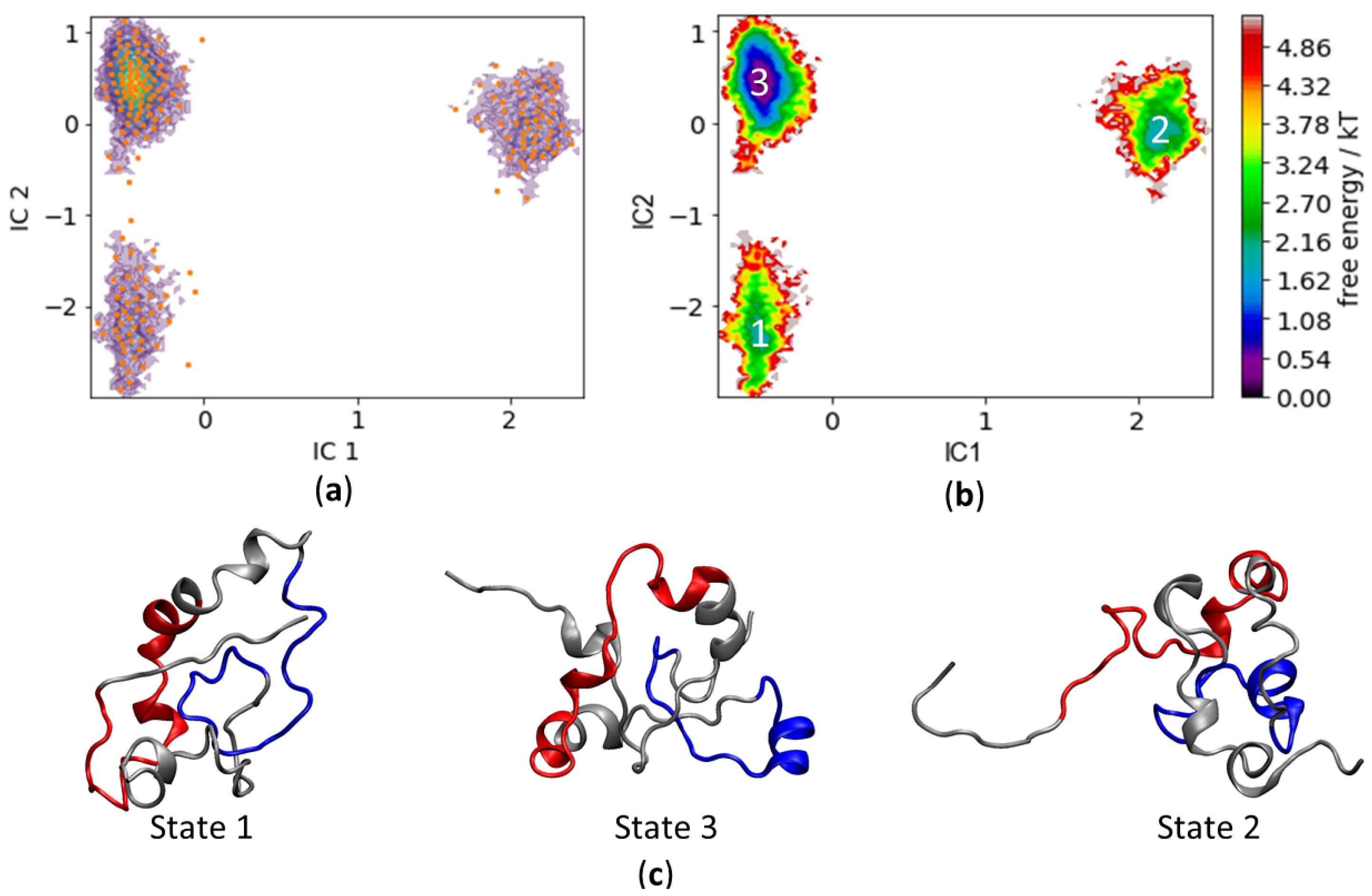
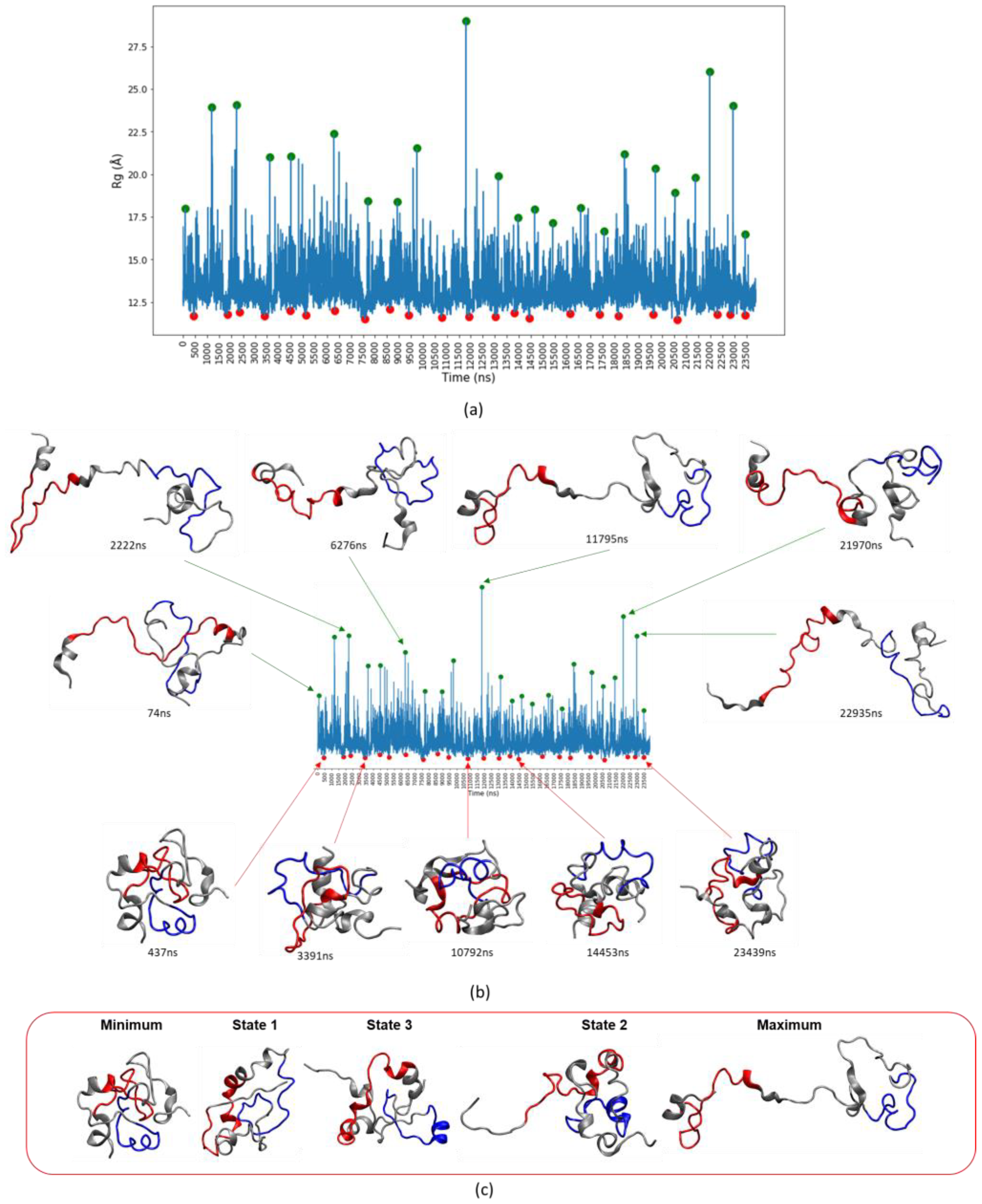
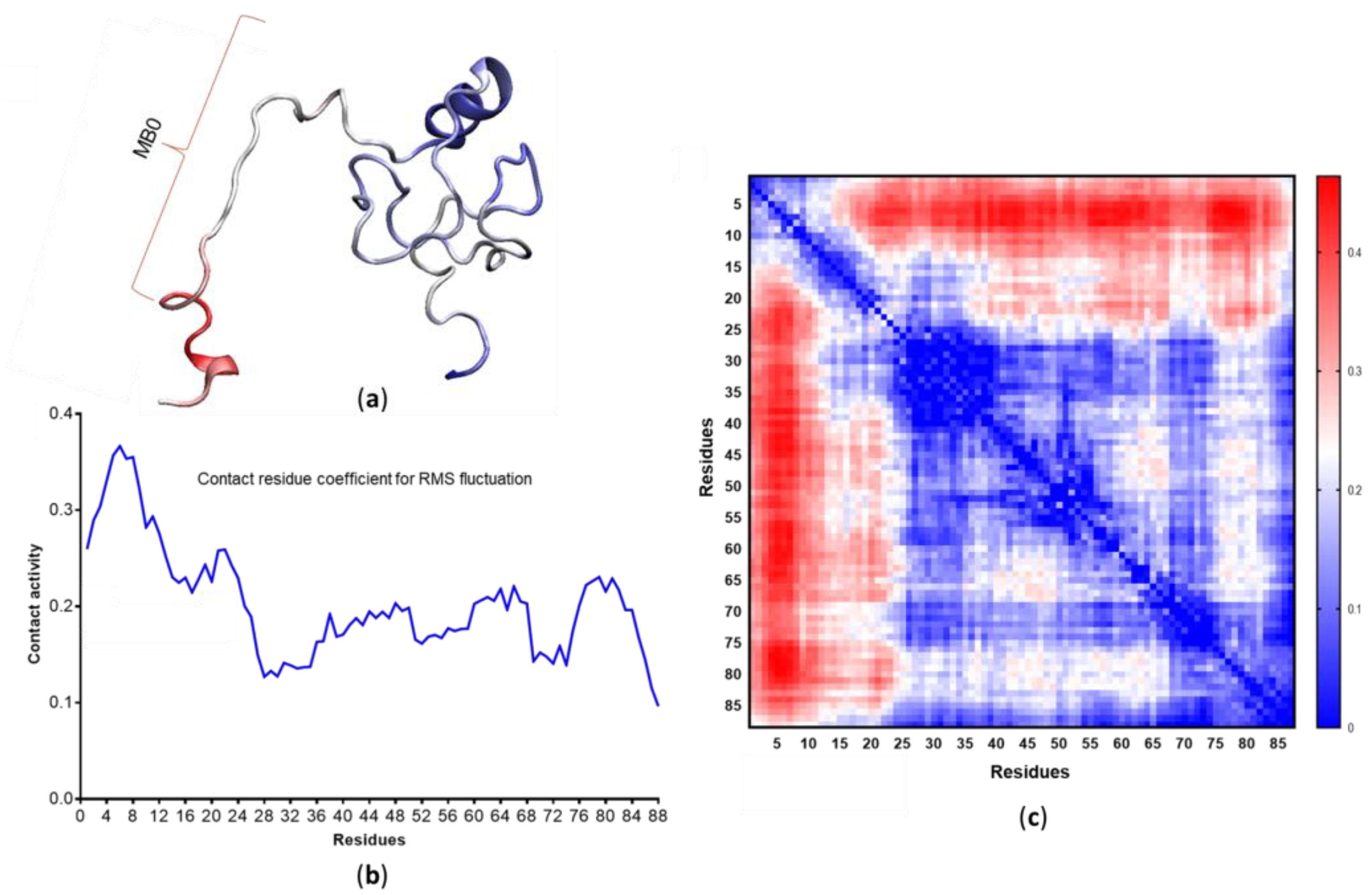
3.4. c-MYC1-88 Trajectory Analysis and Structural Insights
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Levine, Z.A.; Shea, J. Simulations of disordered proteins and systems with conformational heterogeneity. Curr. Opin. Struct. Biol. 2017, 43, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Babu, M.M. The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease. Biochem. Soc. Trans. 2016, 44, 1185–1200. [Google Scholar] [CrossRef] [PubMed]
- Salomon-Ferrer, R.; Gotz, A.W.; Poole, D.; Grand, S.L.; Walker, R.C. Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput. 2013, 9, 3878–3888. [Google Scholar] [CrossRef] [PubMed]
- Stone, J.E.; Hallock, M.J.; Phillips, J.C.; Peterson, J.R.; Schulten, Z.L.; Schulten, K. Evaluation of emerging energy-efficient heterogeneous computing platforms for biomolecular and cellular simulation workloads. In Proceedings of the 2016 IEEE International Parallel and Distributed Processing Symposium Workshops (IPDPSW), Chicago, IL, USA, 23–27 May 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 89–100. [Google Scholar]
- Hollingsworth, S.A.; Dror, R.O. Molecular dynamics simulation for all. Neuron 2018, 99, 1129–1143. [Google Scholar] [CrossRef]
- Beauchamp, K.A.; Lin, Y.-S.; Das, R.; Pande, V.S. Are Protein Force Fields Getting Better? A Systematic Benchmark on 524 Diverse NMR Measurements. J. Chem. Theory Comput. 2012, 8, 1409–1414. [Google Scholar] [CrossRef]
- Chong, S.-H.; Chatterjee, P.; Ham, S. Computer Simulations of Intrinsically Disordered Proteins. Annu. Rev. Phys. Chem. 2017, 68, 117–134. [Google Scholar] [CrossRef]
- Rauscher, S.; Gapsys, V.; Gajda, M.J.; Zweckstetter, M.; De Groot, B.L.; Grubmüller, H. Structural Ensembles of Intrinsically Disordered Proteins Depend Strongly on Force Field: A Comparison to Experiment. J. Chem. Theory Comput. 2015, 11, 5513–5524. [Google Scholar] [CrossRef]
- Henriques, J.; Cragnell, C.; Skepo, M. Molecular dynamics simulations of intrinsically disordered proteins: Force field evaluation and comparison with experiment. J. Chem. Theory Comput. 2015, 11, 3420–3431. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; De Groot, B.L.; Grubmüller, H.; Mackerell, A.D., Jr. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef]
- Piana, S.; Klepeis, J.L.; Shaw, D.E. Assessing the accuracy of physical models used in protein-folding simulations: Quantitative evidence from long molecular dynamics simulations. Curr. Opin. Struct. Biol. 2014, 24, 98–105. [Google Scholar] [CrossRef]
- Best, R.B. Computational and theoretical advances in studies of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 2017, 42, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Piana, S.; Donchev, A.G.; Robustelli, P.; Shaw, D.E. Water Dispersion Interactions Strongly Influence Simulated Structural Properties of Disordered Protein States. J. Phys. Chem. B 2015, 119, 5113–5123. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Wang, W.; Ye, W.; Ji, D.; Luo, R.; Chen, H.-F. ff14IDPs force field improving the conformation sampling of intrinsically disordered proteins. Chem. Biol. Drug Des. 2017, 89, 5–15. [Google Scholar] [CrossRef]
- Song, D.; Luo, R.; Chen, H. The IDP-Specific Force Field ff14IDPSFF Improves the Conformer Sampling of Intrinsically Disordered Proteins. J. Chem. Inf. Modeling 2017, 57, 1166–1178. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Guo, X.; Han, J.; Luo, R.; Chen, H.-F. Order-disorder transition of intrinsically disordered kinase inducible transactivation domain of CREB. J. Chem. Phys. 2018, 148, 225101. [Google Scholar] [CrossRef]
- Mark, P.; Nilsson, L. Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Henriques, J.; Skepo, M. Molecular Dynamics Simulations of Intrinsically Disordered Proteins: On the Accuracy of the TIP4P-D Water Model and the Representativeness of Protein Disorder Models. J. Chem. Theory Comput. 2016, 12, 3407–3415. [Google Scholar] [CrossRef]
- Onufriev, A.V.; Case, D.A. Generalized Born Implicit Solvent Models for Biomolecules. Annu. Rev. Biophys. 2019, 48, 275–296. [Google Scholar] [CrossRef]
- Sawle, L.; Ghosh, K. Convergence of Molecular Dynamics Simulation of Protein Native States: Feasibility vs Self-Consistency Dilemma. J. Chem. Theory Comput. 2016, 12, 861–869. [Google Scholar] [CrossRef]
- Grossfield, A.; Zuckerman, D.M. Quantifying uncertainty and sampling quality in biomolecular simulations. In Annual Reports in Computational Chemistry; Elsevier Science & Technology: Amsterdam, The Netherlands, 2009; Chapter 2; pp. 23–48. [Google Scholar]
- Smith, L.J.; Daura, X.; van Gunsteren, W.F. Assessing equilibration and convergence in biomolecular simulations. Proteins Struct. Funct. Bioinform. 2002, 48, 487–496. [Google Scholar] [CrossRef]
- Lyman, E.; Zuckerman, D.M. Ensemble-Based Convergence Analysis of Biomolecular Trajectories. Biophys. J. 2006, 91, 164–172. [Google Scholar] [CrossRef]
- Hess, B. Similarities between principal components of protein dynamics and random diffusion. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Top. 2000, 62, 8438–8448. [Google Scholar] [CrossRef]
- Hess, B. Convergence of sampling in protein simulations. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2002, 65, 031910. [Google Scholar] [CrossRef] [PubMed]
- Knapp, B.; Frantal, S.; Cibena, M.; Schreiner, W.; Bauer, P. Is an Intuitive Convergence Definition of Molecular Dynamics Simulations Solely Based on the Root Mean Square Deviation Possible? J. Comput. Biol. 2011, 18, 997–1005. [Google Scholar] [CrossRef]
- van Ravenzwaaij, D.; Cassey, P.; Brown, S.D. A simple introduction to Markov Chain Monte-Carlo sampling. Psychon. Bull. Rev. 2018, 25, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Boomsma, W.; Frellsen, J.; Harder, T.; Bottaro, S.; Johansson, K.E.; Tian, P.; Stovgaard, K.; Andreetta, C.; Olsson, S.; Valentin, J.B.; et al. PHAISTOS: A framework for Markov chain Monte Carlo simulation and inference of protein structure. J. Comput. Chem. 2013, 34, 1697–1705. [Google Scholar] [CrossRef]
- Case, D.A. AMBER 2016 Reference Manual; University of California: San Francisco, CA, USA, 2016; pp. 1–923. [Google Scholar]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- The PyMOL Molecular Graphics System, version 1.8.6.0, Computer Software Reference; Schrodinger, LLC: New York, NY, USA, 2010.
- Xu, D.; Zhang, Y. Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins Struct. Funct. Bioinform. 2012, 80, 1715–1735. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Zhang, Y. Toward optimal fragment generations for ab initio protein structure assembly. Proteins Struct. Funct. Bioinform. 2013, 81, 229–239. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Rambo, R. “Scatter”. The SIBYLS Beamline, Version 3.0, Computer Software Reference, 2017. Available online: https://bl1231.als.lbl.gov/scatter/ (accessed on 20 June 2020).
- Petoukhov, M.V.; Franke, D.; Shkumatov, A.V.; Tria, G.; Kikhney, A.G.; Gajda, M.; Gorba, C.; Mertens, H.D.T.; Konarev, P.V.; Svergun, D.I. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012, 45, 342–350. [Google Scholar] [CrossRef]
- Konarev, P.V.; Volkov, V.; Sokolova, A.; Koch, M.H.J.; Svergun, D.I. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003, 36, 1277–1282. [Google Scholar] [CrossRef]
- Svergun, D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992, 25, 495–503. [Google Scholar] [CrossRef]
- Shen, Y.; Bax, A. SPARTA+: A modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J. Biomol. NMR 2010, 48, 13–22. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Tribello, G.A.; Bonomi, M.; Branduardi, D.; Camilloni, C.; Bussi, G. PLUMED 2: New feathers for an old bird. Comput. Phys. Commun. 2014, 185, 604–613. [Google Scholar] [CrossRef]
- Pietrucci, F.; Laio, A. A Collective Variable for the Efficient Exploration of Protein Beta-Sheet Structures: Application to SH3 and GB1. J. Chem. Theory Comput. 2009, 5, 2197–2201. [Google Scholar] [CrossRef] [PubMed]
- Grant, B.J.; Rodrigues, A.P.C.; Sawy, K.M.E.; Cammon, J.A.M.; Caves, L. Bio3d: An R package for the comparative analysis of protein structures. Bioinformatics 2006, 22, 2695–2696. [Google Scholar] [CrossRef]
- Scherer, M.K.; Trendelkamp-Schroer, B.; Paul, F.; Pérez-Hernández, G.; Hoffmann, M.; Plattner, N.; Wehmeyer, C.; Prinz, J.-H.; Noé, F. PyEMMA 2: A Software Package for Estimation, Validation, and Analysis of Markov Models. J. Chem. Theory Comput. 2015, 11, 5525–5542. [Google Scholar] [CrossRef]
- Kovacs, J.A.; Wriggers, W. Spatial Heat Maps from Fast Information Matching of Fast and Slow Degrees of Freedom: Application to Molecular Dynamics Simulations. J. Phys. Chem. B 2016, 120, 8473–8484. [Google Scholar] [CrossRef]
- Wriggers, W.R.; Stafford, K.; Shan, Y.; Piana, S.; Maragakis, P.; Lindorff-Larsen, K.; Miller, P.J.; Gullingsrud, J.; Rendleman, C.A.; Eastwood, M.P.; et al. Automated Event Detection and Activity Monitoring in Long Molecular Dynamics Simulations. J. Chem. Theory Comput. 2009, 5, 2595–2605. [Google Scholar] [CrossRef] [PubMed]
- Cragnell, C.; Durand, M.; Cabane, B.; Skep̦ö, M. Coarse-grained modeling of the intrinsically disordered protein Histatin 5 in solution: Monte Carlo simulations in combination with SAXS. Proteins Struct. Funct. Bioinform. 2016, 84, 777–791. [Google Scholar] [CrossRef] [PubMed]
- Raj, P.A.; Marcus, E.; Sukumaran, D.K. Structure of human salivary histatin 5 in aqueous and nonaqueous solutions. Biopolymers 1998, 45, 51–67. [Google Scholar] [CrossRef]
- Andresen, C.; Helander, S.; Lemak, A.; Farès, C.; Csizmok, V.; Carlsson, J.; Penn, L.Z.; Forman-Kay, J.D.; Arrowsmith, C.H.; Lundström, P.; et al. Transient structure and dynamics in the disordered c-Myc transactivation domain affect Bin1 binding. Nucleic Acids Res. 2012, 40, 6353–6366. [Google Scholar] [CrossRef]
- Wahlström, T.; Henriksson, M.A. Impact of MYC in regulation of tumor cell metabolism. BBA Gene Regul. Mech. 2015, 1849, 563–569. [Google Scholar] [CrossRef]
- Chodera, J.D.; Noé, F. Markov state models of biomolecular conformational dynamics. Curr. Opin. Struct. Biol. 2014, 25, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; West-Osterfield, K.; Spears, E.; Li, Z.; Panaccione, A.; Hann, S.R. MB0 and MBI Are Independent and Distinct Transactivation Domains in MYC that Are Essential for Transformation. Genes 2017, 8, 134. [Google Scholar] [CrossRef]
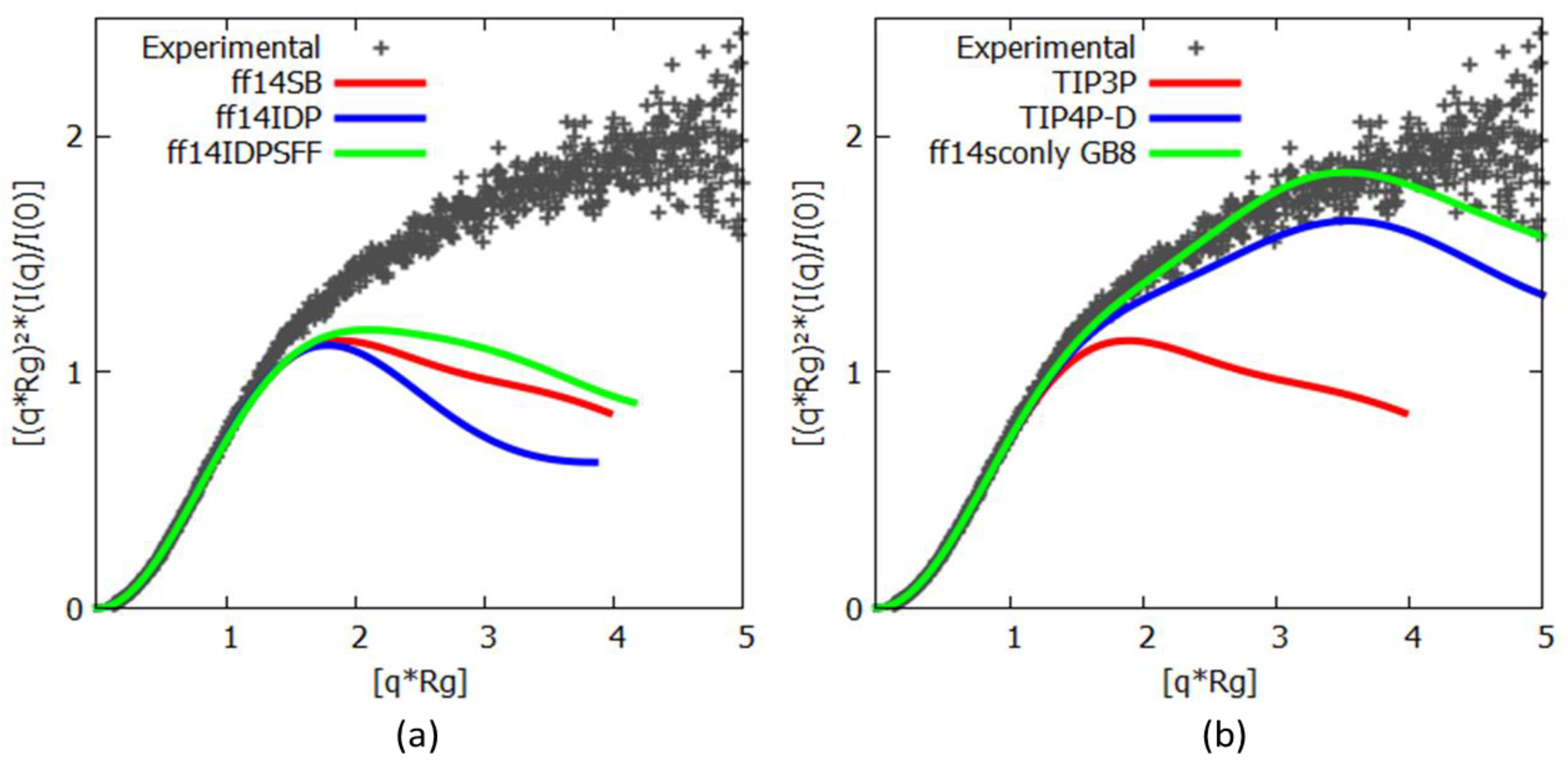
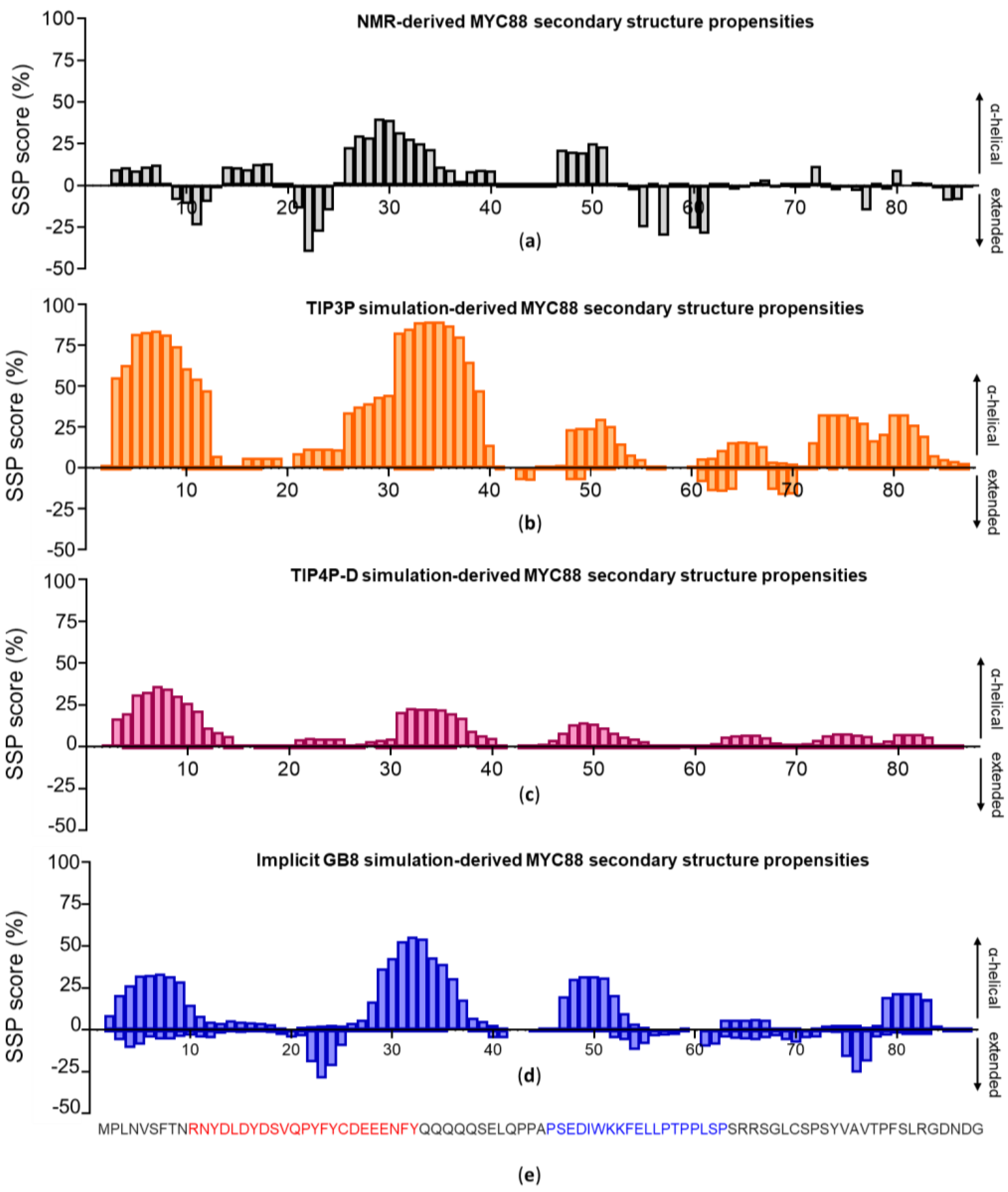
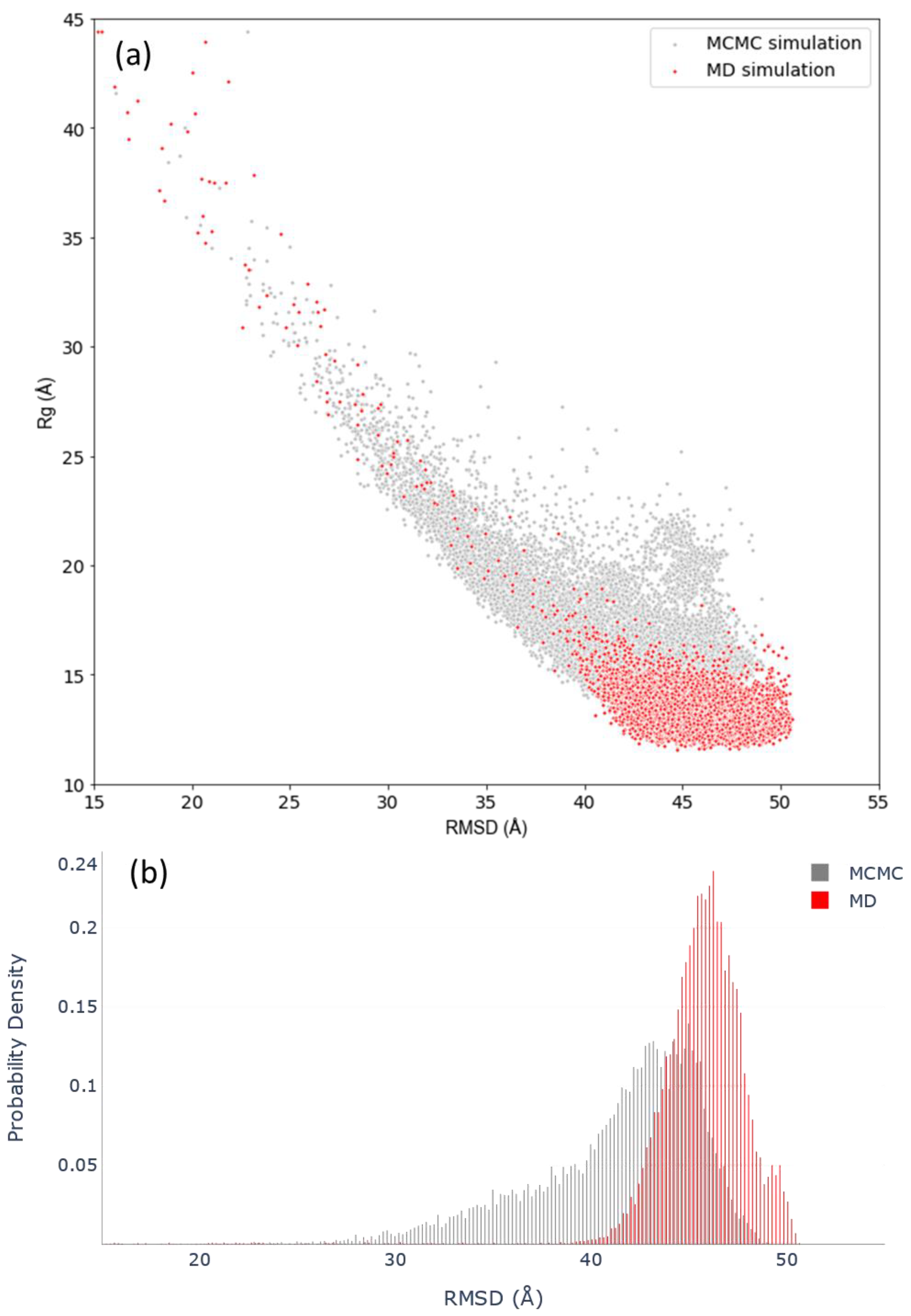
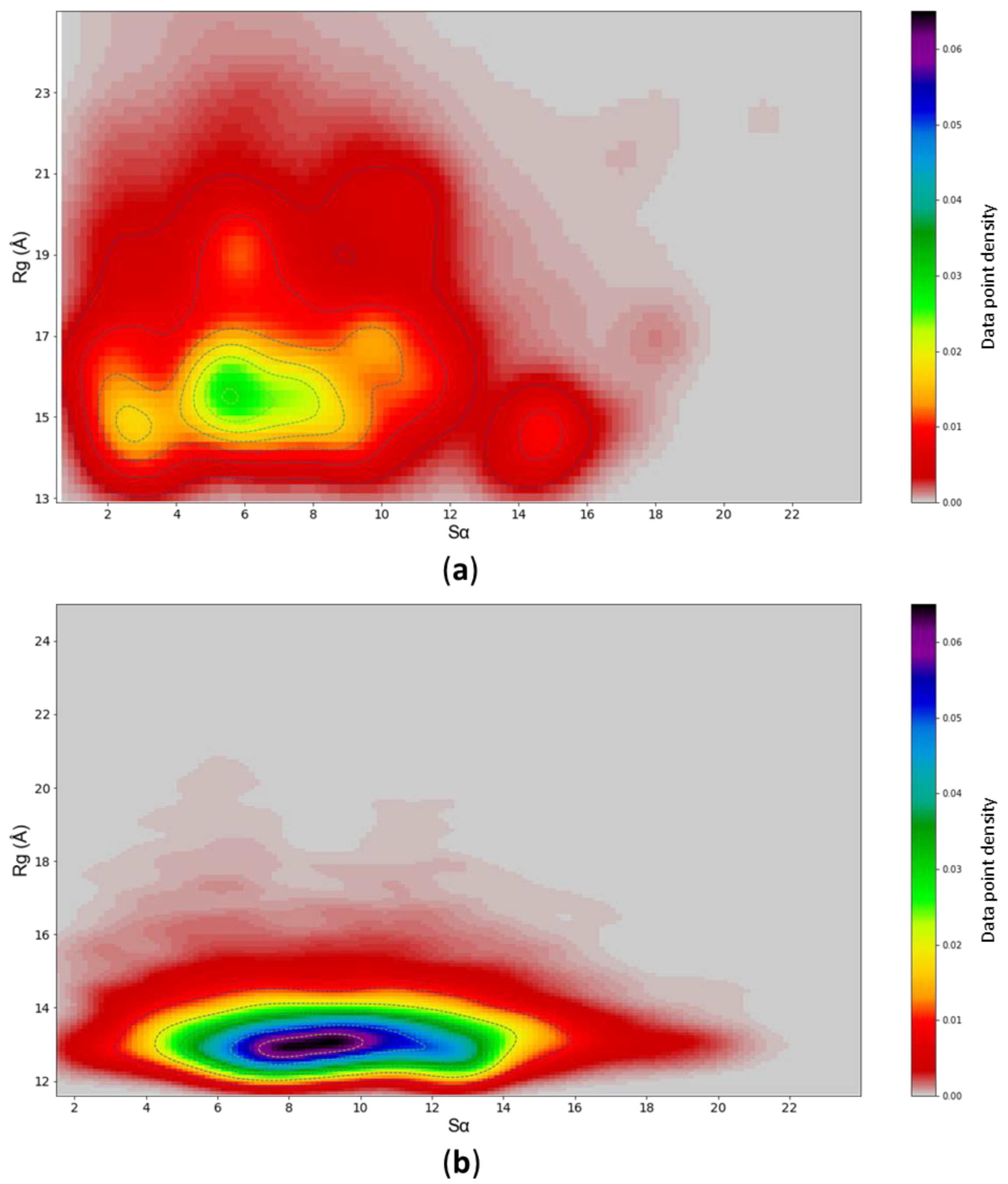

| Force Fields | Cluster 1 | Cluster 2 | Experimental |
|---|---|---|---|
| ff14SB | 9.15 Å | 7.71 Å | |
| ff14IDPs | 7.38 Å | 8.15 Å | 13.8 Å |
| ff14IDPSFF | 7.48 Å | 9.87 Å |
| Water Model | Cluster 1 | Cluster 2 | Experimental |
|---|---|---|---|
| TIP3P | 9.15 Å | 7.71 Å | |
| TIP4P-D | 13.47 Å | 12.12 Å | 13.8 Å |
| Implicit GB8 | 10.68 Å | 14.14 Å |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sullivan, S.S.; Weinzierl, R.O.J. Optimization of Molecular Dynamics Simulations of c-MYC1-88—An Intrinsically Disordered System. Life 2020, 10, 109. https://doi.org/10.3390/life10070109
Sullivan SS, Weinzierl ROJ. Optimization of Molecular Dynamics Simulations of c-MYC1-88—An Intrinsically Disordered System. Life. 2020; 10(7):109. https://doi.org/10.3390/life10070109
Chicago/Turabian StyleSullivan, Sandra S., and Robert O.J. Weinzierl. 2020. "Optimization of Molecular Dynamics Simulations of c-MYC1-88—An Intrinsically Disordered System" Life 10, no. 7: 109. https://doi.org/10.3390/life10070109
APA StyleSullivan, S. S., & Weinzierl, R. O. J. (2020). Optimization of Molecular Dynamics Simulations of c-MYC1-88—An Intrinsically Disordered System. Life, 10(7), 109. https://doi.org/10.3390/life10070109





