On Using Relative Information to Estimate Traits in a Darwinian Evolution Population Dynamics
Abstract
1. Introduction
2. Review of Information Theory
2.1. Fisher’s Information Theory
- A1 :
- The support of G is independent of .
- A2 :
- is nonnegative and for all .
- A3 :
- , the set of continuously and twice differentiable functions of , for all .
2.2. Entropy and Relative Entropy
3. Evolution Population Dynamics and Relative Information
3.1. Single Darwinian Population Model with Multiple Traits
- (H1)
- is the joint distribution of the independent traits , each with mean 0 and variance .
- (H2)
- .
- (H3)
- The density of is given as at .
3.2. Supervised and Unsupervised Learning
3.2.1. Supervised Learning
3.2.2. Unsupervised Learning
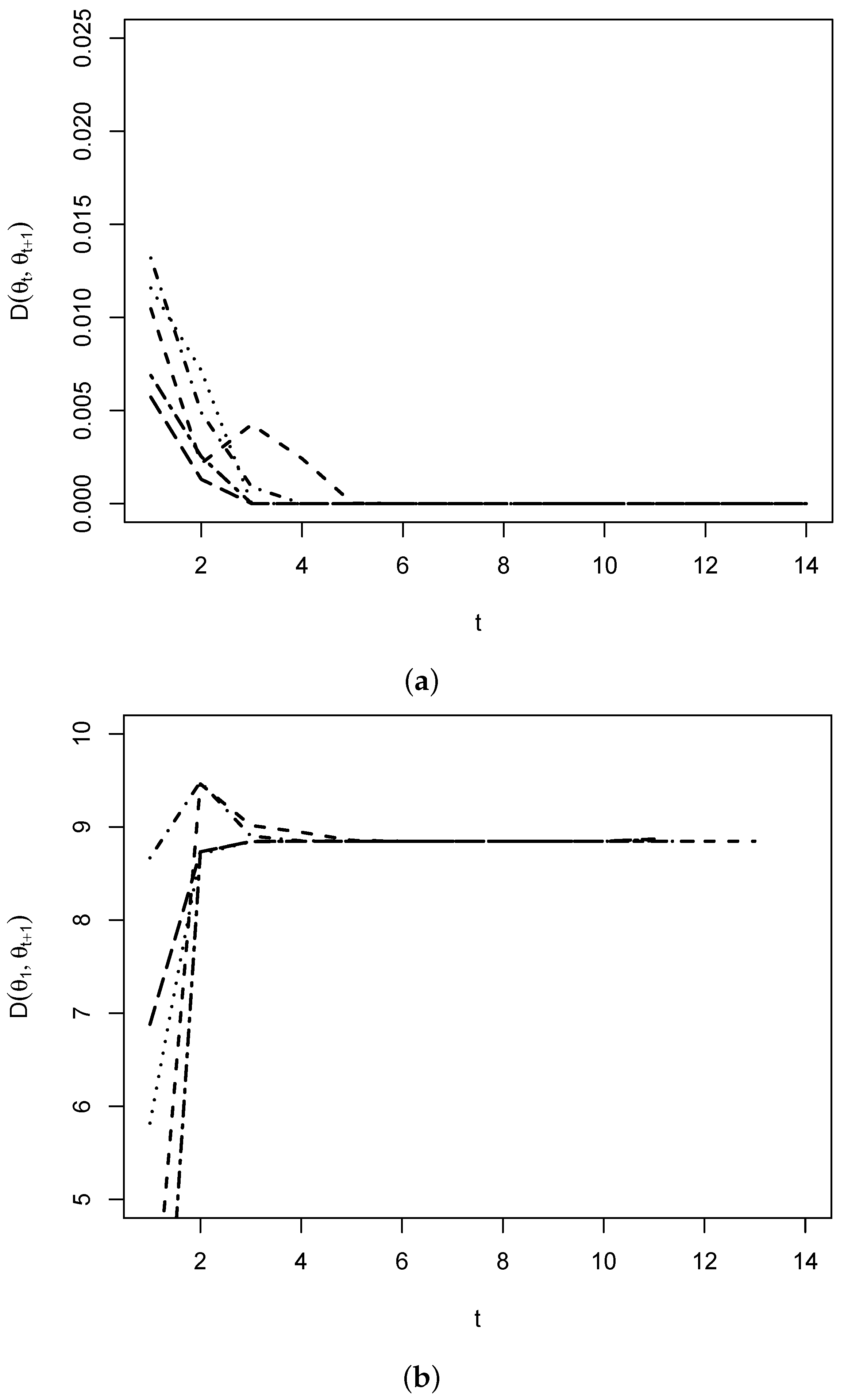
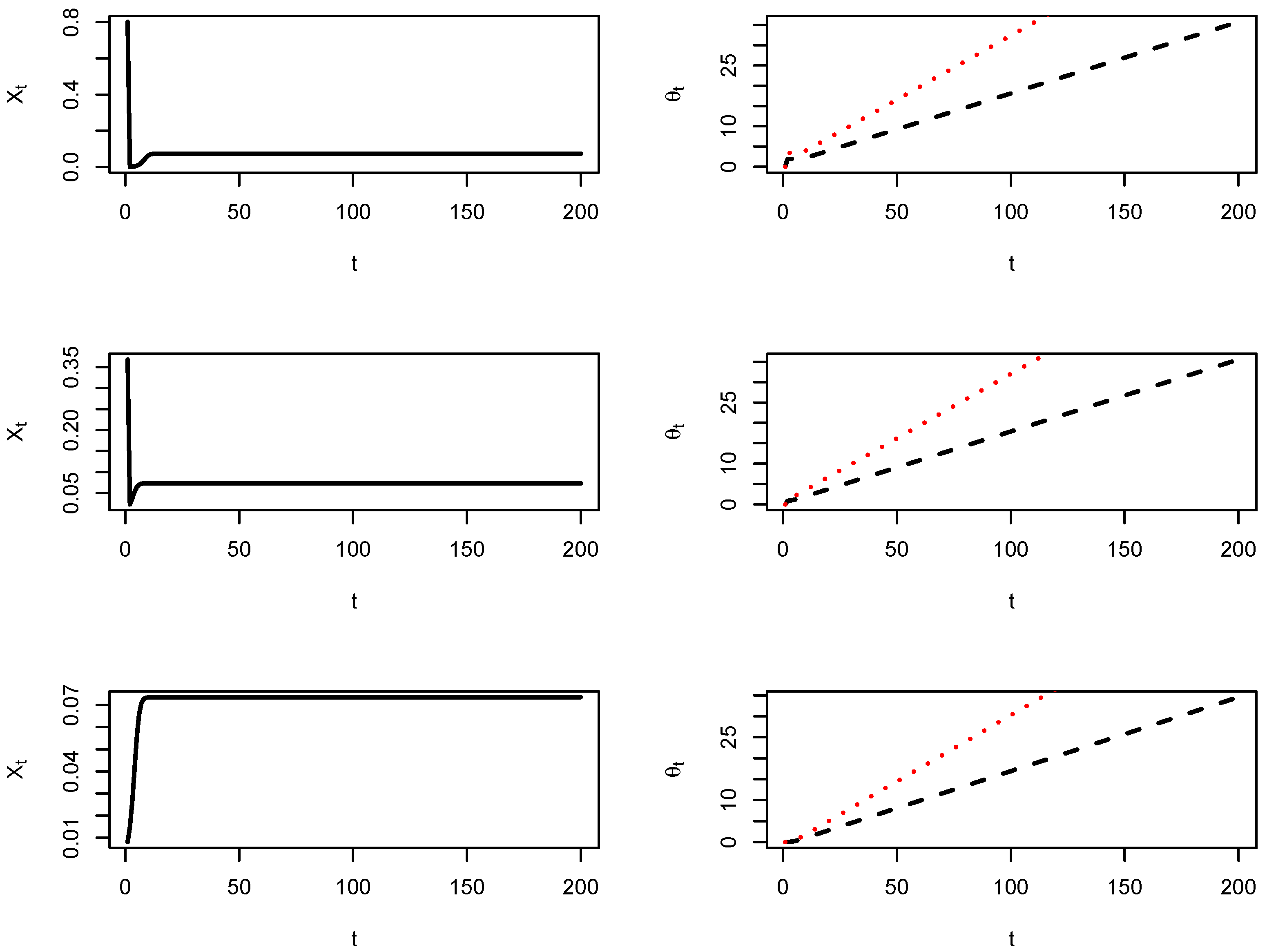
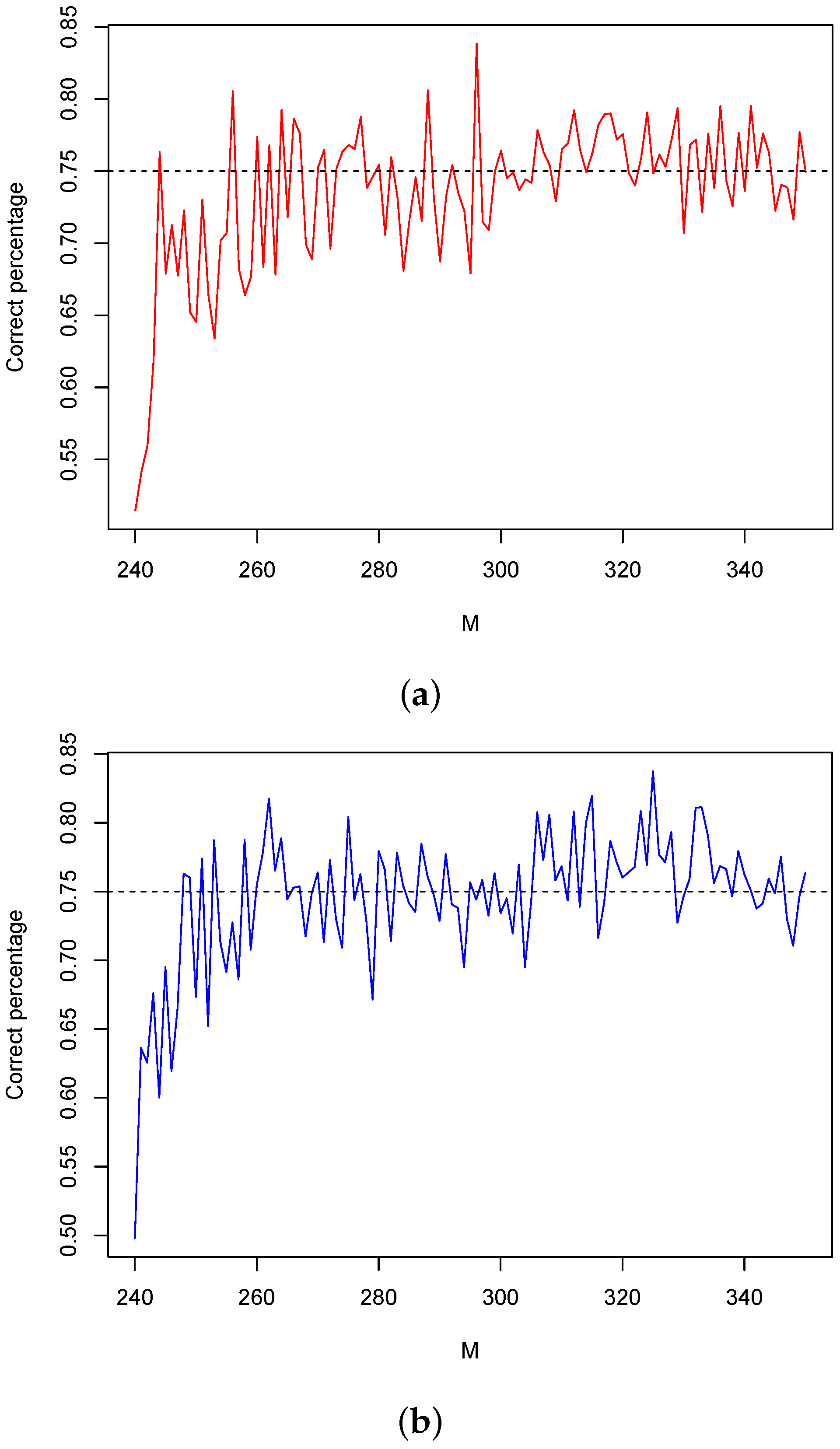
3.3. Simulations
4. Conclusions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Proof of Theorem 1
Appendix A.2. Proof of Theorem 2
References
- Darwin, C. On the Origin of Species by Means of Natural Selection; John Murray: London, UK, 1859. [Google Scholar]
- Vincent, T.L.; Vincent, T.L.S.; Cohen, Y. Darwinian dynamics and evolutionary game theory. J. Biol. Dyn. 2011, 5, 215–226. [Google Scholar] [CrossRef]
- Ackleh, A.S.; Cushing, J.M.; Salceneau, P.L. On the dynamics of evolutionary competition models. Nat. Resour. Model. 2015, 28, 380–397. [Google Scholar] [CrossRef]
- Cushing, J.M. Difference equations as models of evolutionary population dynamics. J. Biol. Dyn. 2019, 13, 103–127. [Google Scholar] [CrossRef] [PubMed]
- Cushing, J.M.; Park, J.; Farrell, A.; Chitnis, N. Treatment of outocme in an si model with evolutionary resistance: A darwinian model for the evolutionary resistance. J. Biol. Dyn. 2023, 17, 2255061. [Google Scholar] [CrossRef] [PubMed]
- Elaydi, S.; Kang, Y.; Luis, R. The effects of evolution on the stability of competing species. J. Biol. Dyn. 2022, 16, 816–839. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.A. On the mathematical foundation of theoretical statistics. Philos. Trans. R. Soc. Lond. Ser. 1922, 222, 594–604. [Google Scholar]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 623–656. [Google Scholar] [CrossRef]
- Kwessi, E. Information theory in a darwinian evolution population dynamics model. arXiv 2024, arXiv:2403.05044. [Google Scholar]
- Shun-ich, A.; Horishi, N. Methods of Information Geometry; chapter Chentsov Theorem and Some Historical Remarks; Oxford University Press: Oxford, UK, 2000; pp. 37–40. [Google Scholar]
- Dowty, J.G. Chentsov theorem for exponential families. Inf. Geom. 2018, 1, 117–135. [Google Scholar] [CrossRef]
- Harper, M. Information geometry and evolution game theory. arXiv 2009, arXiv:0911.1383. [Google Scholar] [CrossRef]
- Kimura, M. On the change of population fitness by natural selection. Heredity 1958, 12, 145–167. [Google Scholar] [CrossRef]
- Hinton, G.E.; Sejnowski, T.J. Optimal perceptual inference. In Proceedings of the IEEE conference on Computer Vision and Pattern Recognition, Washington, DC, USA, 19–23 June 1983; pp. 448–453. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwessi, E. On Using Relative Information to Estimate Traits in a Darwinian Evolution Population Dynamics. Axioms 2024, 13, 406. https://doi.org/10.3390/axioms13060406
Kwessi E. On Using Relative Information to Estimate Traits in a Darwinian Evolution Population Dynamics. Axioms. 2024; 13(6):406. https://doi.org/10.3390/axioms13060406
Chicago/Turabian StyleKwessi, Eddy. 2024. "On Using Relative Information to Estimate Traits in a Darwinian Evolution Population Dynamics" Axioms 13, no. 6: 406. https://doi.org/10.3390/axioms13060406
APA StyleKwessi, E. (2024). On Using Relative Information to Estimate Traits in a Darwinian Evolution Population Dynamics. Axioms, 13(6), 406. https://doi.org/10.3390/axioms13060406






