Selection and Characterization of a Nanobody Biosensor of GTP-Bound RHO Activities
Abstract
:1. Introduction
2. Material and Methods
2.1. Plasmids
2.2. Cell Lines, Transfection Method, and Reagents
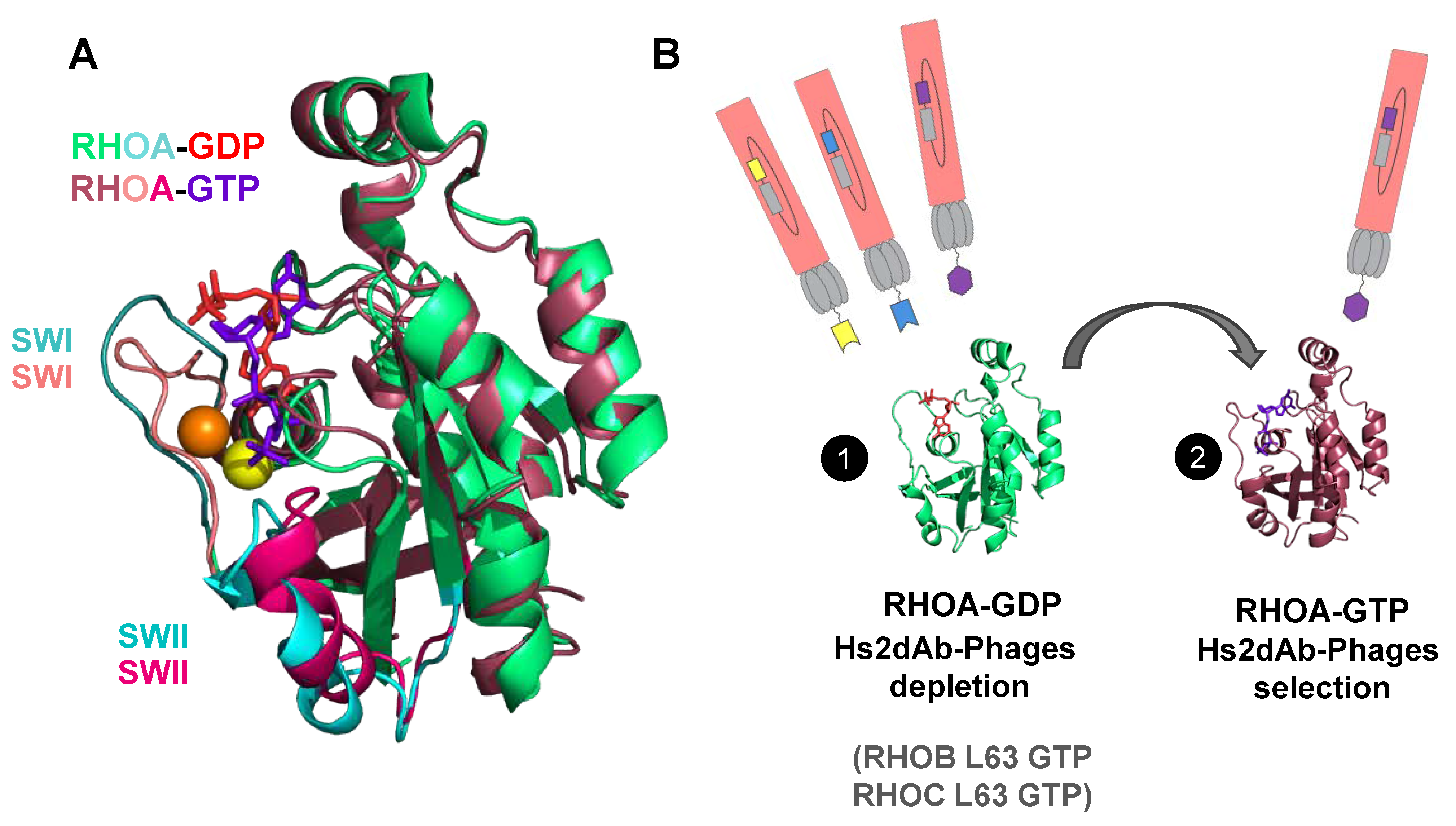
2.3. Subtractive Phage Display Panning for Isolating RHO-GTP Specific hs2dAb
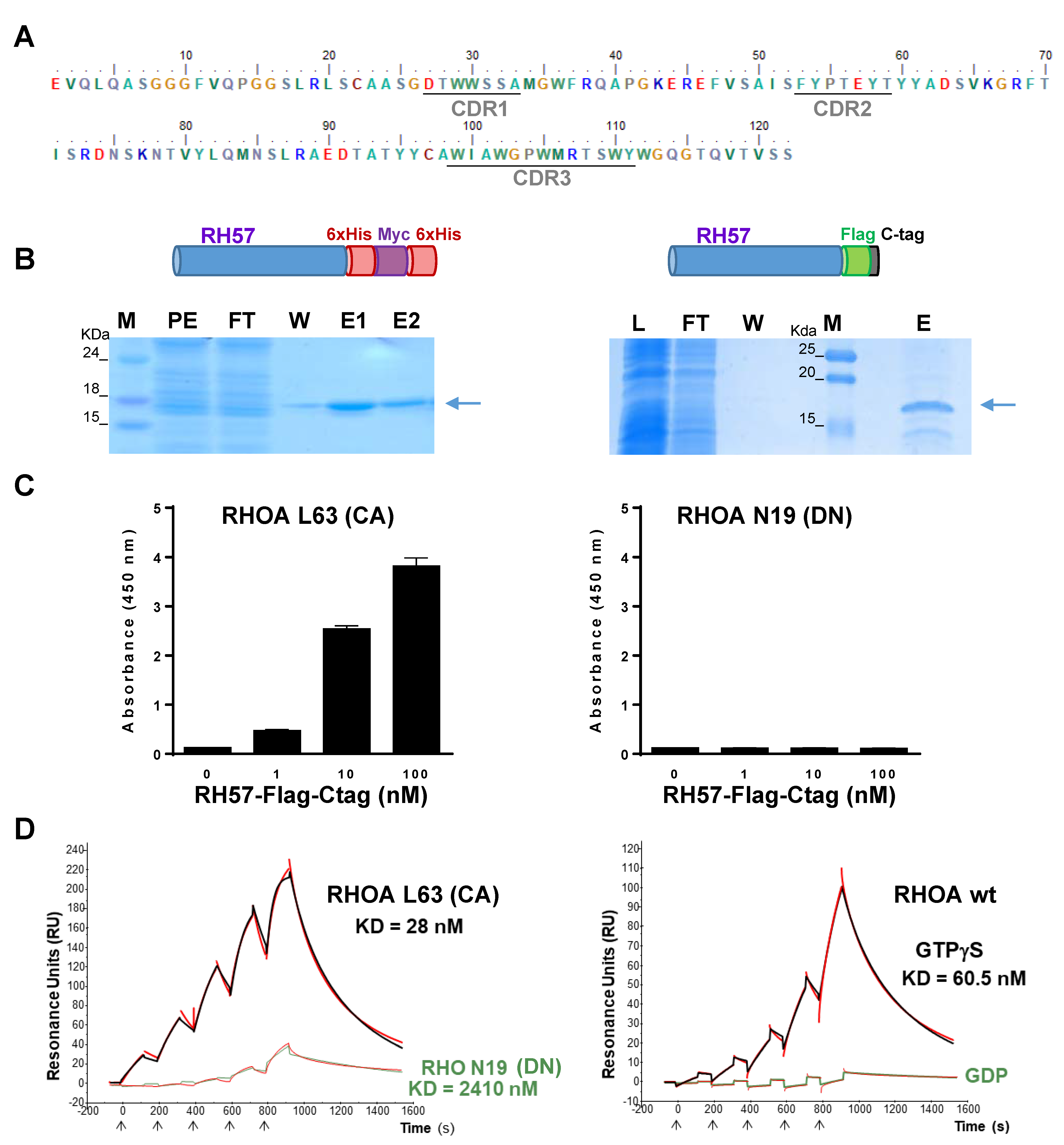
2.4. Hs2dAb Purification
2.5. RHO GTPases Purification
2.6. ELISA Assays
2.7. Loading Recombinant Proteins with GTPγS/GDP
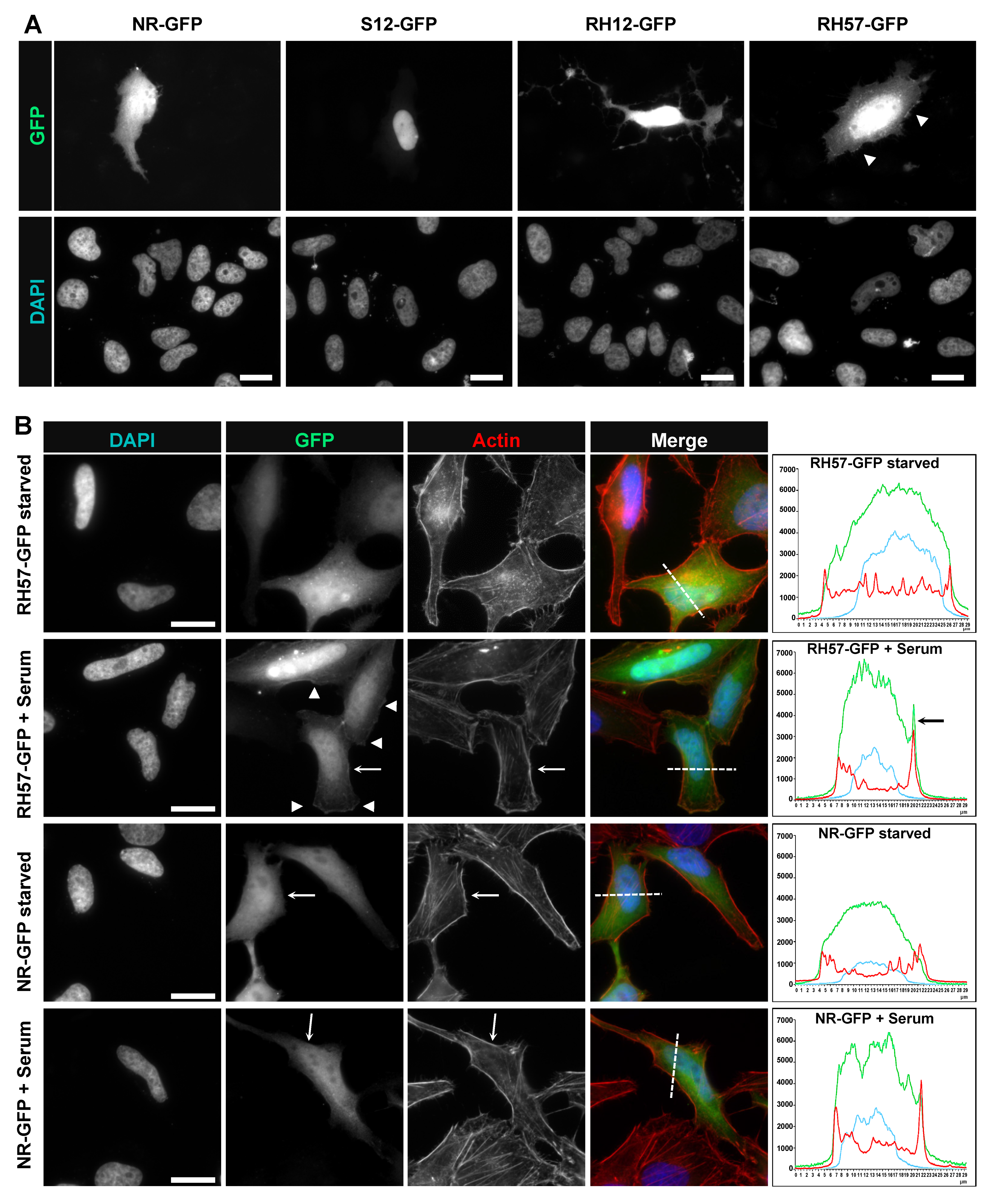
2.8. Immunofluorescence Staining
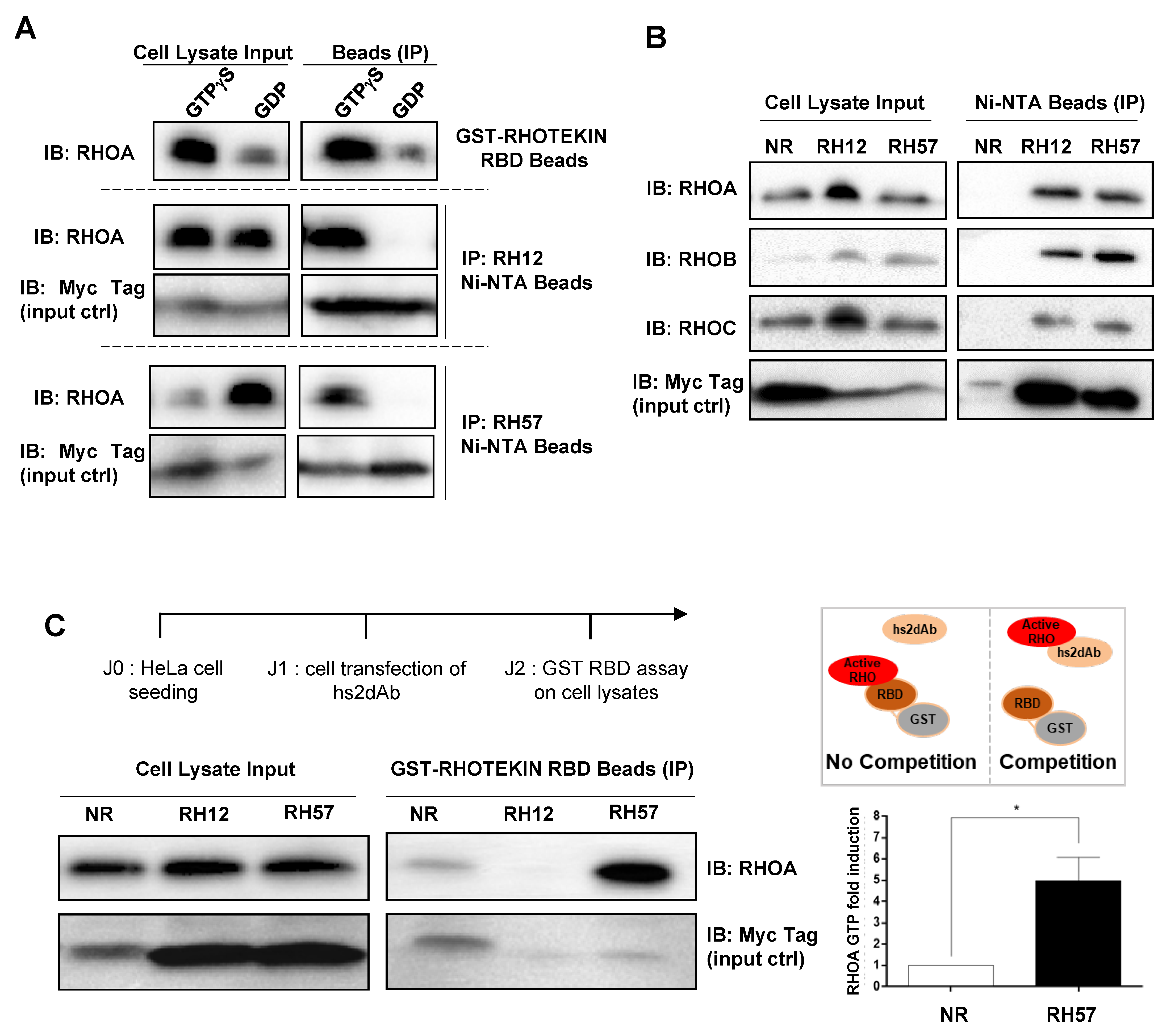
2.9. Endogenous RHO Proteins Intracellular Antibodies Co-Immunoprecipitation Assays
2.10. GST-RBD Assay
2.11. Affinity Measurement
2.12. BRET
2.12.1. Molecular Cloning
2.12.2. BRET2 Titration Curves and Stimulation Assays
2.12.3. BRET2 Measurements
3. Results
3.1. Phage Display Selection of a New High Affinity GTP-Bound RHO Conformational hs2dAb
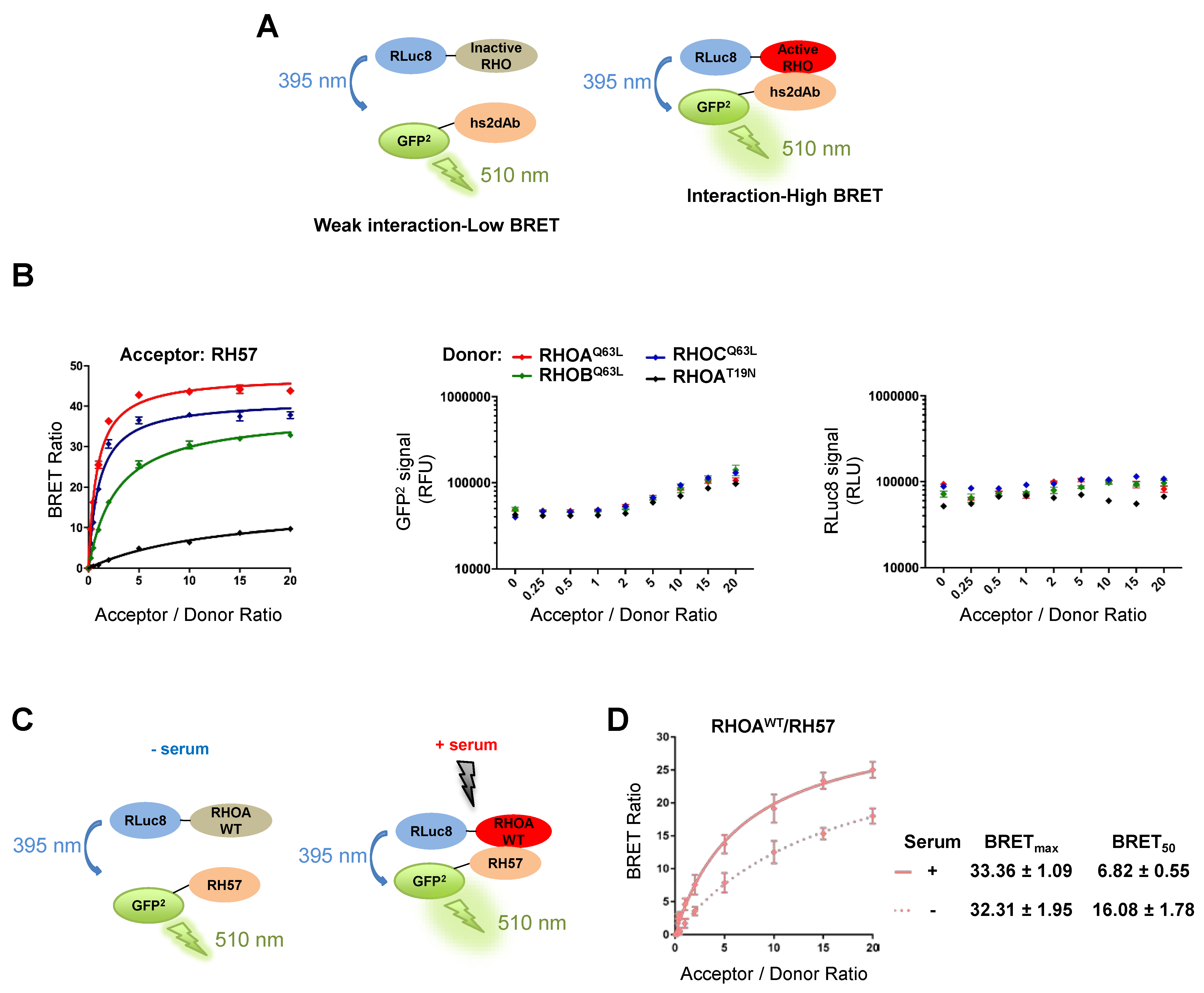
3.2. Characteriation of the RH57 Intracellular Antibody Binding Properties
3.3. Evaluating the RH57 intracellular antibody as a biosensor of RHOA-GTP.
4. Discussion
5. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wennerberg, K. The Ras superfamily at a glance. J. Cell Sci. 2005, 118, 843–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bishop, A.L.; Hall, A. Rho GTPases and their effector proteins. Biochem. J. 2000, 348 Pt 2, 241–255. [Google Scholar] [CrossRef]
- Satoh, T.; Kaziro, Y. Ras in signal transduction. Semin. Cancer Biol. 1992, 3, 169–177. [Google Scholar] [PubMed]
- Ren, X.D.; Schwartz, M.A. Determination of GTP loading on Rho. Methods Enzymol. 2000, 325, 264–272. [Google Scholar] [PubMed]
- Sahai, E.; Marshall, C.J. RHO—GTPases and cancer. Nat. Rev. Cancer 2002, 2, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-G.; Islam, R.; Cho, J.Y.; Jeong, H.; Cap, K.-C.; Park, Y.; Hossain, A.J.; Park, J.-B. Regulation of RhoA GTPase and various transcription factors in the RhoA pathway. J. Cell. Physiol. 2018, 233, 6381–6392. [Google Scholar] [CrossRef]
- Valon, L.; Etoc, F.; Remorino, A.; di Pietro, F.; Morin, X.; Dahan, M.; Coppey, M. Predictive Spatiotemporal Manipulation of Signaling Perturbations Using Optogenetics. Biophys. J. 2015, 109, 1785–1797. [Google Scholar] [CrossRef] [Green Version]
- Adini, I.; Rabinovitz, I.; Sun, J.F.; Prendergast, G.C.; Benjamin, L.E. RhoB controls Akt trafficking and stage-specific survival of endothelial cells during vascular development. Genes Dev. 2003, 17, 2721–2732. [Google Scholar] [CrossRef] [Green Version]
- Calvayrac, O.; Mazières, J.; Figarol, S.; Marty-Detraves, C.; Raymond-Letron, I.; Bousquet, E.; Farella, M.; Clermont-Taranchon, E.; Milia, J.; Rouquette, I.; et al. The RAS-related GTPase RHOB confers resistance to EGFR-tyrosine kinase inhibitors in non-small-cell lung cancer via an AKT-dependent mechanism. EMBO Mol. Med. 2017, 9, 238–250. [Google Scholar] [CrossRef]
- Vega, F.M.; Ridley, A.J. Rho GTPases in cancer cell biology. FEBS Lett. 2008, 582, 2093–2101. [Google Scholar] [CrossRef] [Green Version]
- Zandvakili, I.; Lin, Y.; Morris, J.C.; Zheng, Y. Rho GTPases: Anti- or pro-neoplastic targets? Oncogene 2017, 36, 3213–3222. [Google Scholar] [CrossRef]
- Delmas, A.; Cherier, J.; Pohorecka, M.; Medale-Giamarchi, C.; Meyer, N.; Casanova, A.; Sordet, O.; Lamant, L.; Savina, A.; Pradines, A.; et al. The c-Jun/RHOB/AKT pathway confers resistance of BRAF-mutant melanoma cells to MAPK inhibitors. Oncotarget 2015, 6, 15250–15264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shang, X.; Marchioni, F.; Sipes, N.; Evelyn, C.R.; Jerabek-Willemsen, M.; Duhr, S.; Seibel, W.; Wortman, M.; Zheng, Y. Rational design of small molecule inhibitors targeting RhoA subfamily Rho GTPases. Chem. Biol. 2012, 19, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Reinhard, N.R.; van Helden, S.F.; Anthony, E.C.; Yin, T.; Wu, Y.I.; Goedhart, J.; Gadella, T.W.J.; Hordijk, P.L. Spatiotemporal analysis of RhoA/B/C activation in primary human endothelial cells. Sci. Rep. 2016, 6, 25502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertz, O.; Hodgson, L.; Klemke, R.L.; Hahn, K.M. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature 2006, 440, 1069–1072. [Google Scholar] [CrossRef] [PubMed]
- Koraïchi, F.; Gence, R.; Bouchenot, C.; Grosjean, S.; Lajoie-Mazenc, I.; Favre, G.; Cabantous, S. High-content tripartite split-GFP cell-based assays to screen for modulators of small GTPase activation. J. Cell Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Stofega, M.; DerMardirossian, C.; Bokoch, G.M. Affinity-Based Assay of Rho Guanosine Triphosphatase Activation. In Transmembrane Signaling Protocols; Humana Press: Totowa, NJ, USA, 2006; Volume 332, pp. 269–280. ISBN 978-1-59745-048-5. [Google Scholar]
- Pellegrin, S.; Mellor, H. Rho GTPase activation assays. Curr. Protoc. Cell Biol. 2008. [Google Scholar] [CrossRef] [PubMed]
- Rothbauer, U.; Zolghadr, K.; Tillib, S.; Nowak, D.; Schermelleh, L.; Gahl, A.; Backmann, N.; Conrath, K.; Muyldermans, S.; Cardoso, M.C.; et al. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods 2006, 3, 887–889. [Google Scholar] [CrossRef]
- Tanaka, T.; Lobato, M.N.; Rabbitts, T.H. Single domain intracellular antibodies: A minimal fragment for direct in vivo selection of antigen-specific intrabodies. J. Mol. Biol. 2003, 331, 1109–1120. [Google Scholar] [CrossRef]
- Tanaka, T.; Rabbitts, T.H. Intrabodies based on intracellular capture frameworks that bind the RAS protein with high affinity and impair oncogenic transformation. EMBO J. 2003, 22, 1025–1035. [Google Scholar] [CrossRef] [Green Version]
- Galli, V.; Sebastian, R.; Moutel, S.; Ecard, J.; Perez, F.; Roux, A. Uncoupling of dynamin polymerization and GTPase activity revealed by the conformation-specific nanobody dynab. eLife 2017, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moutel, S.; Bery, N.; Bernard, V.; Keller, L.; Lemesre, E.; de Marco, A.; Ligat, L.; Rain, J.-C.; Favre, G.; Olichon, A.; et al. NaLi-H1: A universal synthetic library of humanized nanobodies providing highly functional antibodies and intrabodies. eLife 2016, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olichon, A.; Surrey, T. Selection of genetically encoded fluorescent single domain antibodies engineered for efficient expression in Escherichia coli. J. Biol. Chem. 2007, 282, 36314–36320. [Google Scholar] [CrossRef] [PubMed]
- Bery, N.; Cruz-Migoni, A.; Bataille, C.J.; Quevedo, C.E.; Tulmin, H.; Miller, A.; Russell, A.; Phillips, S.E.; Carr, S.B.; Rabbitts, T.H. BRET-based RAS biosensors that show a novel small molecule is an inhibitor of RAS-effector protein-protein interactions. eLife 2018, 7. [Google Scholar] [CrossRef]
- Kaiser, P.D.; Maier, J.; Traenkle, B.; Emele, F.; Rothbauer, U. Recent progress in generating intracellular functional antibody fragments to target and trace cellular components in living cells. Biochim. Biophys. Acta 2014. [Google Scholar] [CrossRef] [PubMed]
- Jullien, D.; Vignard, J.; Fedor, Y.; Béry, N.; Olichon, A.; Crozatier, M.; Erard, M.; Cassard, H.; Ducommun, B.; Salles, B.; et al. Chromatibody, a novel non-invasive molecular tool to explore and manipulate chromatin in living cells. J. Cell Sci. 2016, 129, 2673–2683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ridley, A.J. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006, 16, 522–529. [Google Scholar] [CrossRef]
- Machacek, M.; Hodgson, L.; Welch, C.; Elliott, H.; Pertz, O.; Nalbant, P.; Abell, A.; Johnson, G.L.; Hahn, K.M.; Danuser, G. Coordination of Rho GTPase activities during cell protrusion. Nature 2009, 461, 99–103. [Google Scholar] [CrossRef] [Green Version]
- Pfleger, K.D.G.; Seeber, R.M.; Eidne, K.A. Bioluminescence resonance energy transfer (BRET) for the real-time detection of protein-protein interactions. Nat. Protoc. 2006, 1, 337–345. [Google Scholar] [CrossRef] [Green Version]
- Mercier, J.-F.; Salahpour, A.; Angers, S.; Breit, A.; Bouvier, M. Quantitative assessment of beta 1- and beta 2-adrenergic receptor homo- and heterodimerization by bioluminescence resonance energy transfer. J. Biol. Chem. 2002, 277, 44925–44931. [Google Scholar] [CrossRef]
- Haque, A.; Andersen, J.N.; Salmeen, A.; Barford, D.; Tonks, N.K. Conformation-sensing antibodies stabilize the oxidized form of PTP1B and inhibit its phosphatase activity. Cell 2011, 147, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Nizak, C.; Monier, S.; del Nery, E.; Moutel, S.; Goud, B.; Perez, F. Recombinant antibodies to the small GTPase Rab6 as conformation sensors. Science 2003, 300, 984–987. [Google Scholar] [CrossRef] [PubMed]
- Dimitrov, A.; Quesnoit, M.; Moutel, S.; Cantaloube, I.; Poüs, C.; Perez, F. Detection of GTP-tubulin conformation in vivo reveals a role for GTP remnants in microtubule rescues. Science 2008, 322, 1353–1356. [Google Scholar] [CrossRef] [PubMed]
- Fritz, R.D.; Pertz, O. The dynamics of spatio-temporal Rho GTPase signaling: Formation of signaling patterns. F1000Research 2016, 5, 749. [Google Scholar] [CrossRef] [PubMed]
- Poulter, N.S.; Pitkeathly, W.T.E.; Smith, P.J.; Rappoport, J.Z. The Physical Basis of Total Internal Reflection Fluorescence (TIRF) Microscopy and Its Cellular Applications. In Advanced Fluorescence Microscopy; Verveer, P.J., Ed.; Springer: New York, NY, USA, 2015; Volume 1251, pp. 1–23. ISBN 978-1-4939-2079-2. [Google Scholar]
- Nalbant, P.; Hodgson, L.; Kraynov, V.; Toutchkine, A.; Hahn, K.M. Activation of endogenous Cdc42 visualized in living cells. Science 2004, 305, 1615–1619. [Google Scholar] [CrossRef] [PubMed]
- Namkung, Y.; Le Gouill, C.; Lukashova, V.; Kobayashi, H.; Hogue, M.; Khoury, E.; Song, M.; Bouvier, M.; Laporte, S.A. Monitoring G protein-coupled receptor and β-arrestin trafficking in live cells using enhanced bystander BRET. Nat. Commun. 2016, 7, 12178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beautrait, A.; Paradis, J.S.; Zimmerman, B.; Giubilaro, J.; Nikolajev, L.; Armando, S.; Kobayashi, H.; Yamani, L.; Namkung, Y.; Heydenreich, F.M.; et al. A new inhibitor of the β-arrestin/AP2 endocytic complex reveals interplay between GPCR internalization and signalling. Nat. Commun. 2017, 8, 15054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellot, M.; Galandrin, S.; Boularan, C.; Matthies, H.J.; Despas, F.; Denis, C.; Javitch, J.; Mazères, S.; Sanni, S.J.; Pons, V.; et al. Dual agonist occupancy of AT1-R-α2C-AR heterodimers results in atypical Gs-PKA signaling. Nat. Chem. Biol. 2015, 11, 271–279. [Google Scholar] [CrossRef]
- Quevedo, C.E.; Cruz-Migoni, A.; Bery, N.; Miller, A.; Tanaka, T.; Petch, D.; Bataille, C.J.R.; Lee, L.Y.W.; Fallon, P.S.; Tulmin, H.; et al. Small molecule inhibitors of RAS-effector protein interactions derived using an intracellular antibody fragment. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keller, L.; Bery, N.; Tardy, C.; Ligat, L.; Favre, G.; Rabbitts, T.H.; Olichon, A. Selection and Characterization of a Nanobody Biosensor of GTP-Bound RHO Activities. Antibodies 2019, 8, 8. https://doi.org/10.3390/antib8010008
Keller L, Bery N, Tardy C, Ligat L, Favre G, Rabbitts TH, Olichon A. Selection and Characterization of a Nanobody Biosensor of GTP-Bound RHO Activities. Antibodies. 2019; 8(1):8. https://doi.org/10.3390/antib8010008
Chicago/Turabian StyleKeller, Laura, Nicolas Bery, Claudine Tardy, Laetitia Ligat, Gilles Favre, Terence H. Rabbitts, and Aurélien Olichon. 2019. "Selection and Characterization of a Nanobody Biosensor of GTP-Bound RHO Activities" Antibodies 8, no. 1: 8. https://doi.org/10.3390/antib8010008
APA StyleKeller, L., Bery, N., Tardy, C., Ligat, L., Favre, G., Rabbitts, T. H., & Olichon, A. (2019). Selection and Characterization of a Nanobody Biosensor of GTP-Bound RHO Activities. Antibodies, 8(1), 8. https://doi.org/10.3390/antib8010008




