A Robust, High-Titer, Semi-Automated, and In-Culture Antibody-Capturing Transient CHO Platform Technology
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Transient Transfection
2.2. DNA Complexation Using the Tecan Fluent Automation System
2.3. Transient CHO4Tx® Production at 500 mL Scale
2.4. Transient Transfection of CHO4Tx® Cells at 100 mL Scale
2.5. Post-Transfection Data Acquisition and Determination of mAb Concentration
2.6. Evaluating Effects of Incubation Time and Elution Buffers on Control IgG In-Culture Capturing with Magnetic ProA Beads
2.7. Semi-Automatic Purification of mAb Using SA Plus and Magnetic ProA Beads
2.8. CHO Host Cell Protein (HCP) Content Measurement
2.9. Sephadex G-25 Column Chromatography
2.10. Statistical Analysis
3. Results
3.1. The CHO4Tx® System Is a High-Titer Transient CHO Expression System
3.2. Production with 100 mL Flasks Could Be Further Optimized Through Cultivation in Different Vessels
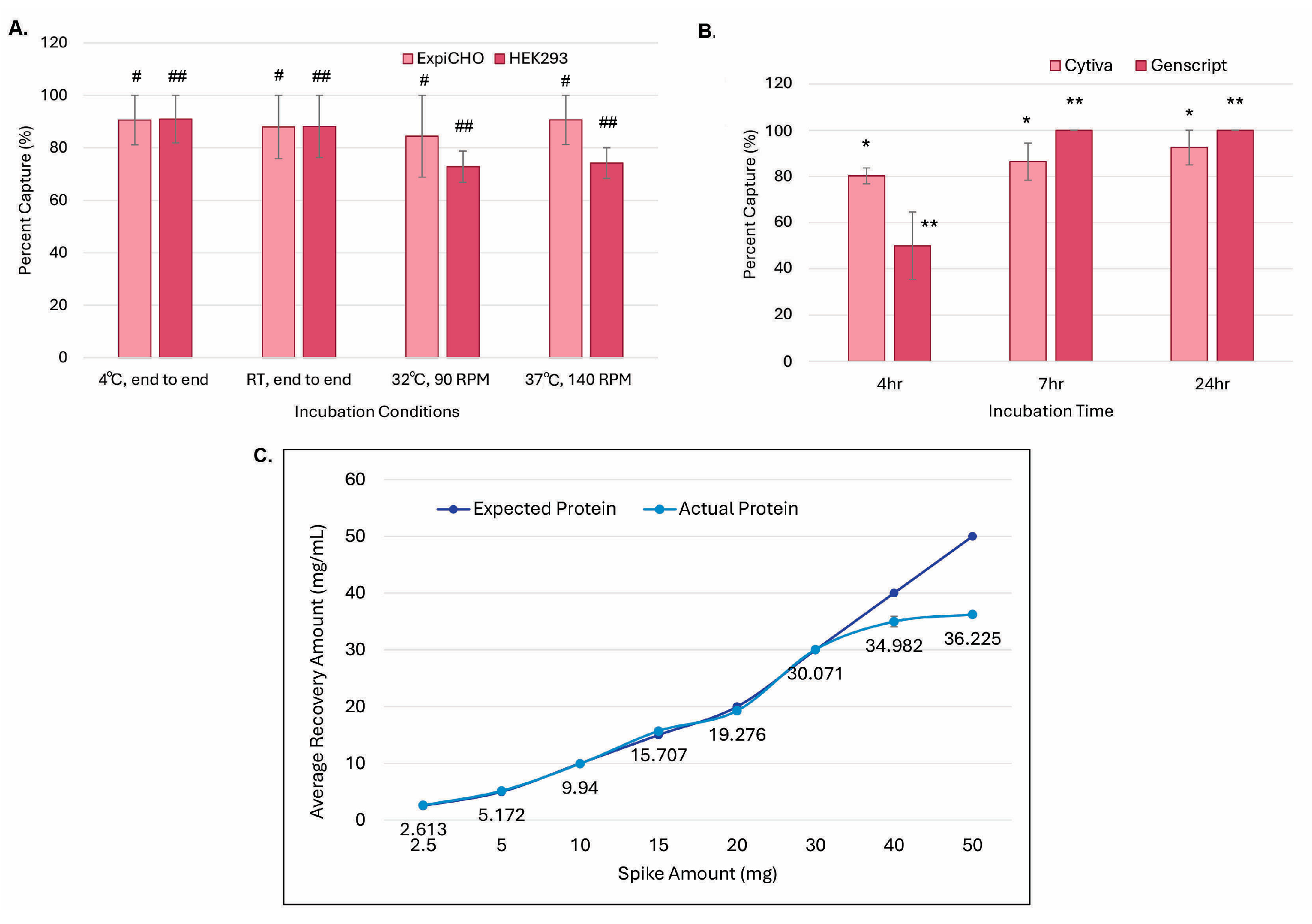
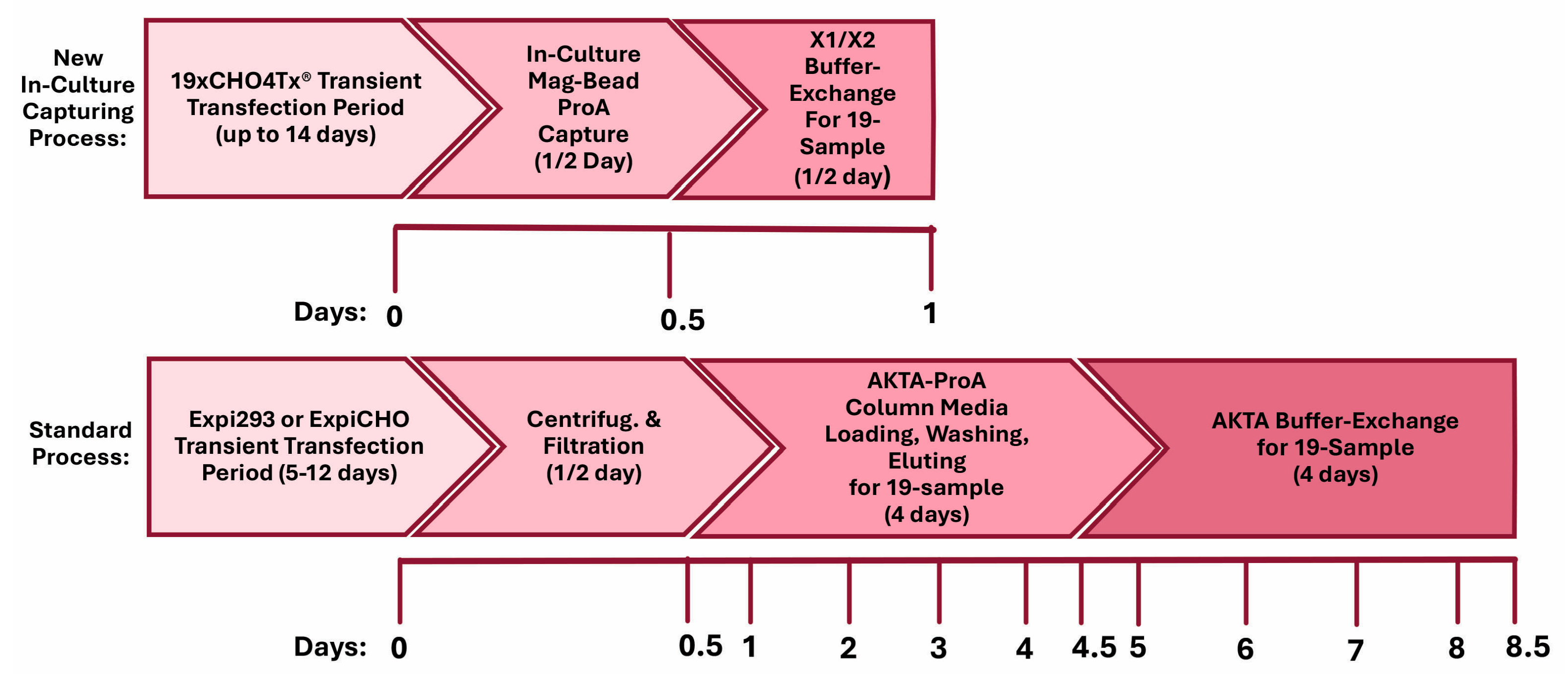
3.3. Antibody Protein Could Be Captured Through In-Culture Magnetic ProA Bead Incubation
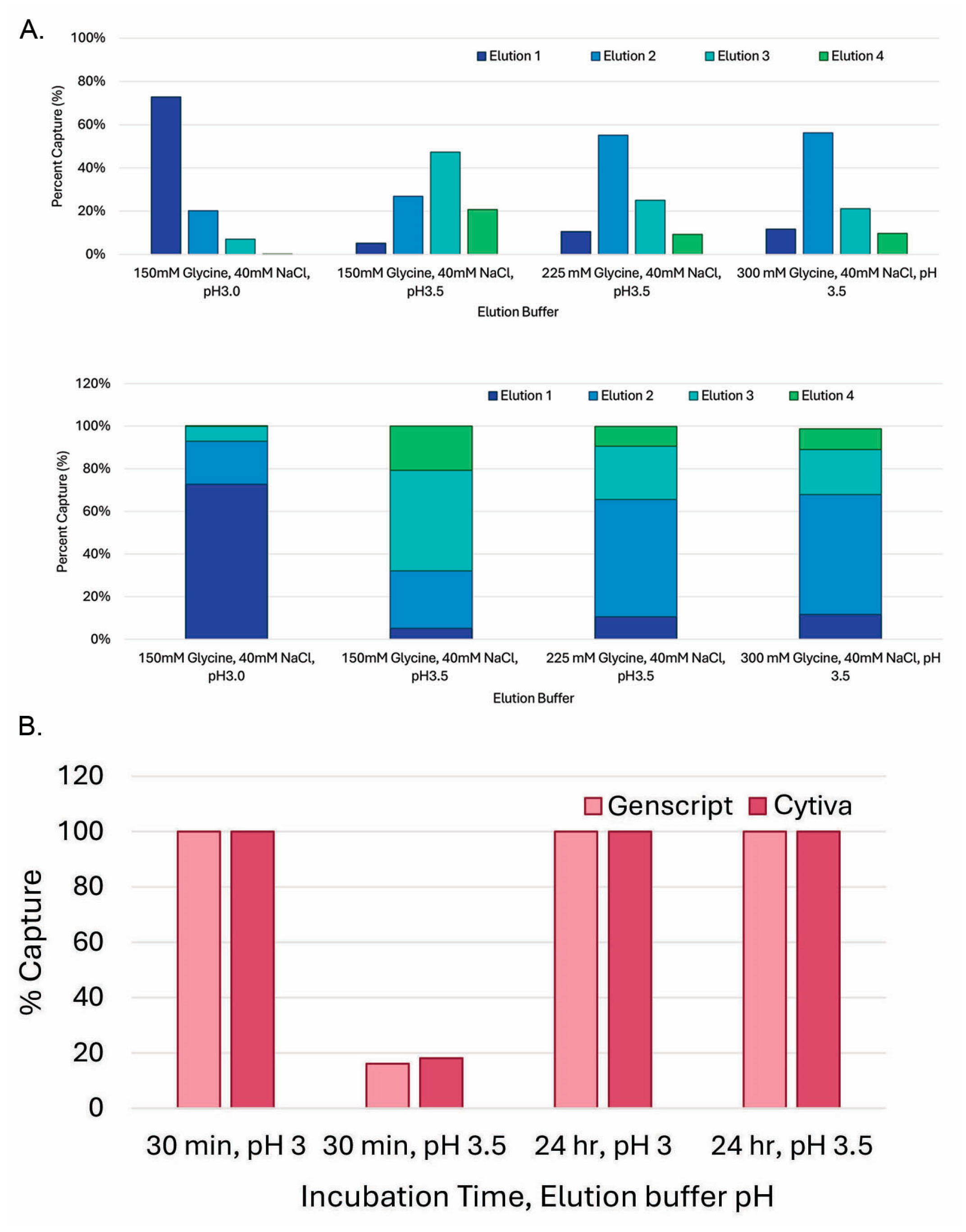
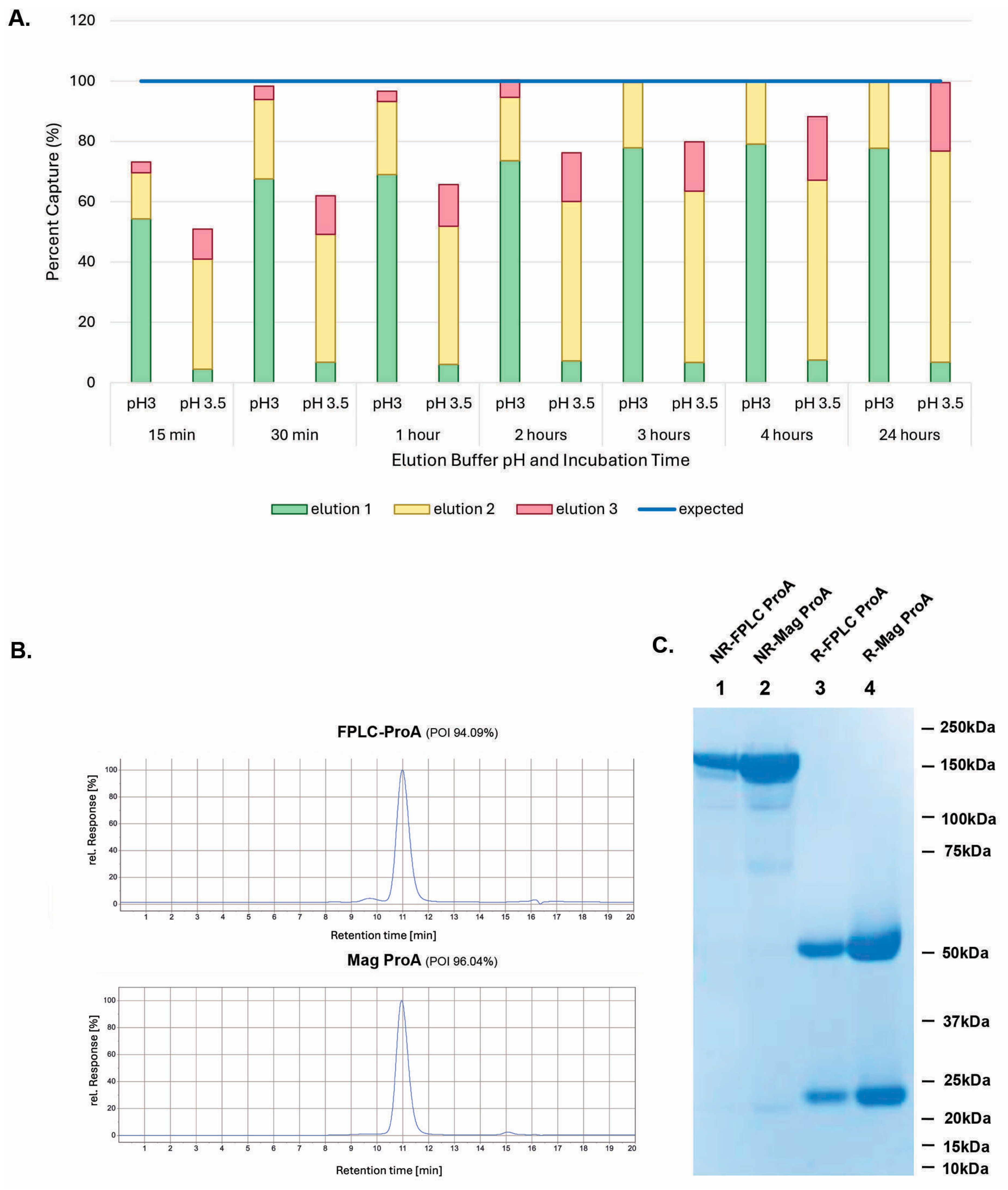
3.4. Decreasing Elution pH to 3.0 Dramatically Increased Elution Efficiency and Reduced In-Culture Capturing Time
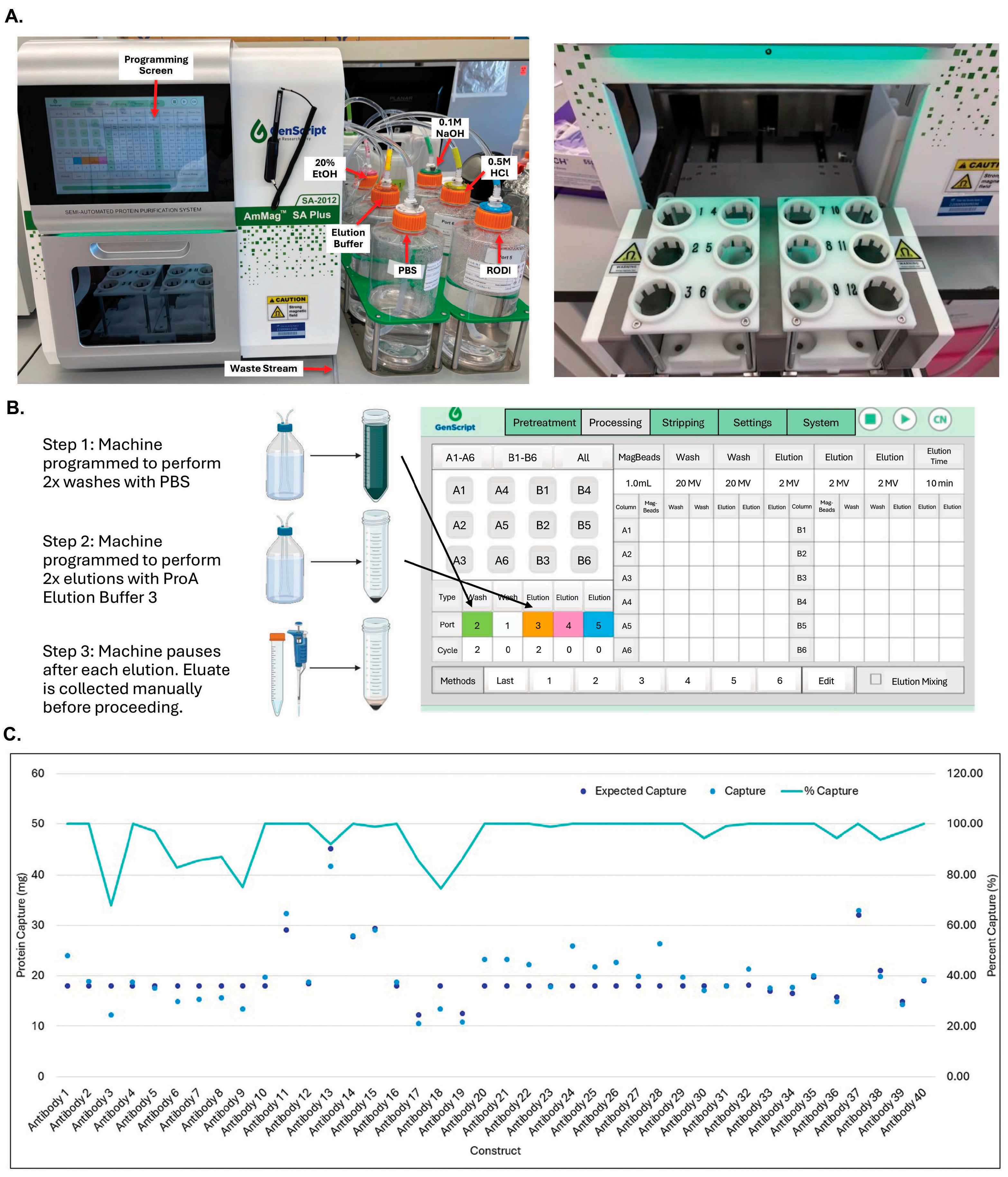
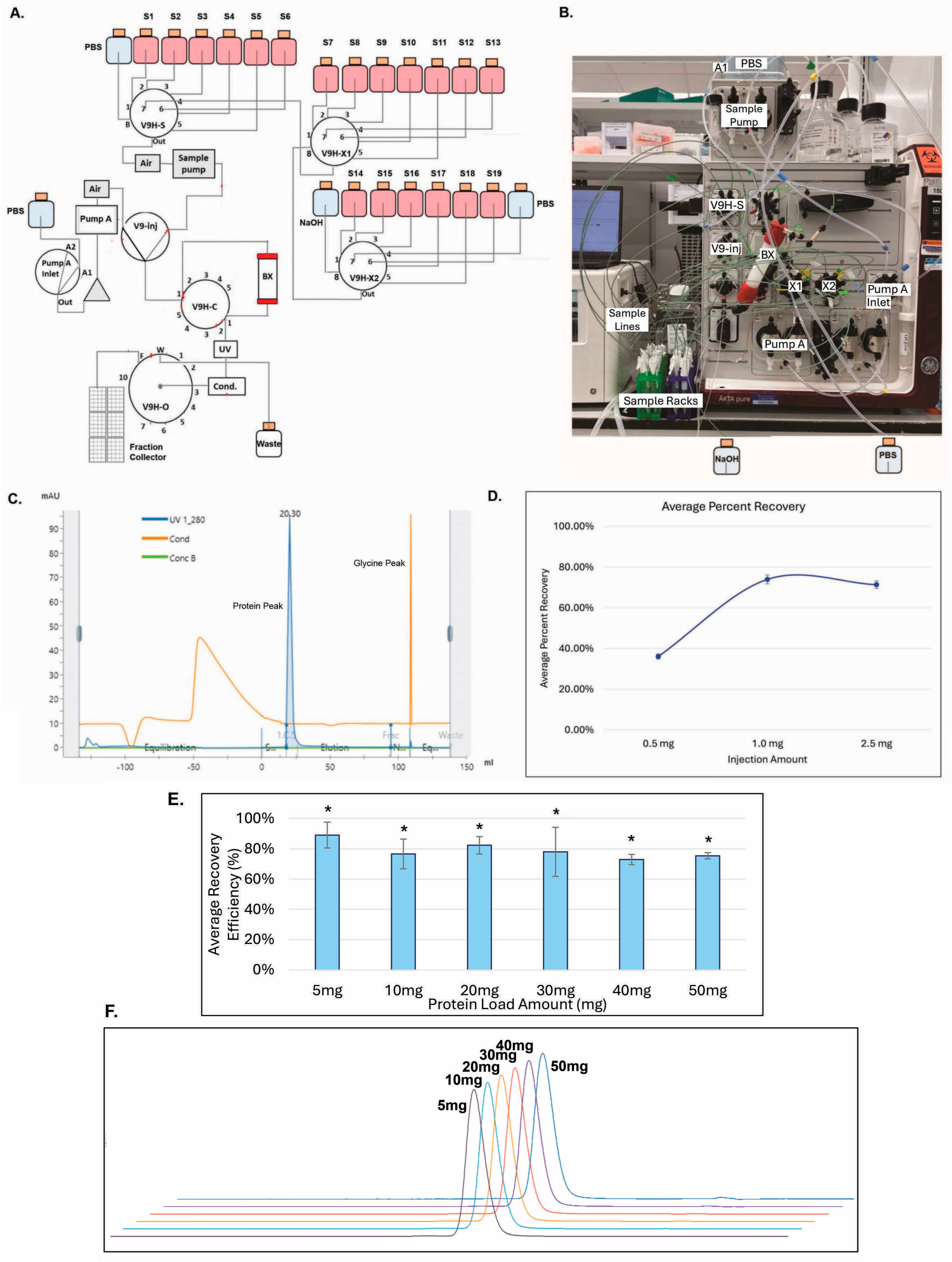
3.5. GenScript AmMag™ SA Plus System Enabled Automated Elution with 12 Constructs per Round in Under 1 h
3.6. Building V9HX1 and V9HX2 Values Facilitated Automated Buffer Exchange for 19 Constructs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lu, R.M.; Hwang, Y.C.; Liu, I.J.; Lee, C.C.; Tsai, H.Z.; Li, H.J.; Wu, H.C. Development of therapeutic antibodies for the treatment of diseases. J. Biomed. Sci. 2020, 27, 1. [Google Scholar] [CrossRef]
- Pedrioli, A.; Oxenius, A. Singsle B cell technologies for monoclonal antibody discovery. Trends Immunol. 2021, 42, 1143–1158. [Google Scholar] [CrossRef]
- Teixeira, A.A.R.; Erasmus, M.F.; D’Angelo, S.; Naranjo, L.; Ferrara, F.; Leal-Lopes, C.; Durrant, O.; Galmiche, C.; Morelli, A.; Scott-Tucker, A.; et al. Drug-like antibodies with high affinity, diversity and developability directly from next-generation antibody libraries. MAbs 2021, 13, 1980942. [Google Scholar] [CrossRef]
- Carter, P.J.; Rajpal, A. Designing antibodies as therapeutics. Cell 2022, 185, 2789–2805. [Google Scholar] [CrossRef]
- Sulea, T.; Kumar, S.; Kuroda, D. Editorial: Progress and challenges in computational structure-based design and development of biologic drugs. Front. Mol. Biosci. 2024, 11, 1360267. [Google Scholar] [CrossRef]
- Graves, J.; Byerly, J.; Priego, E.; Makkapati, N.; Parish, S.V.; Medellin, B.; Berrondo, M. A Review of Deep Learning Methods for Antibodies. Antibodies 2020, 9, 12. [Google Scholar] [CrossRef]
- Erasmus, M.F.; Spector, L.; Ferrara, F.; DiNiro, R.; Pohl, T.J.; Perea-Schmittle, K.; Wang, W.; Tessier, P.M.; Richardson, C.; Turner, L.; et al. AIntibody: An experimentally validated in silico antibody discovery design challenge. Nat. Biotechnol. 2024, 42, 1637–1642. [Google Scholar] [CrossRef] [PubMed]
- Bennett, N.R.; Coventry, B.; Goreshnik, I.; Huang, B.; Allen, A.; Vafeados, D.; Peng, Y.P.; Dauparas, J.; Baek, M.; Stewart, L.; et al. Improving de novo protein binder design with deep learning. Nat. Commun. 2023, 14, 2625. [Google Scholar] [CrossRef] [PubMed]
- Hie, B.L.; Shanker, V.R.; Xu, D.; Bruun, T.U.J.; Weidenbacher, P.A.; Tang, S.; Wu, W.; Pak, J.E.; Kim, P.S. Efficient evolution of human antibodies from general protein language models. Nat. Biotechnol. 2024, 42, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Pham, P.L.; Kamen, A.; Durocher, Y. Large-scale transfection of mammalian cells for the fast production of recombinant protein. Mol. Biotechnol. 2006, 34, 225–237. [Google Scholar] [CrossRef]
- Geisse, S.; Voedisch, B. Transient expression technologies: Past, present, and future. Methods Mol. Biol. 2012, 899, 203–219. [Google Scholar] [CrossRef]
- Gutierrez-Granados, S.; Cervera, L.; Kamen, A.A.; Godia, F. Advancements in mammalian cell transient gene expression (TGE) technology for accelerated production of biologics. Crit. Rev. Biotechnol. 2018, 38, 918–940. [Google Scholar] [CrossRef] [PubMed]
- Jager, V.; Bussow, K.; Schirrmann, T. Transient Recombinant Protein Expression in Mammalian Cells; Springer Interantional Publishing: Cham, Switzerland, 2015; Volume 9, p. 27. [Google Scholar]
- Dumont, J.; Euwart, D.; Mei, B.; Estes, S.; Kshirsagar, R. Human cell lines for biopharmaceutical manufacturing: History, status, and future perspectives. Crit. Rev. Biotechnol. 2016, 36, 1110–1122. [Google Scholar] [CrossRef]
- Van den Nieuwenhof, I.M.; Koistinen, H.; Easton, R.L.; Koistinen, R.; Kamarainen, M.; Morris, H.R.; Van Die, I.; Seppala, M.; Dell, A.; Van den Eijnden, D.H. Recombinant glycodelin carrying the same type of glycan structures as contraceptive glycodelin-A can be produced in human kidney 293 cells but not in chinese hamster ovary cells. Eur. J. Biochem. 2000, 267, 4753–4762. [Google Scholar] [CrossRef] [PubMed]
- Croset, A.; Delafosse, L.; Gaudry, J.P.; Arod, C.; Glez, L.; Losberger, C.; Begue, D.; Krstanovic, A.; Robert, F.; Vilbois, F.; et al. Differences in the glycosylation of recombinant proteins expressed in HEK and CHO cells. J. Biotechnol. 2012, 161, 336–348. [Google Scholar] [CrossRef]
- Gaudry, J.P.; Arod, C.; Sauvage, C.; Busso, S.; Dupraz, P.; Pankiewicz, R.; Antonsson, B. Purification of the extracellular domain of the membrane protein GlialCAM expressed in HEK and CHO cells and comparison of the glycosylation. Protein Expr. Purif. 2008, 58, 94–102. [Google Scholar] [CrossRef]
- Zhong, X.; Jagarlapudi, S.; Weng, Y.; Ly, M.; Rouse, J.C.; McClure, K.; Ishino, T.; Zhang, Y.; Sousa, E.; Cohen, J.; et al. Structure-function relationships of the soluble form of the antiaging protein Klotho have therapeutic implications for managing kidney disease. J. Biol. Chem. 2020, 295, 3115–3133. [Google Scholar] [CrossRef]
- Zhong, X.; Yan, G.G.; Chaturvedi, A.; Li, X.; Gao, Y.; Girgenrath, M.; Corcoran, C.J.; Diblasio-Smith, L.; LaVallie, E.R.; de Rham, T.; et al. Metabolic Engineering of Glycofusion Bispecific Antibodies for alpha-Dystroglycanopathies. Antibodies 2024, 13, 83. [Google Scholar] [CrossRef]
- Nelson, P.N.; Carnegie, P.R.; Martin, J.; Davari Ejtehadi, H.; Hooley, P.; Roden, D.; Rowland-Jones, S.; Warren, P.; Astley, J.; Murray, P.G. Demystified. Human endogenous retroviruses. Mol. Pathol. 2003, 56, 11–18. [Google Scholar] [CrossRef]
- Liu, C.; Dalby, B.; Chen, W.; Kilzer, J.M.; Chiou, H.C. Transient transfection factors for high-level recombinant protein production in suspension cultured mammalian cells. Mol. Biotechnol. 2008, 39, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Kober, V.; Tellers, M.; Naji, Z.; Salmon, P.; Markusen, J.F. High-level protein expression in scalable CHO transient transfection. Biotechnol. Bioeng. 2009, 103, 542–551. [Google Scholar] [CrossRef]
- Daramola, O.; Stevenson, J.; Dean, G.; Hatton, D.; Pettman, G.; Holmes, W.; Field, R. A high-yielding CHO transient system: Coexpression of genes encoding EBNA-1 and GS enhances transient protein expression. Biotechnol. Prog. 2014, 30, 132–141. [Google Scholar] [CrossRef]
- Rajendra, Y.; Hougland, M.D.; Alam, R.; Morehead, T.A.; Barnard, G.C. A high cell density transient transfection system for therapeutic protein expression based on a CHO GS-knockout cell line: Process development and product quality assessment. Biotechnol. Bioeng. 2015, 112, 977–986. [Google Scholar] [CrossRef]
- Zhong, X.; Ma, W.; Meade, C.L.; Tam, A.S.; Llewellyn, E.; Cornell, R.; Cote, K.; Scarcelli, J.J.; Marshall, J.K.; Tzvetkova, B.; et al. Transient CHO expression platform for robust antibody production and its enhanced N-glycan sialylation on therapeutic glycoproteins. Biotechnol. Prog. 2019, 35, e2724. [Google Scholar] [CrossRef]
- Stuible, M.; Burlacu, A.; Perret, S.; Brochu, D.; Paul-Roc, B.; Baardsnes, J.; Loignon, M.; Grazzini, E.; Durocher, Y. Optimization of a high-cell-density polyethylenimine transfection method for rapid protein production in CHO-EBNA1 cells. J. Biotechnol. 2018, 281, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.G.; White, R.N.; Barnard, G.C. Development of a high cell density transient CHO platform yielding mAb titers greater than 2 g/L in only 7 days. Biotechnol. Prog. 2020, 36, e3047. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Conde, S.; Inman, S.; Lindo, V.; Amery, L.; Tang, A.; Okorji-Obike, U.; Du, W.; Bosch, B.J.; Wichgers Schreur, P.J.; Kortekaas, J.; et al. Suitability of transiently expressed antibodies for clinical studies: Product quality consistency at different production scales. MAbs 2022, 14, 2052228. [Google Scholar] [CrossRef]
- Steger, K.; Brady, J.; Wang, W.; Duskin, M.; Donato, K.; Peshwa, M. CHO-S antibody titers >1 gram/liter using flow electroporation-mediated transient gene expression followed by rapid migration to high-yield stable cell lines. J. Biomol. Screen. 2015, 20, 545–551. [Google Scholar] [CrossRef]
- Zhou, J.; Yan, G.G.; Cluckey, D.; Meade, C.; Ruth, M.; Sorm, R.; Tam, A.S.; Lim, S.; Petridis, C.; Lin, L.; et al. Exploring Parametric and Mechanistic Differences between Expi293F(TM) and ExpiCHO-S(TM) Cells for Transient Antibody Production Optimization. Antibodies 2023, 12, 53. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, D.E.; Mahan, E.R.; Ma, W.; Bitzas, G.; Zhong, X.; Zollner, R.; D’Antona, A.M. Parallel loading and complete automation of a 3-step mAb purification process for multiple samples using a customized preparative chromatography instrument with networked pumps. J. Chromatogr. A 2018, 1542, 50–60. [Google Scholar] [CrossRef]
- Tang, S.; Tao, J.; Li, Y. Challenges and solutions for the downstream purification of therapeutic proteins. Antib. Ther. 2024, 7, 1–12. [Google Scholar] [CrossRef]
- Li, Q.; Zhao, H.; Liang, X.; He, Q.; Wang, Z.; Qin, G.; Li, G.; Xu, D. The downstream purification of bispecific antibodies. Anal. Biochem. 2025, 696, 115692. [Google Scholar] [CrossRef]
- Franke, B.; Frigard, T.; Grzesiek, S.; Isogai, S. Versatile modules enable automated multi-column purifications on the AKTA pure chromatography system. J. Chromatogr. A 2020, 1618, 460846. [Google Scholar] [CrossRef]
- Yoo, D.; Provchy, J.; Park, C.; Schulz, C.; Walker, K. Automated high-throughput protein purification using an AKTApurifier and a CETAC autosampler. J. Chromatogr. A 2014, 1344, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, S.; Schonfeld, D.L.; Freitag, B.; Gotzberger-Schad, C.; Fischer, M.; Linden, L. Keeping pace with the increasing demand for high quality drug candidates in pharmaceutical research: Development of a new two-step preparative tandem high performance chromatographic system for the purification of antibodies. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2019, 1104, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Wurm, F.M. CHO quasispecies-implications for manufacturing processes. Processes 2013, 1, 296–311. [Google Scholar] [CrossRef]
- Wurm, F.M.; Wurm, M.J.; Rodrigues, M.L.; Kiseljak, D.; Burki, C.; Pugin, C.; Raussin, G.; Heymoz, J. Eukaryotic Cell Transfections Systems and Related Methods. U.S. Patent 11,746,360 B2, 5 September 2023. [Google Scholar]
- Zhu, L.; Monteil, D.T.; Wang, Y.; Song, B.; Hacker, D.L.; Wurm, M.J.; Li, X.; Wang, Z.; Wurm, F.M. Fluid dynamics of flow fields in a disposable 600-mL orbitally shaken bioreactor. Biochem. Eng. J. 2018, 129, 84–95. [Google Scholar] [CrossRef]
- Stuible, M.; van Lier, F.; croughan, M.S.; Durocher, Y. Beyond preclinical research: Production of CHO-derived biotherapeutics for toxicology and early-phase trials by transient gene expression or stable pools. Curr. Opin. Chem. Eng. 2018, 22, 145–151. [Google Scholar] [CrossRef]
- Borlido, L.; Azevedo, A.M.; Roque, A.C.; Aires-Barros, M.R. Magnetic separations in biotechnology. Biotechnol. Adv. 2013, 31, 1374–1385. [Google Scholar] [CrossRef]
- Yavuz, C.T.; Prakash, A.; Mayo, J.T.; Colvin, V.L. Magnetic separations: From steel plants to biotechnology. Chem. Eng. Sci. 2009, 64, 2510–2521. [Google Scholar] [CrossRef]
- Brechmann, N.A.; Schwarz, H.; Eriksson, P.O.; Eriksson, K.; Shokri, A.; Chotteau, V. Antibody capture process based on magnetic beads from very high cell density suspension. Biotechnol. Bioeng. 2021, 118, 3499–3510. [Google Scholar] [CrossRef] [PubMed]
- Brechmann, N.A.; Eriksson, P.O.; Eriksson, K.; Oscarsson, S.; Buijs, J.; Shokri, A.; Hjalm, G.; Chotteau, V. Pilot-scale process for magnetic bead purification of antibodies directly from non-clarified CHO cell culture. Biotechnol. Prog. 2019, 35, e2775. [Google Scholar] [CrossRef] [PubMed]
- Ebeler, M.; Lind, O.; Norrman, N.; Palmgren, R.; Franzreb, M. One-step integrated clarification and purification of a monoclonal antibody using Protein A Mag Sepharose beads and a cGMP-compliant high-gradient magnetic separator. New Biotechnol. 2018, 42, 48–55. [Google Scholar] [CrossRef]
- Winters, D.; Tran, M.; Yoo, D.; Walker, K.W. Development of BioRad NGC and GE AKTA pure systems for highly automated three column protein purification employing tandem affinity, buffer exchange and size exclusion chromatography. Protein Expr. Purif. 2020, 165, 105497. [Google Scholar] [CrossRef]
- Holenstein, F.; Eriksson, C.; Erlandsson, I.; Norrman, N.; Simon, J.; Danielsson, A.; Milicov, A.; Schindler, P.; Schlaeppi, J.M. Automated harvesting and 2-step purification of unclarified mammalian cell-culture broths containing antibodies. J. Chromatogr. A 2015, 1418, 103–109. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gebhardt, L.; Abel, M.; Zhou, J.; Vogt, A.M.; Shin, B.H.; Herrick Wagman, S.L.; Santos, A.; Puginier, J.; Wurm, F.M.; Wurm, M.J.; et al. A Robust, High-Titer, Semi-Automated, and In-Culture Antibody-Capturing Transient CHO Platform Technology. Antibodies 2025, 14, 87. https://doi.org/10.3390/antib14040087
Gebhardt L, Abel M, Zhou J, Vogt AM, Shin BH, Herrick Wagman SL, Santos A, Puginier J, Wurm FM, Wurm MJ, et al. A Robust, High-Titer, Semi-Automated, and In-Culture Antibody-Capturing Transient CHO Platform Technology. Antibodies. 2025; 14(4):87. https://doi.org/10.3390/antib14040087
Chicago/Turabian StyleGebhardt, Lauren, Molica Abel, Jing Zhou, Audrey M. Vogt, Bo Hee Shin, Sarah L. Herrick Wagman, Ana Santos, Jerome Puginier, Florian M. Wurm, Maria J. Wurm, and et al. 2025. "A Robust, High-Titer, Semi-Automated, and In-Culture Antibody-Capturing Transient CHO Platform Technology" Antibodies 14, no. 4: 87. https://doi.org/10.3390/antib14040087
APA StyleGebhardt, L., Abel, M., Zhou, J., Vogt, A. M., Shin, B. H., Herrick Wagman, S. L., Santos, A., Puginier, J., Wurm, F. M., Wurm, M. J., Yan, G. G., Adeniyi, A., Lim, S. K. H., Somers, W., Lin, L., D’Antona, A. M., & Zhong, X. (2025). A Robust, High-Titer, Semi-Automated, and In-Culture Antibody-Capturing Transient CHO Platform Technology. Antibodies, 14(4), 87. https://doi.org/10.3390/antib14040087






