Detection of Viable Bacteria during Sludge Ozonation by the Combination of ATP Assay with PMA-Miseq Sequencing
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
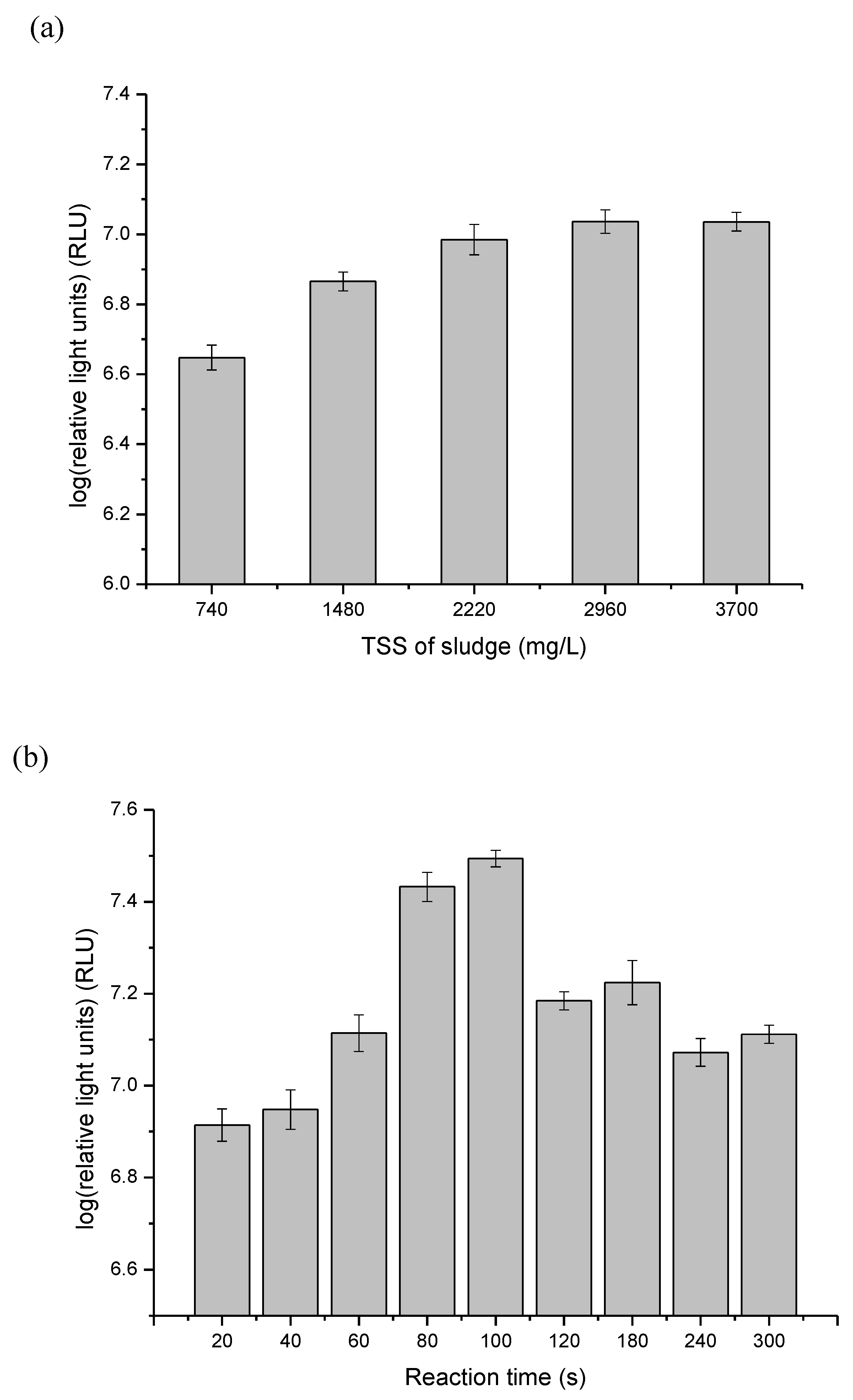
3.1. Optimize ATP Assay for Fast Detection of the Quantity of Viable Biomass during Sludge Ozonation
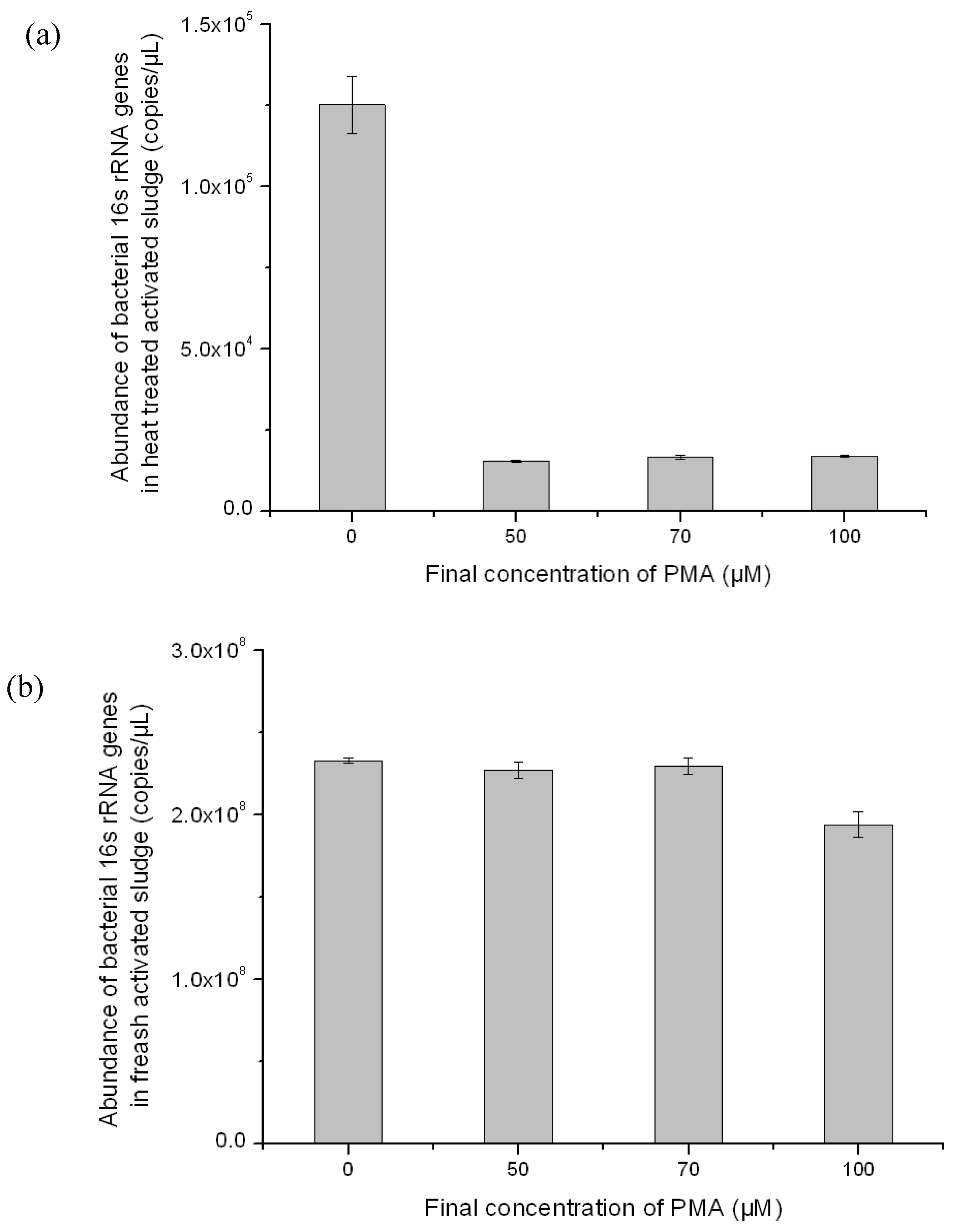
3.2. Optimization of PMA-Modified PCR for Selectively Amplifying the 16S rRNA Genes of Viable Bacteria in Activated Sludge
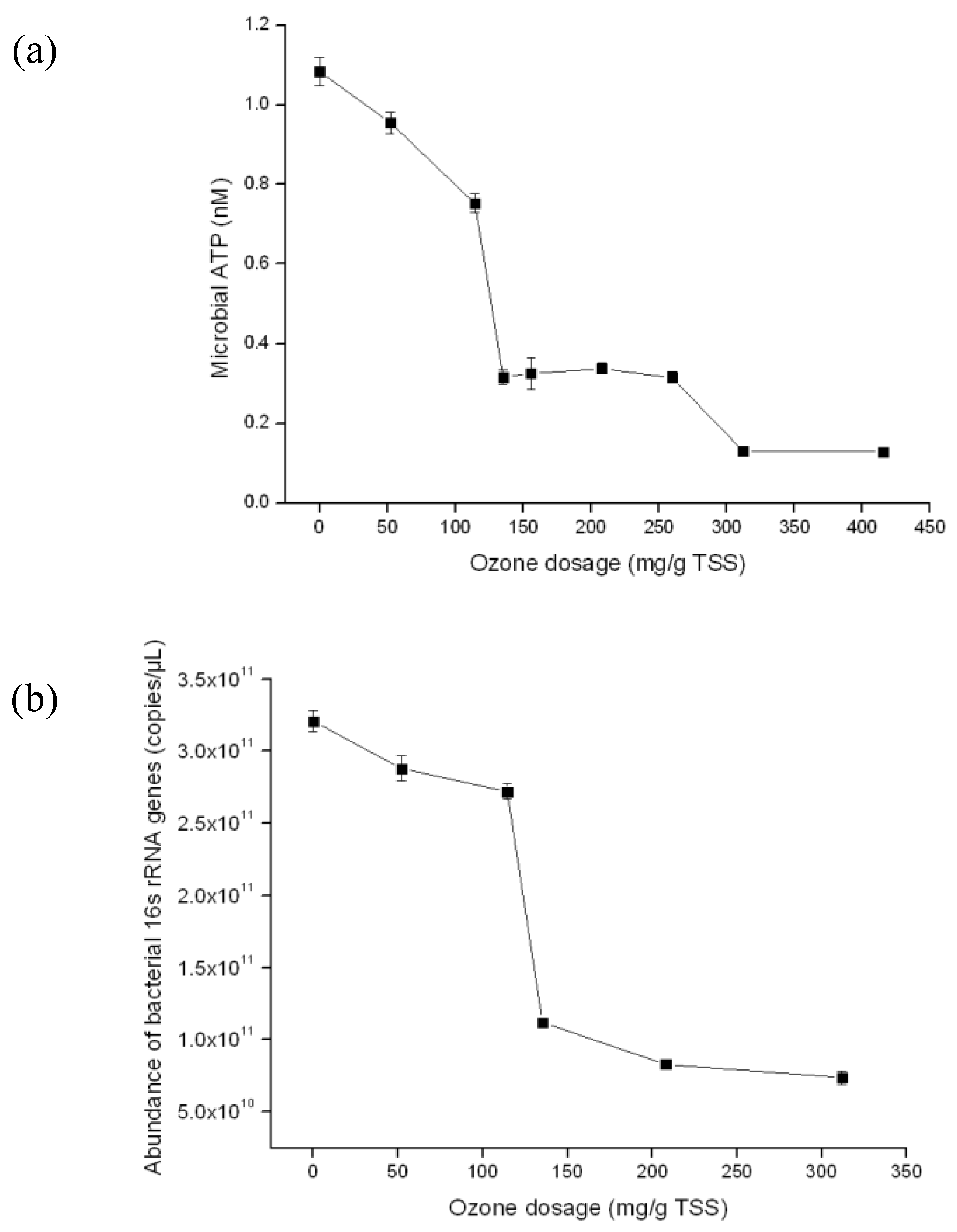
3.3. Detection of the Quantity of Viable Bacterial Populations during Sludge Ozonation Using Optimized ATP Assay and PMA-qPCR
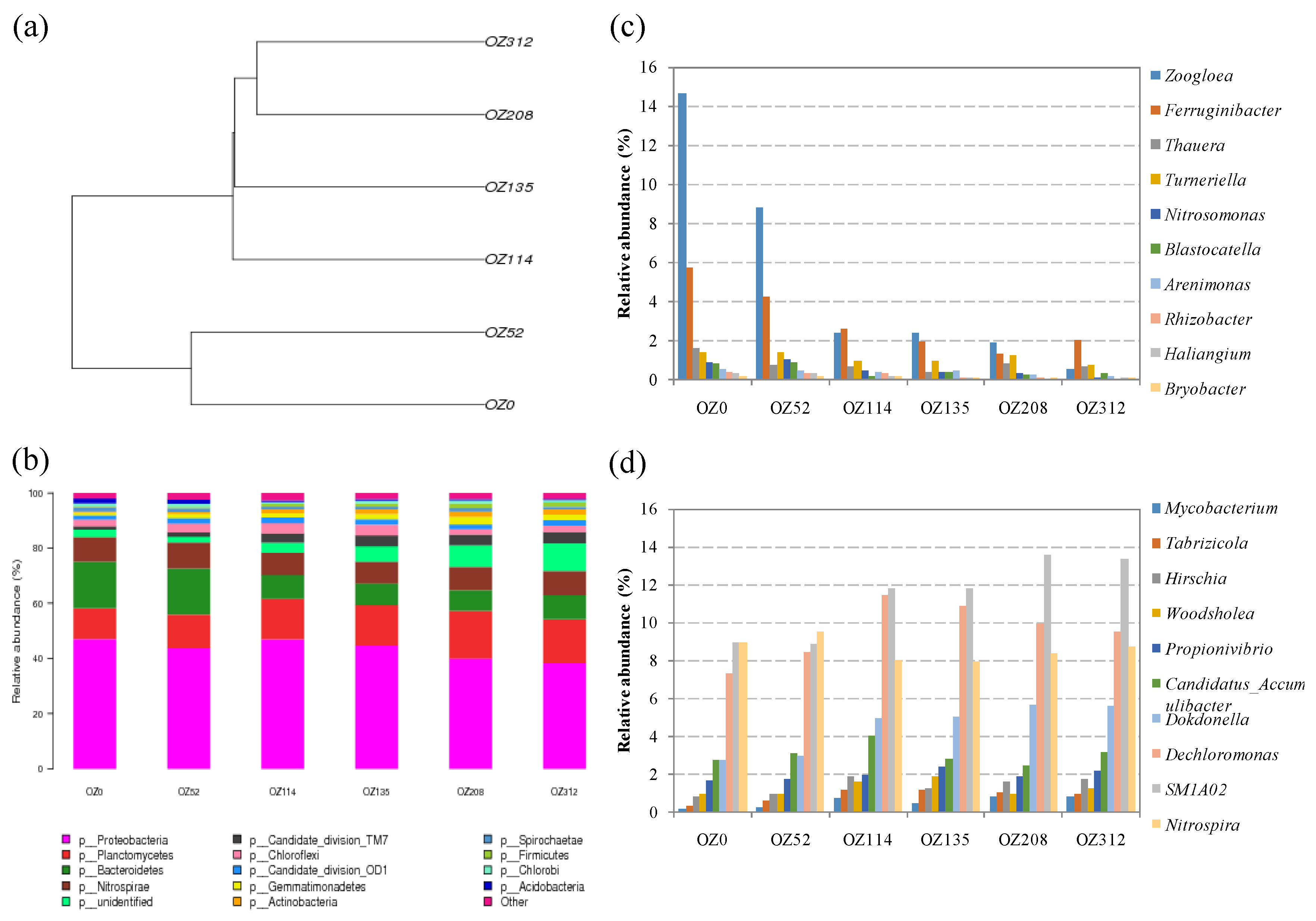
3.4. Detection of the Composition of Viable Bacterial Populations during Sludge Ozonation Using Optimized PMA-Miseq Sequencing
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Spellman, F. Wastewater Biosolids to Compost; Technomic Publishing Company: Lancaster, PA, USA, 1997; pp. 223–235. [Google Scholar]
- Wei, Y.S.; Van Houten, R.T.; Borger, A.R.; Eikelboom, D.H.; Fan, Y.B. Minimization of excess sludge production for biological wastewater treatment. Water Res. 2003, 37, 4453–4467. [Google Scholar] [CrossRef]
- Yan, S.; Miyanaga, K.; Xing, X.-H.; Tanji, Y. Succession of bacterial community and enzymatic activities of activated sludge by heat-treatment for reduction of excess sludge. Biochem. Eng. J. 2008, 39, 598–603. [Google Scholar] [CrossRef]
- Yasui, H.; Shibata, M. An innovative approach to reduce excess sludge production in the activated-sludge process. Water Sci. Technol. 1994, 30, 11–20. [Google Scholar]
- Yan, S.-T.; Chu, L.-B.; Xing, X.-H.; Yu, A.-F.; Sun, X.-L.; Jurcik, B. Analysis of the mechanism of sludge ozonation by a combination of biological and chemical approaches. Water Res. 2009, 43, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Komanapalli, I.R.; Lau, B.H.S. Ozone-induced damage of Escherichia coli K-12. Appl. Microbiol. Biotechnol. 1996, 46, 610–614. [Google Scholar] [CrossRef] [PubMed]
- Hysert, D.W.; Knudsen, F.B.; Morrison, N.M.; Vangheluwe, G.; Lom, T. Application of a bioluminescence ATP assay in brewery wastewater-treatment studies. Biotechnol. Bioeng. 1979, 21, 1301–1314. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, W.; Liu, R.; Li, Q.; Li, B.; Wang, S.; Song, C.; Qiao, C.; Mulchandani, A. Phylogenetic diversity and metabolic potential of activated sludge microbial communities in full-scale wastewater treatment plants. Environ. Sci. Technol. 2011, 45, 7408–7415. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Martinez, A.; Rodriguez-Sanchez, A.; Lotti, T.; Garcia-Ruiz, M.-J.; Osorio, F.; Gonzalez-Lopez, J.; van Loosdrecht, M.C.M. Comparison of bacterial communities of conventional and A-stage activated sludge systems. Sci. Rep. 2016, 6, 18786. [Google Scholar] [CrossRef] [PubMed]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in-situ detection of individual microbial-cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [PubMed]
- Vang, O.K.; Corfitzen, C.B.; Smith, C.; Albrechtsen, H.-J. Evaluation of ATP measurements to detect microbial ingress by wastewater and surface water in drinking water. Water Res. 2014, 64, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Van der Wielen, P.W.J.J.; van der Kooij, D. Effect of water composition, distance and season on the adenosine triphosphate concentration in unchlorinated drinking water in The Netherlands. Water Res. 2010, 44, 4860–4867. [Google Scholar] [CrossRef] [PubMed]
- Deininger, R.A.; Lee, J. Rapid determination of bacteria in drinking water using an ATP assay. Field Anal. Chem. Technol. 2001, 5, 185–189. [Google Scholar] [CrossRef]
- Delahaye, E.; Welte, B.; Levi, Y.; Leblon, G.; Montiel, A. An ATP-based method for monitoring the microbiological drinking water quality in a distribution network. Water Res. 2003, 37, 3689–3696. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Shao, M.-F.; Ye, L. 454 pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J. 2012, 6, 1137–1147. [Google Scholar] [CrossRef] [PubMed]
- Nocker, A.; Cheung, C.-Y.; Camper, A.K. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 2006, 67, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Rogers, G.; Cuthbertson, L.; Hoffman, L.; Wing, P.; Pope, C.; Hooftman, D.; Lilley, A.; Oliver, A.; Carroll, M.; Bruce, K.; et al. Reducing bias in bacterial community analysis of lower respiratory infections. ISME J. 2013, 7, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Zhang, T. Detecting the nonviable and heat-tolerant bacteria in activated sludge by minimizing DNA from dead cells. Microbial Ecol. 2014, 67, 829–836. [Google Scholar] [CrossRef] [PubMed]
- Nocker, A.; Richter-Heitmann, T.; Montijn, R.; Schuren, F.; Kort, R. Discrimination between live and dead cells in bacterial communities from environmental water samples analyzed by 454 pyrosequencing. Int. Microbiol. 2010, 13, 59–65. [Google Scholar] [PubMed]
- Ahn, K.H.; Park, K.Y.; Maeng, S.K.; Hwang, J.H.; Lee, J.W.; Song, K.G.; Choi, S. Ozonation of wastewater sludge for reduction and recycling. Water Sci. Technol. 2002, 46, 71–77. [Google Scholar] [PubMed]
- Chu, L.-B.; Yan, S.-T.; Xing, X.-H.; Yu, A.-F.; Sun, X.-L.; Jurcik, B. Enhanced sludge solubilization by microbubble ozonation. Chemosphere 2008, 72, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Dytczak, M.A.; Londry, K.L.; Siegrist, H.; Oleszkiewicz, J.A. Ozonation reduces sludge production and improves denitrification. Water Res. 2007, 41, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Goel, R.; Komatsu, K.; Yasui, H.; Harada, H. Process performance and change in sludge characteristics during anaerobic digestion of sewage sludge with ozonation. Water Sci. Technol. 2004, 49, 105–113. [Google Scholar] [PubMed]
- Pires, M.; Carvalho, L. An artifact in air carbonyls sampling using C-18 DNPH-coated cartridge. Anal. Chim. Acta 1998, 367, 223–231. [Google Scholar] [CrossRef]
- Abzazou, T.; Salvado, H.; Bruguera-Casamada, C.; Simon, P.; Lardin, C.; Araujo, R.M. Assessment of total bacterial cells in extended aeration activated sludge plants using flow cytometry as a microbial monitoring tool. Environ. Sci. Pollut. Res. 2015, 22, 11446–11455. [Google Scholar] [CrossRef] [PubMed]
- Hammes, F.; Goldschmidt, F.; Vital, M.; Wang, Y.; Egli, T. Measurement and interpretation of microbial adenosine tri-phosphate (ATP) in aquatic environments. Water Res. 2010, 44, 3915–3923. [Google Scholar] [CrossRef] [PubMed]
- Taskin, B.; Gozen, A.G.; Duran, M. Selective quantification of viable Escherichia coli bacteria in biosolids by quantitative PCR with propidium monoazide modification. Appl. Environ. Microbiol. 2011, 77, 4329–4335. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhang, Y.; Yang, M.; Tian, Z.; Ren, L.; Zhang, S. Abundance and distribution of tetracycline resistance genes and mobile elements in an oxytetracycline production wastewater treatment system. Environ. Sci. Technol. 2012, 46, 7551–7557. [Google Scholar] [CrossRef] [PubMed]
- Pei, R.; Kim, S.-C.; Carlson, K.H.; Pruden, A. Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res. 2006, 40, 2427–2435. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Liu, Y.; Lin, X.; Zhang, H.; Zeng, J.; Hou, J.; Yang, Y.; Yao, T.; Knight, R.; Chu, H. Geographic distance and pH drive bacterial distribution in alkaline lake sediments across Tibetan Plateau. Environ. Microbiol. 2012, 14, 2457–2466. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. Qiime allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
- Boe-Hansen, R.; Albrechtsen, H.J.; Arvin, E.; Jorgensen, C. Bulk water phase and biofilm growth in drinking water at low nutrient conditions. Water Res. 2002, 36, 4477–4486. [Google Scholar] [CrossRef]
- Ikonen, J.; Pitkanen, T.; Miettinen, I.T. Suitability of optical, physical and chemical measurements for detection of changes in bacterial drinking water quality. Int. J. Environ. Res. Public Health 2013, 10, 5349–5363. [Google Scholar] [CrossRef] [PubMed]
- Kramer, M.; Obermajer, N.; Matijasic, B.B.; Rogelj, I.; Kmetec, V. Quantification of live and dead probiotic bacteria in lyophilised product by real-time PCR and by flow cytometry. Appl. Microbiol. Biotechnol. 2009, 84, 1137–1147. [Google Scholar] [CrossRef] [PubMed]
- Cawthorn, D.M.; Witthuhn, R.C. Selective PCR detection of viable enterobacter sakazakii cells utilizing propidium monoazide or ethidium bromide monoazide. J. Appl. Microbiol. 2008, 105, 1178–1185. [Google Scholar] [CrossRef] [PubMed]
- Fittipaldi, M.; Pino Rodriguez, N.J.; Codony, F.; Adrados, B.; Penuela, G.A.; Morato, J. Discrimination of infectious bacteriophage t4 virus by propidium monoazide real-time PCR. J. Virol. Methods 2010, 168, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Oethinger, M.; Tuohy, M.J.; Hall, G.S.; Bauer, T.W. Improving clinical significance of PCR: Use of propidium monoazide to distinguish viable from dead staphylococcus aureus and staphylococcus epidermidis. J. Orthop. Res. 2009, 27, 1243–1247. [Google Scholar] [CrossRef] [PubMed]
- Rawsthorne, H.; Dock, C.N.; Jaykus, L.A. PCR-based method using propidium monoazide to distinguish viable from nonviable bacillus subtilis spores. Appl. Environ. Microbiol. 2009, 75, 2936–2939. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.-C.; Xi, J.-Y.; Xu, Y.; Huo, Z.-Y.; Hu, H.-Y. Shifts of live bacterial community in secondary effluent by chlorine disinfection revealed by Miseq high-throughput sequencing combined with propidium monoazide treatment. Appl. Microbiol. Biotechnol. 2016, 100, 6435–6446. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jin, J.-H.; Liu, H.-C.; Liu, Z.-P. Dokdonella immobilis sp. nova, isolated from a batch reactor for the treatment of triphenylmethane dye effluent. Int. J. Syst. Evolut. Microbiol. 2013, 63, 1557–1561. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.K.; Hiraishi, A.; Sugiyama, J. Molecular systematics of the genus Zoogloea and emendation of the genus. Int. J. Syst. Bacteriol. 1993, 43, 826–831. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, T.R.; Kong, Y.; Nielsen, P.H. Ecophysiology of abundant denitrifying bacteria in activated sludge. Fems Microbiol. Ecol. 2007, 60, 370–382. [Google Scholar] [CrossRef] [PubMed]
- Pal, P.; Khairnar, K.; Paunikar, W.N. Causes and remedies for filamentous foaming in activated sludge treatment plant. Glob. Nest J. 2014, 16, 762–772. [Google Scholar]
- Chu, Z.-R.; Wang, K.; Li, X.-K.; Zhu, M.-T.; Yang, L.; Zhang, J. Microbial characterization of aggregates within a one-stage nitritation-anammox system using high-throughput amplicon sequencing. Chem. Eng. J. 2015, 262, 41–48. [Google Scholar] [CrossRef]
- Wei, C.; He, W.; Wei, L.; Li, C.; Ma, J. The analysis of a microbial community in the UV/O3-anaerobic/aerobic integrated process for petrochemical nanofiltration concentrate (NFC) treatment by 454-pyrosequencing. PLoS ONE 2015, 10, e0139991. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.-W.; Zhang, H.-Y.; Lu, P.-F.; Peng, Y.-Z. Effects of carbon sources on the enrichment of halophilic polyhydroxyalkanoate-storing mixed microbial culture in an aerobic dynamic feeding process. Sci. Rep. 2016, 6, 30766. [Google Scholar] [CrossRef] [PubMed]
- Seviour, R.J.; Mino, T.; Onuki, M. The microbiology of biological phosphorus removal in activated sludge systems. Fems Microbiol. Rev. 2003, 27, 99–127. [Google Scholar] [CrossRef]
- Lee, J.W.; Cha, H.Y.; Park, K.Y.; Song, K.G.; Ahn, K.H. Operational strategies for an activated sludge process in conjunction with ozone oxidation for zero excess sludge production during winter season. Water Res. 2005, 39, 1199–1204. [Google Scholar] [CrossRef] [PubMed]
- Prabakaran, M.; Tamil Selvi, S.; Merinal, S.; Panneerselvam, A. Effect of ozonation on pathogenic bacteria. Adv. Appl. Sci. Res. 2012, 3, 299–302. [Google Scholar]
- Crocetti, G.; Hugenholtz, P.; Bond, P.L.; Schuler, A.; Keller, J.; Jenkins, D.; Blackall, L. Identification of polyphosphate-accumulating organisms and design of 16S rRNA-directed probes for their detection and quantitation. Appl. Environ. Microbiol. 2000, 66, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- Dziurla, M.A.; Salhi, M.; Leroy, P.; Paul, E.; Ginestet, P.; Block, J.C. Variations of respiratory activity and glutathione in activated sludges exposed to low ozone doses. Water Res. 2005, 39, 2591–2598. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, S.; Tian, Z.; Yang, H.; Yang, M.; Zhang, Y. Detection of Viable Bacteria during Sludge Ozonation by the Combination of ATP Assay with PMA-Miseq Sequencing. Water 2017, 9, 166. https://doi.org/10.3390/w9030166
Tian S, Tian Z, Yang H, Yang M, Zhang Y. Detection of Viable Bacteria during Sludge Ozonation by the Combination of ATP Assay with PMA-Miseq Sequencing. Water. 2017; 9(3):166. https://doi.org/10.3390/w9030166
Chicago/Turabian StyleTian, Shaonan, Zhe Tian, Hong Yang, Min Yang, and Yu Zhang. 2017. "Detection of Viable Bacteria during Sludge Ozonation by the Combination of ATP Assay with PMA-Miseq Sequencing" Water 9, no. 3: 166. https://doi.org/10.3390/w9030166
APA StyleTian, S., Tian, Z., Yang, H., Yang, M., & Zhang, Y. (2017). Detection of Viable Bacteria during Sludge Ozonation by the Combination of ATP Assay with PMA-Miseq Sequencing. Water, 9(3), 166. https://doi.org/10.3390/w9030166





