Contrasting Fauna in Two Neighboring Territories of the African Horn: A Case of the Genus Moina Baird, 1850 (Cladocera: Moinidae)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Morphological Analysis
2.2. Genetic Analysis
3. Results
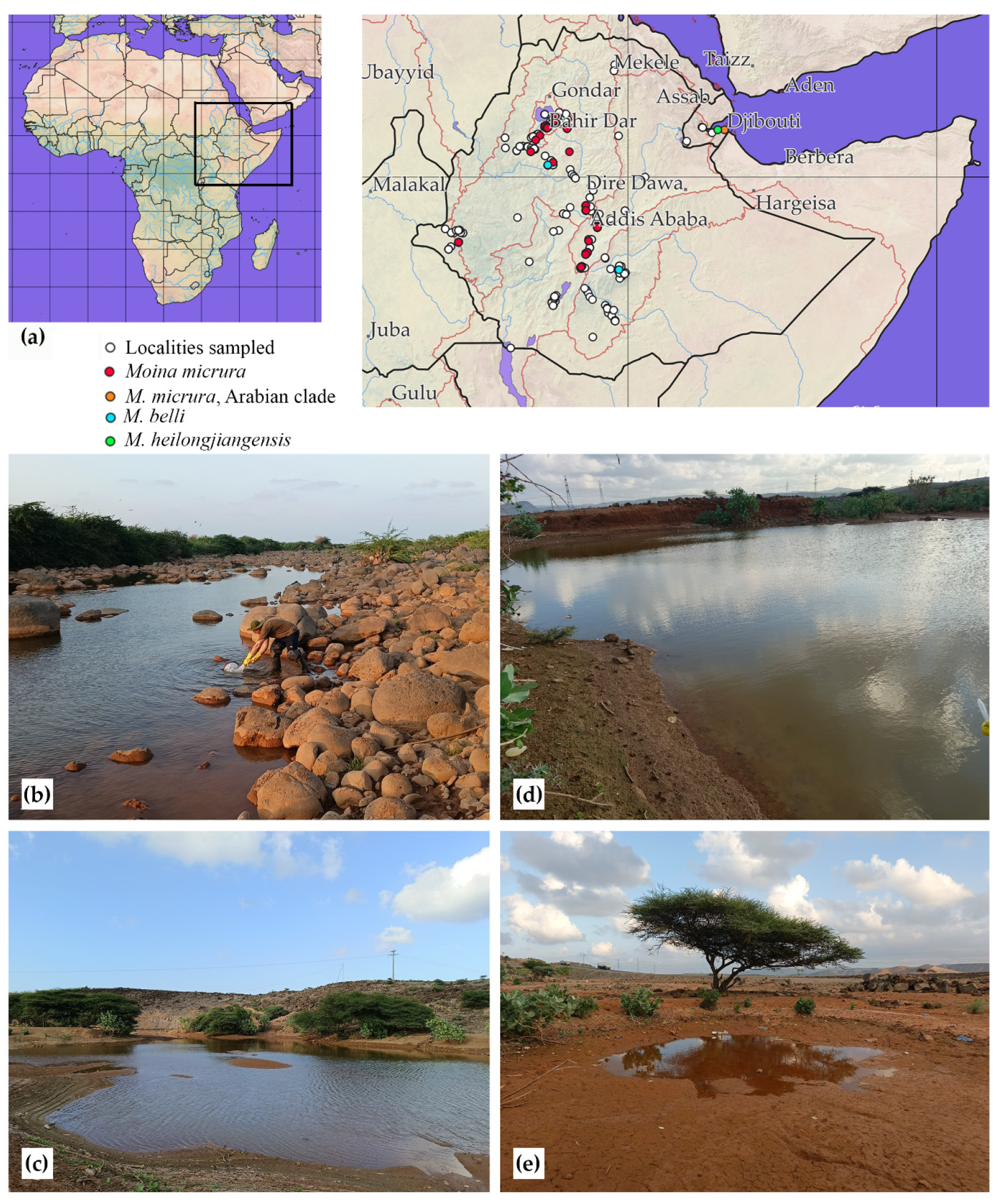
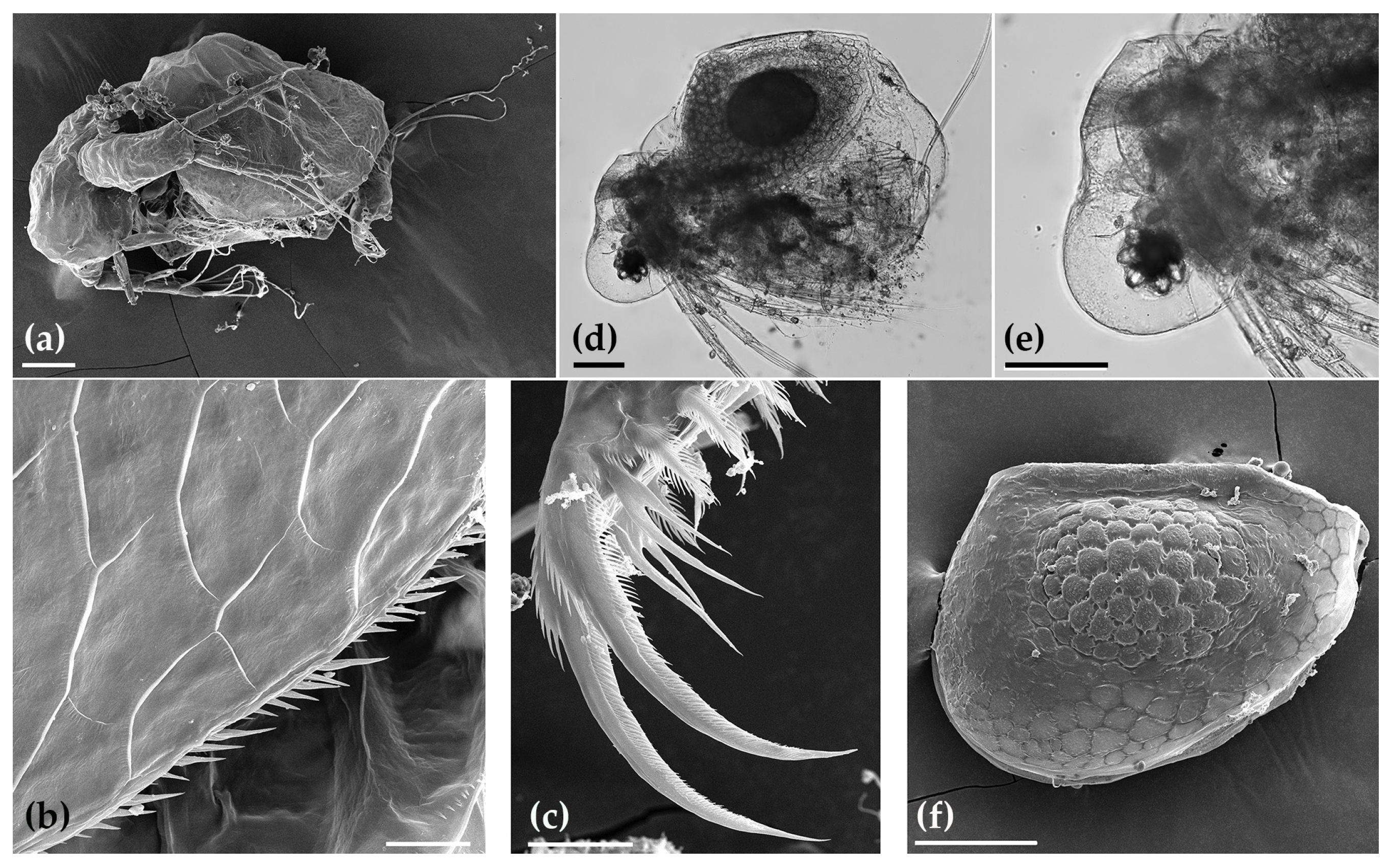
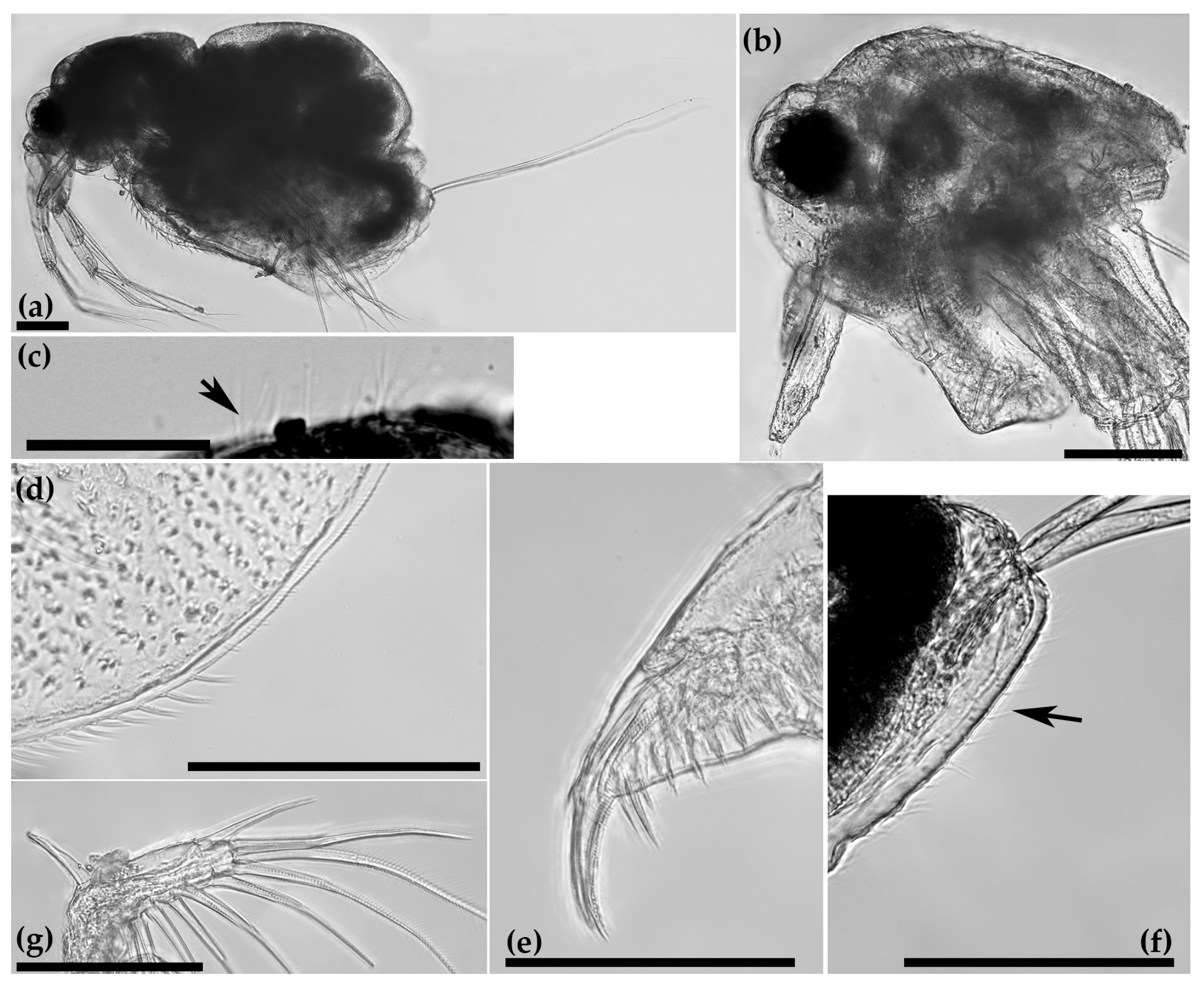
3.1. Faunistic and Taxonomic Account
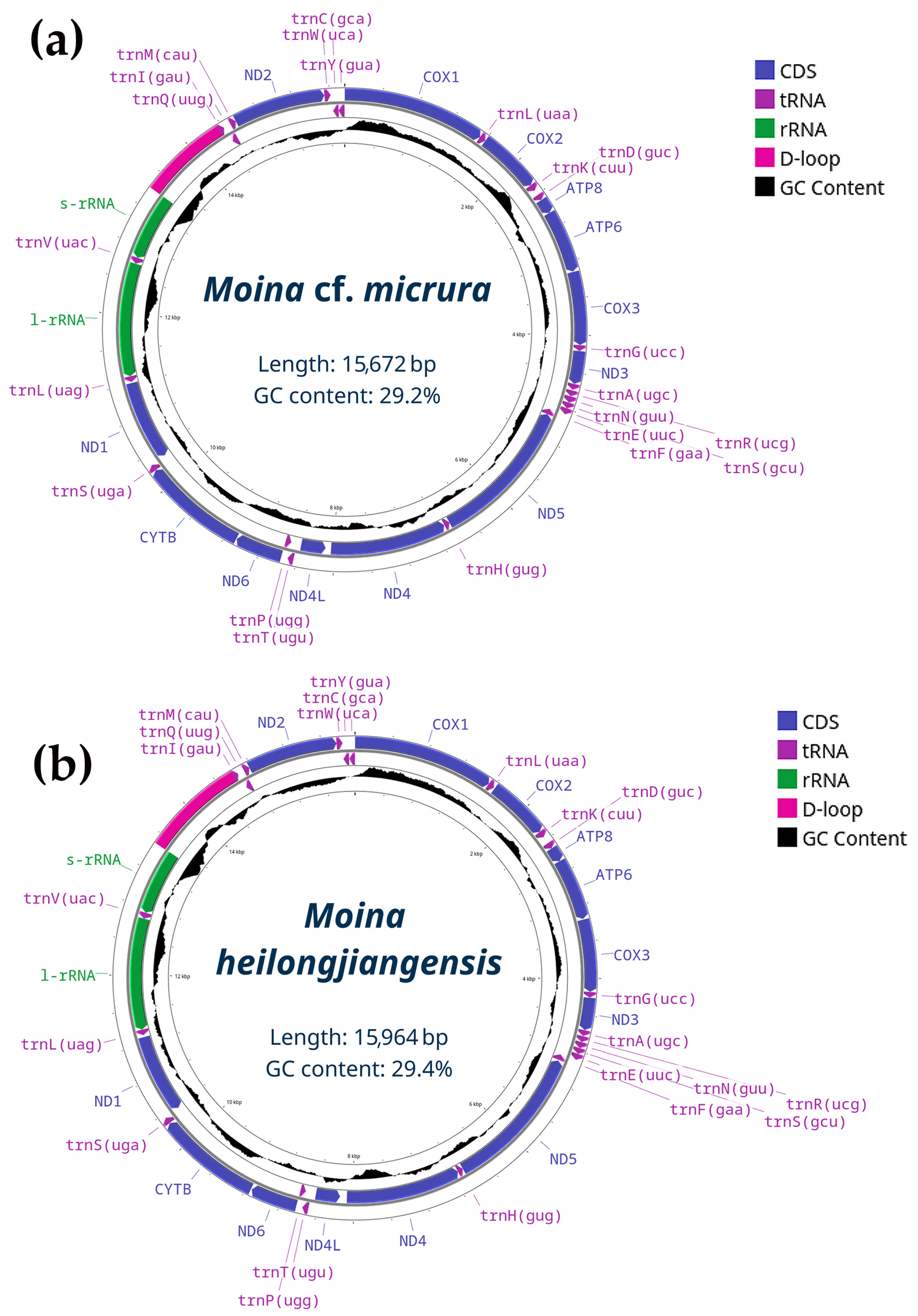
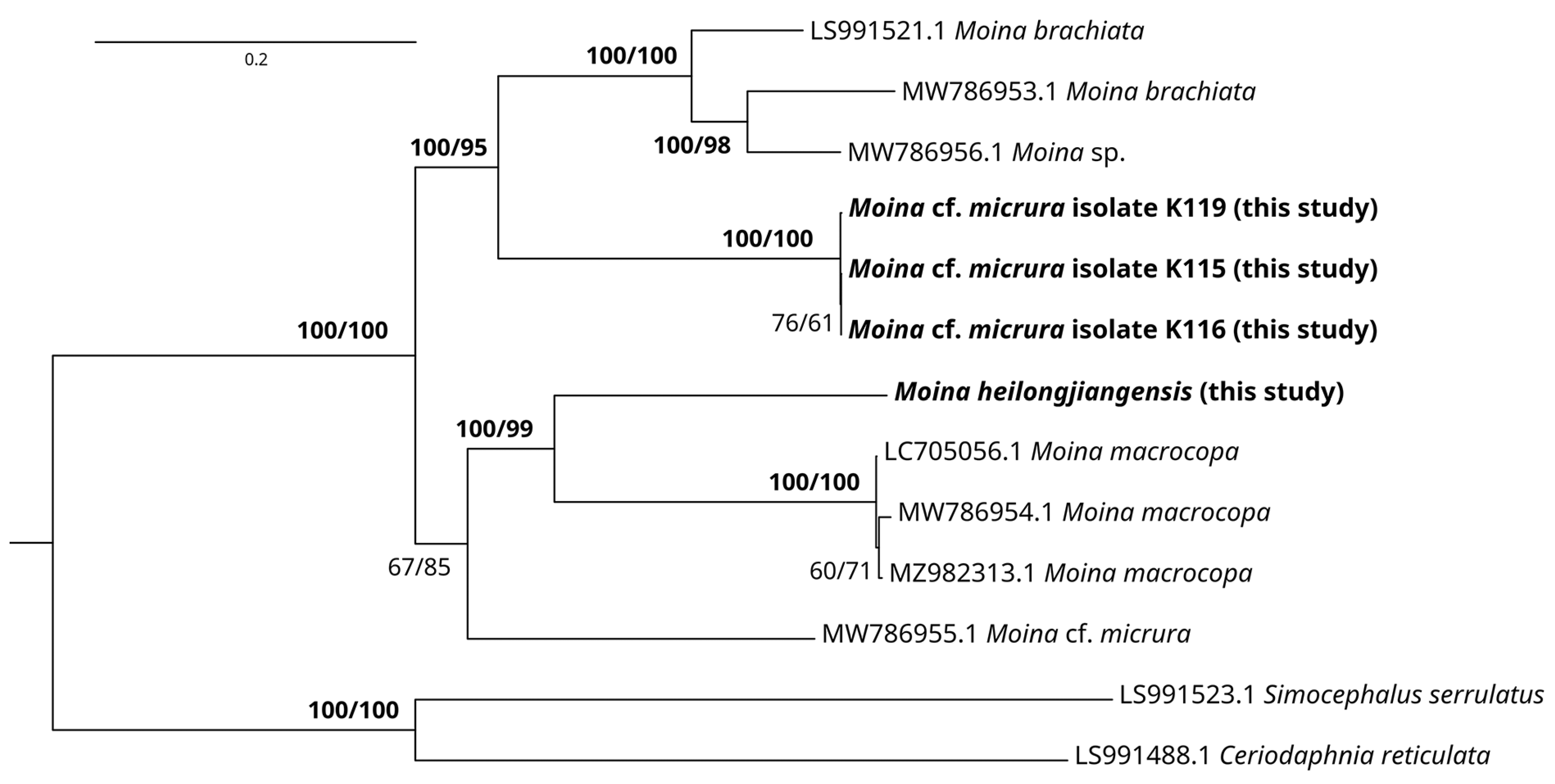
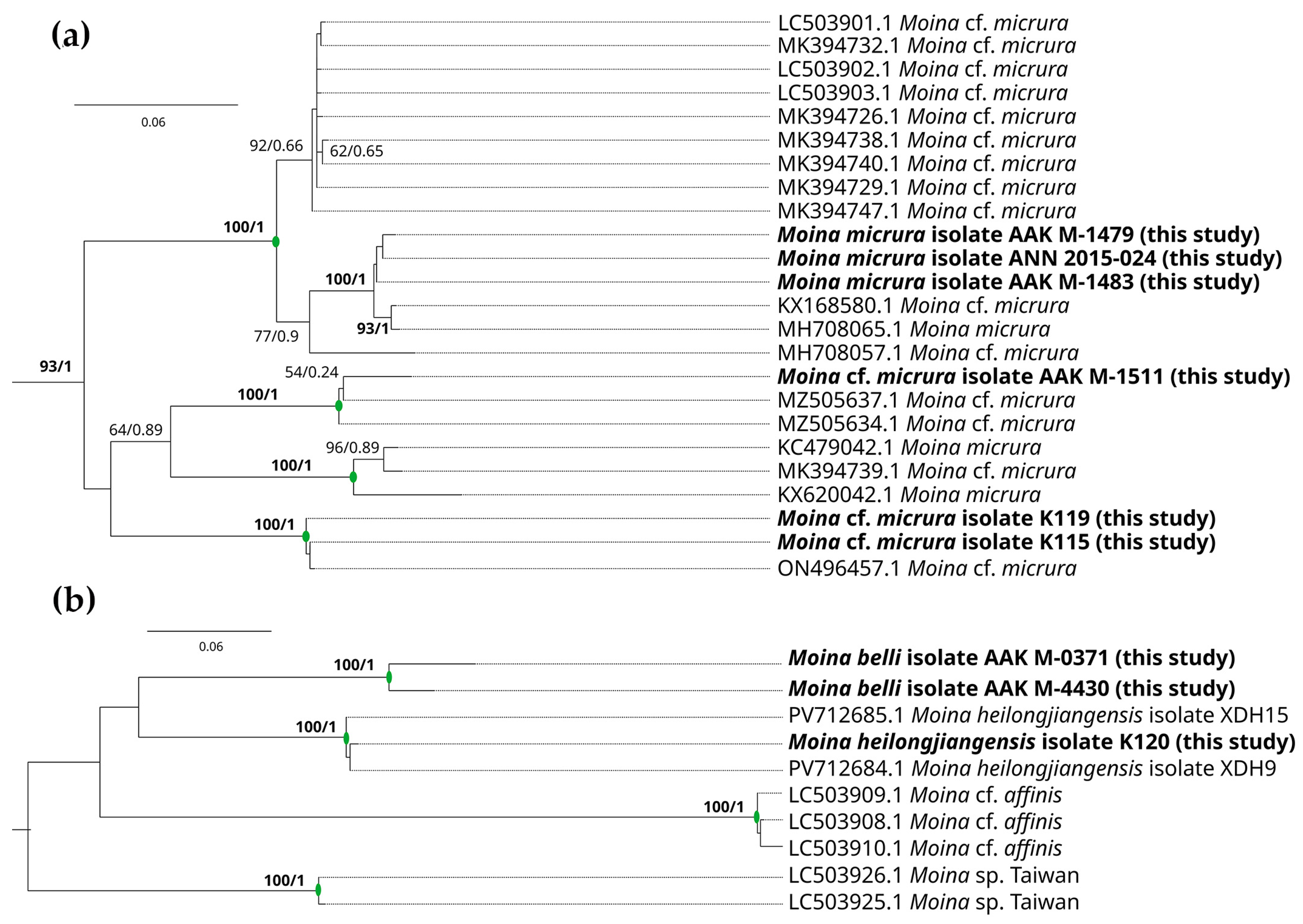
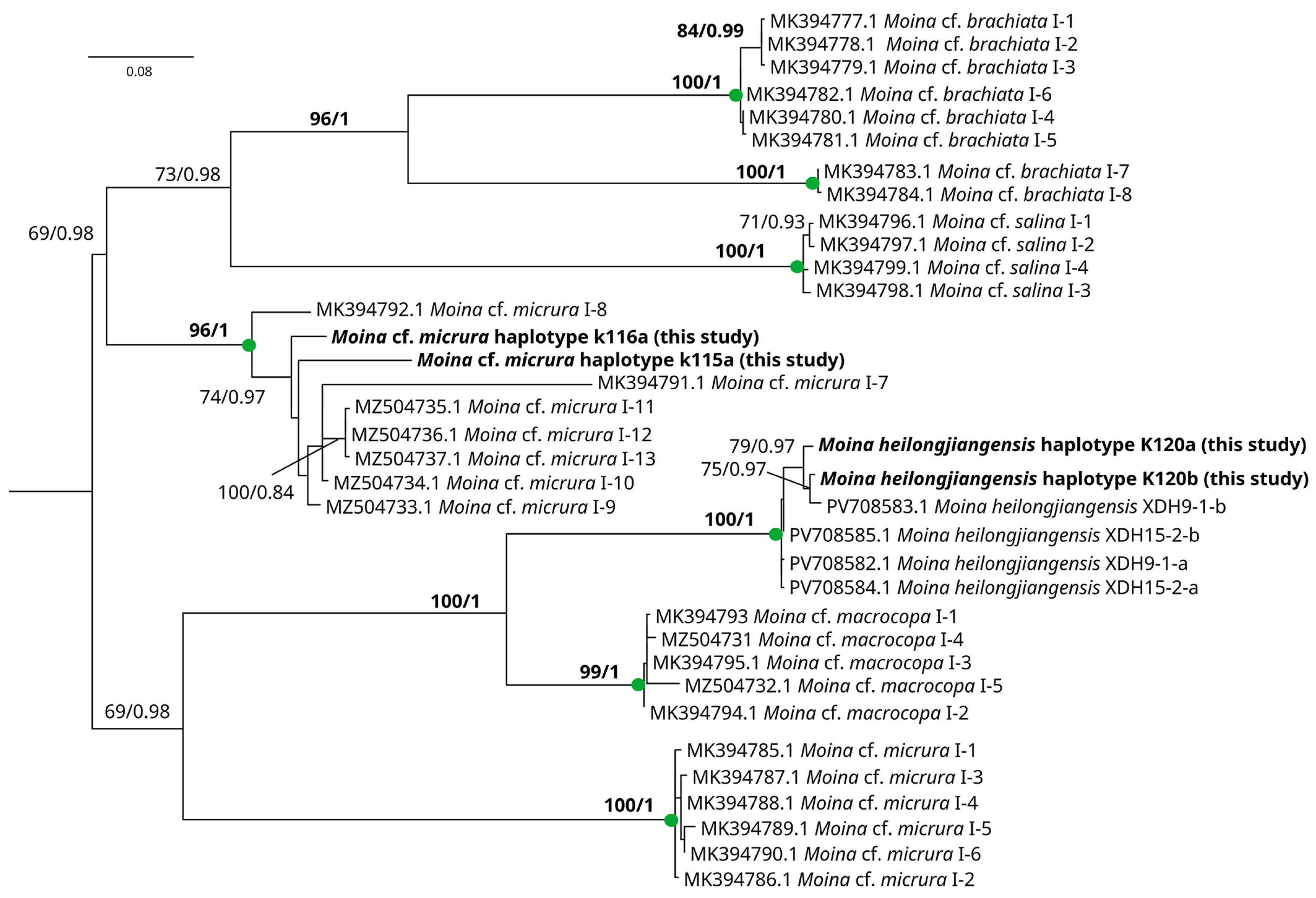
3.2. Genetic Account
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goulden, C.E. The Systematics and Evolution of the Moinidae. Trans. Am. Philos. Soc. 1968, 58, 1–101. [Google Scholar] [CrossRef]
- Streletskaya, E.A. Review of the fauna of Rotatoria, Cladocera, and Copepoda of the basin of the Anadyr’ River. Contemp. Probl. Ecol. 2010, 3, 469–480. [Google Scholar] [CrossRef]
- Jeffery, N.W.; Elías-Gutiérrez, M.; Adamowicz, S.J. Species diversity and phylogeographical affinities of the Branchiopoda (Crustacea) of Churchill, Manitoba, Canada. PLoS ONE 2011, 6, e18364. [Google Scholar] [CrossRef]
- Smirnov, N.N. Mesozoic Anomopoda (Crustacea) from Mongolia. Zool. J. Linn. Soc. 1992, 104, 97–116. [Google Scholar] [CrossRef]
- Smirnov, N.N. Macrothricidae and Moinidae of the World Fauna; Nauka: Leningrad, Russia, 1976. [Google Scholar]
- Korovchinsky, N.M.; Kotov, A.A.; Sinev, A.Y.; Neretina, A.N.; Garibian, P.G. Water fleas (Crustacea: Cladocera) of North Eurasia; KMK Press: Moscow, Russia, 2021; Volume 2. [Google Scholar]
- Dumont, H.J.; Rietzler, A.C.; Kalapothakis, E. Micromoina arboricola n. gen., n. spec. (Crustacea: Cladocera), a new moinid living in a forest tree-hole in Minas Gerais, Brazil. Zootaxa 2013, 3652, 533–546. [Google Scholar] [CrossRef]
- Dumont, H.J.; Negrea, Ș. Branchiopoda; Backhuys: Leiden, The Netherlands, 2002; ISBN 9789057821127. [Google Scholar]
- Kotov, A.A.; Neretina, A.N.; Al Neyadi, S.E.S.; Karabanov, D.P.; Hamza, W. Cladocera (Crustacea: Branchiopoda) of Man-Made Lakes at the Northeast Part of the United Arab Emirates with a Hypothesis on Their Origin. Diversity 2022, 14, 688. [Google Scholar] [CrossRef]
- Dumont, H.J. On a collection of zooplankton from Somalia, with a description of three new species of Copepoda. Monitore Zoologico Italiano. Supplemento 1981, 14, 103–111. [Google Scholar] [CrossRef]
- van Damme, K.; Dumont, H.J. A new species of Moina Baird, 1950 (Crustacea: Anomopoda) from Socotra Island, Yemen. Zootaxa 2008, 1721, 24–34. [Google Scholar] [CrossRef]
- Fetahi, T.; Mengistou, S.; Schagerl, M. Zooplankton community structure and ecology of the tropical-highland Lake Hayq, Ethiopia. Limnologica 2011, 41, 389–397. [Google Scholar] [CrossRef]
- Fetahi, T. Plankton Communities and Ecology of Tropical Lakes Hayq and Awasa. Ethiopia. Ph.D. Thesis, Universitat Wien, Vienna, Austria, 2010. [Google Scholar]
- Dagne, A.; Herzig, A.; Jersabek, C.D.; Tadesse, Z. Abundance, Species Composition and Spatial Distribution of Planktonic Rotifers and Crustaceans in Lake Ziway (Rift Valley, Ethiopia). Int. Rev. Hydrobiol. 2008, 93, 210–226. [Google Scholar] [CrossRef]
- Vijverberg, J.; Dejen, E.; Getahun, A.; Nagelkerke, L.A.J. Zooplankton, fish communities and the role of planktivory in nine Ethiopian lakes. Hydrobiologia 2014, 722, 45–60. [Google Scholar] [CrossRef]
- Neretina, A.N. Fauna of the water fleas (Crustacea: Cladocera) of Ethiopia. Ph.D. Thesis, A.N. Severtsov Institute of Ecology and Evolution, Moscow, Russia, 2018. [Google Scholar]
- Dumont, H.J.; Maas, S.; Martens, K. Cladocera, Copepoda and Ostracoda (Crustacea) from Fresh Waters in South Yemen. Fauna Saudi Arab. 1986, 8, 12–19. [Google Scholar]
- Ghaouaci, S.; Amarouayache, M.; Sinev, A.Y.; Korovchinsky, N.M.; Kotov, A.A. An annotated checklist of the Algerian Cladocera (Crustacea: Branchiopoda). Zootaxa 2018, 4377, 412–430. [Google Scholar] [CrossRef] [PubMed]
- Ekman, S. Cladoceren und freilebende Copepoden aus Ägypten und dem Sudan. Results Swed. Zool. Exped. Egypt White Nile 1901 Under Dir. L. A. Jägerskiöld 1904, 1, 1–18. [Google Scholar]
- Dumont, H.J.; Pensaert, J.; el Moghraby, A.I. Cladocera from the Sudan: Red Sea Hills, Jebel Marra and valley of the main Nile. Hydrobiologia 1984, 110, 163–169. [Google Scholar] [CrossRef]
- Dumont, H.J. Limnologie van Sahara en Sahel. Ph.D. Thesis; University of Ghent: Ghent, Belgium, 1979. [Google Scholar]
- Bromley, H.J. A checklist of the Cladocera of Israel and Eastern Sinai. Hydrobiologia 1993, 257, 21–28. [Google Scholar] [CrossRef]
- Abell, R.; Thieme, M.L.; Revenga, C.; Bryer, M.; Kottelat, M.; Bogutskaya, N.; Coad, B.; Mandrak, N.; Balderas, S.C.; Bussing, W.; et al. Freshwater Ecoregions of the World: A New Map of Biogeographic Units for Freshwater Biodiversity Conservation. Bioscience 2008, 58, 403–414. [Google Scholar] [CrossRef]
- Bi, R.; Wei, J.; Deng, Z.; Blair, D.; Hu, W.; Yin, M. Moina heilongjiangensis sp. nov. (Crustacea, Cladocera) from the northeast of China. ZSE 2025, 101, 1325–1337. [Google Scholar] [CrossRef]
- Chen, S. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. Imeta 2023, 2, e107. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Jin, J.-J.; Yu, W.-B.; Yang, J.-B.; Song, Y.; dePamphilis, C.W.; Yi, T.-S.; Li, D.-Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Birney, E.; Clamp, M.; Durbin, R. GeneWise and Genomewise. Genome Res. 2004, 14, 988–995. [Google Scholar] [CrossRef] [PubMed]
- Gertz, E.M.; Yu, Y.-K.; Agarwala, R.; Schäffer, A.A.; Altschul, S.F. Composition-based statistics and translated nucleotide searches: Improving the TBLASTN module of BLAST. BMC Biol. 2006, 4, 41. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef]
- Pereboev, D.D.; Garibian, P.G.; Karabanov, D.P.; Efeykin, B.D.; Galimov, Y.R.; Petrusek, A.; Kotov, A.A. A non-monophyly of ‘crowned’ Daphnia (Ctenodaphnia) Dybowski et Grochowski, 1895 (Cladocera: Daphniidae): From genomes to morphology. Zool. Scr. 2025, 54, 69–90. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.-Y.; Graham, M.; van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Chernomor, O.; von Haeseler, A.; Minh, B.Q. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef]
- Bekker, E.I.; Karabanov, D.P.; Galimov, Y.R.; Kotov, A.A. DNA barcoding reveals high cryptic diversity in the North Eurasian Moina species (Crustacea: Cladocera). PLoS ONE 2016, 11, e0161737. [Google Scholar] [CrossRef]
- Zimin, A.V.; Marçais, G.; Puiu, D.; Roberts, M.; Salzberg, S.L.; Yorke, J.A. The MaSuRCA genome assembler. Bioinformatics 2013, 29, 2669–2677. [Google Scholar] [CrossRef]
- Puillandre, N.; Brouillet, S.; Achaz, G. ASAP: Assemble species by automatic partitioning. Mol. Ecol. Resour. 2021, 21, 609–620. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Moreno, M.A.; Holder, M.T.; Sukumaran, J. DendroPy 5: A mature Python library for phylogenetic computing. JOSS 2024, 9, 6943. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef] [PubMed]
- Montoliu Elena, L.; Elias-Gutierrez, M.; Miracle Sole, M.R.; Korinek, V. Who is Moina micrura? An example of how barcodes can help to clarify highly confused species. Genome 2015, 58, 215. [Google Scholar]
- Elías-Gutiérrez, M.; JURAČKA, P.J.; Montoliu-Elena, L.; Miracle, M.R.; Petrusek, A.; Kořínek, V. Who is Moina micrura? Redescription of one of the most confusing cladocerans from terra typica, based on integrative taxonomy. Limnetica 2019, 38, 227–252. [Google Scholar] [CrossRef]
- Neretina, A.N.; Alonso, M.; Kotov, A.A. Investigation of the distribution patterns in moinids (Crustacea: Cladocera: Moinidae) forming ephippia with two resting eggs. Zootaxa 2024, 5512, 451–490. [Google Scholar] [CrossRef]
- Gurney, R. List of Entomostraca collected in Seistan and the Baluch Desert. Rec. Indian Mus. 1920, 18, 145–146. [Google Scholar] [CrossRef]
- Muñoz-Pedreros, A.; De Los Ríos-Escalante, P.; Möller, P. Zooplankton of the highland bogs of Putana, a desert wetland of the high puna, northern Chile. Crustaceana 2015, 88, 1235–1244. [Google Scholar] [CrossRef]
- Hamza, W.; Ramadan, G.; AlKaabi, M. Morphological and molecular identification of first recorded Cladoceran organisms in the desert of Abu Dhabi, UAE. MOJ Ecol. Environ. Sci. 2018, 3, 220–224. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, H.; Hu, L.; Leppänen, J.J. Cladoceran communities in soda lakes of the Badain Jaran desert, NW China. J. Arid. Environ. 2020, 177, 104133. [Google Scholar] [CrossRef]
- Korinek, V. Cladocera. In Hydrobiological Survey of Lake Bangweulu and Luapulu River Basin; Symoens, J.-J., Ed.; Cercle Hydrobiologique de Bruxelles: Bruxelles, Belgium, 1984; pp. 1–117. [Google Scholar]
- Kim, K.; Kotov, A.A.; Taylor, D.J. Hormonal induction of undescribed males resolves cryptic species of cladocerans. Proc. Biol. Sci. 2006, 273, 141–147. [Google Scholar] [CrossRef]
- Soesbergen, M. A preliminary investigation of plankton organisms of fresh and brackish inland waters in the northern United Arab Emirates. Tribulus 2018, 26, 46–58. [Google Scholar]
- Deng, Z.; Yao, Y.; Blair, D.; Hu, W.; Yin, M. Ceriodaphnia (Cladocera: Daphniidae) in China: Lineage diversity, phylogeography and possible interspecific hybridization. Mol. Phylogenet. Evol. 2022, 175, 107586. [Google Scholar] [CrossRef] [PubMed]
- Dumont, H.J. Relict distribution patterns of aquatic animals: Another tool in evaluating late Pleistocene climate changes in the Sahara and Sahel. Palaeoecol. Afr. Surround. Isl. 1982, 14, 1–24. [Google Scholar]
- Korovchinsky, N.M. The Cladocera (Crustacea: Branchiopoda) as a relict group. Zool. J. Linn. Soc. 2006, 147, 109–124. [Google Scholar] [CrossRef]
- Grandcolas, P.; Nattier, R.; Trewick, S. Relict species: A relict concept? Trends Ecol. Evol. 2014, 29, 655–663. [Google Scholar] [CrossRef]
- Buechley, E.R.; de La Cruz Muñoza, A.; Roman, J.; Caucala, G.; Rayaleh, H. Notable bird observations for Djibouti, including the first record of Semicollared Flycatcher Ficedula semitorquata. Bull. Afr. Bird. Cl. 2019, 26, 179–185. [Google Scholar]
- Trading Economics. China Exports to Djibouti. Available online: https://tradingeconomics.com/china/imports/djibouti (accessed on 12 November 2025).
- Kotov, A.A.; Karabanov, D.P.; van Damme, K. Non-Indigenous Cladocera (Crustacea: Branchiopoda): From a Few Notorious Cases to a Potential Global Faunal Mixing in Aquatic Ecosystems. Water 2022, 14, 2806. [Google Scholar] [CrossRef]
- Mergeay, J.; Verschuren, D.; de Meester, L. Invasion of an asexual American water flea clone throughout Africa and rapid displacement of a native sibling species. Proc. Biol. Sci. 2006, 273, 2839–2844. [Google Scholar] [CrossRef]
| Isolate | Species | Country | Folmer Fragment of the COI Gene Variant | ITS1 Alleles |
|---|---|---|---|---|
| K115 | Moina cf. micrura | Djibouti | K115 | K115a |
| K116 | Moina cf. micrura | Djibouti | K115 | K115a K116a |
| K119 | Moina cf. micrura | Djibouti | K119 | K115a K116a |
| K120 | Moina heilongjiangensis | Djibouti | K120 | K120a K120b |
| AAK M-0371 | Moina belli | Ethiopia | AAK M-0371 | N/A |
| AAK M-4430 | Moina belli | South Africa | AAK M-4430 | N/A |
| AAK M-1479 | Moina cf. micrura | Ethiopia | AAK M-1479 | N/A |
| ANN 2015-024 | Moina cf. micrura | Ethiopia | ANN 2015-024 | N/A |
| AAK M-1483 | Moina cf. micrura | Ethiopia | AAK M-1483 | N/A |
| AAK M-1511 | Moina cf. micrura | Ethiopia | AAK M-1511 | N/A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pereboev, D.D.; Neretina, A.N.; Garibian, P.G.; Efeykin, B.D.; Waais, I.O.; Kotov, A.A. Contrasting Fauna in Two Neighboring Territories of the African Horn: A Case of the Genus Moina Baird, 1850 (Cladocera: Moinidae). Water 2025, 17, 3312. https://doi.org/10.3390/w17223312
Pereboev DD, Neretina AN, Garibian PG, Efeykin BD, Waais IO, Kotov AA. Contrasting Fauna in Two Neighboring Territories of the African Horn: A Case of the Genus Moina Baird, 1850 (Cladocera: Moinidae). Water. 2025; 17(22):3312. https://doi.org/10.3390/w17223312
Chicago/Turabian StylePereboev, Dmitry D., Anna N. Neretina, Petr G. Garibian, Boris D. Efeykin, Idriss Okiye Waais, and Alexey A. Kotov. 2025. "Contrasting Fauna in Two Neighboring Territories of the African Horn: A Case of the Genus Moina Baird, 1850 (Cladocera: Moinidae)" Water 17, no. 22: 3312. https://doi.org/10.3390/w17223312
APA StylePereboev, D. D., Neretina, A. N., Garibian, P. G., Efeykin, B. D., Waais, I. O., & Kotov, A. A. (2025). Contrasting Fauna in Two Neighboring Territories of the African Horn: A Case of the Genus Moina Baird, 1850 (Cladocera: Moinidae). Water, 17(22), 3312. https://doi.org/10.3390/w17223312







