A Comparative Evaluation of eDNA Metabarcoding Primers in Fish Community Monitoring in the East Lake
Abstract
1. Introduction
2. Methods
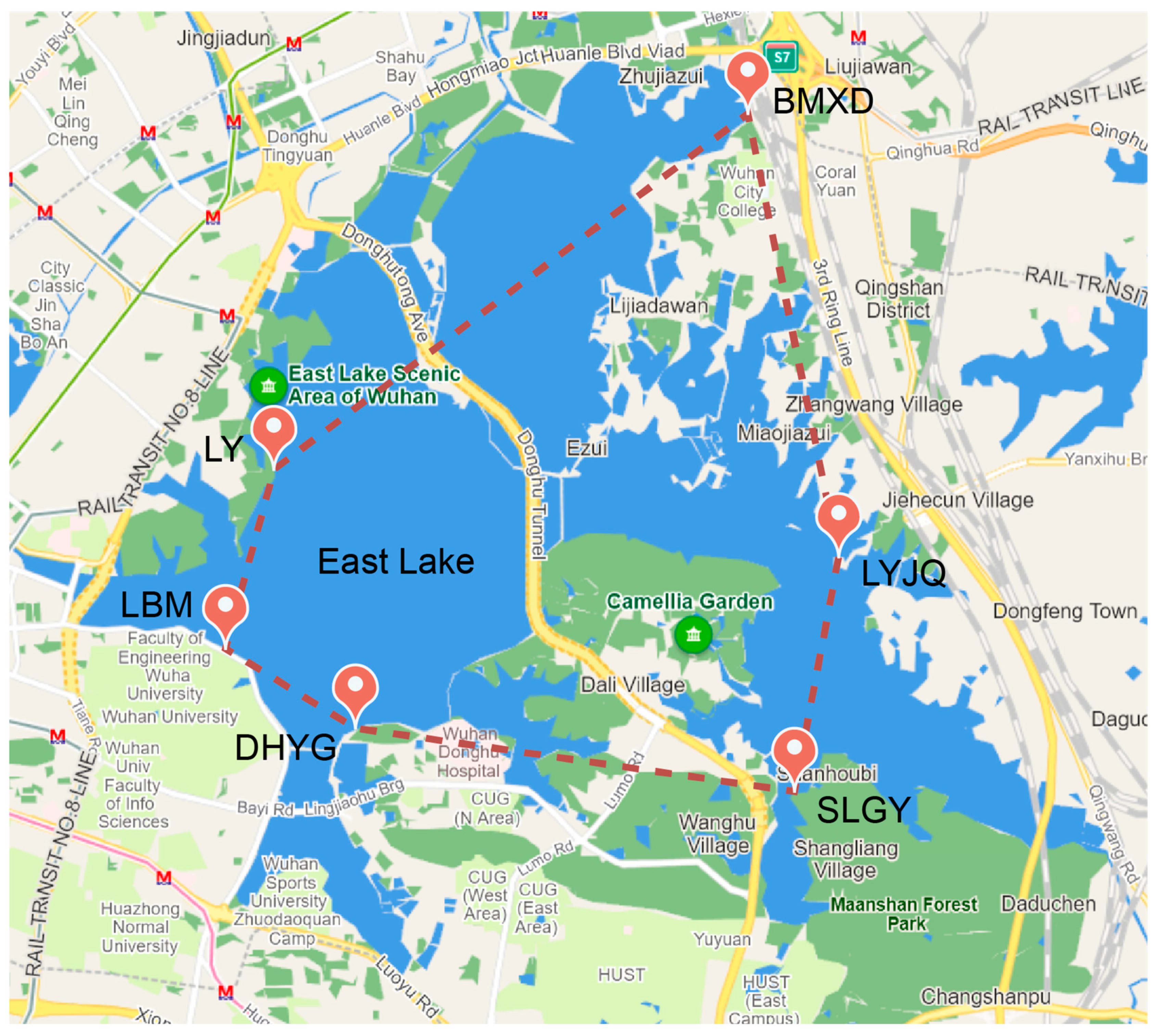
2.1. Sample Collection
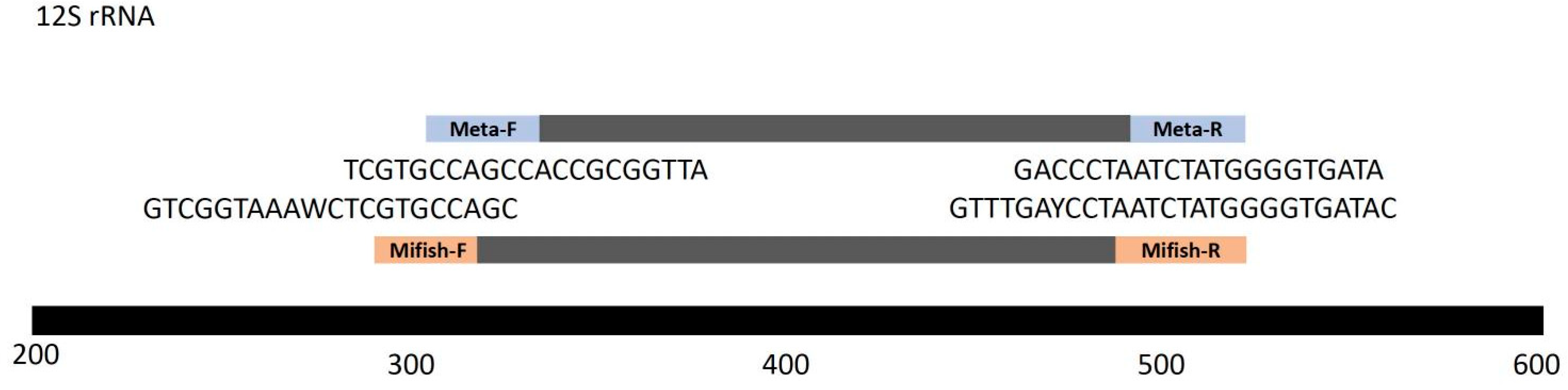
2.2. Metabarcoding of eDNA Samples
2.3. Bioinformatics and Statistical Analyses
3. Results
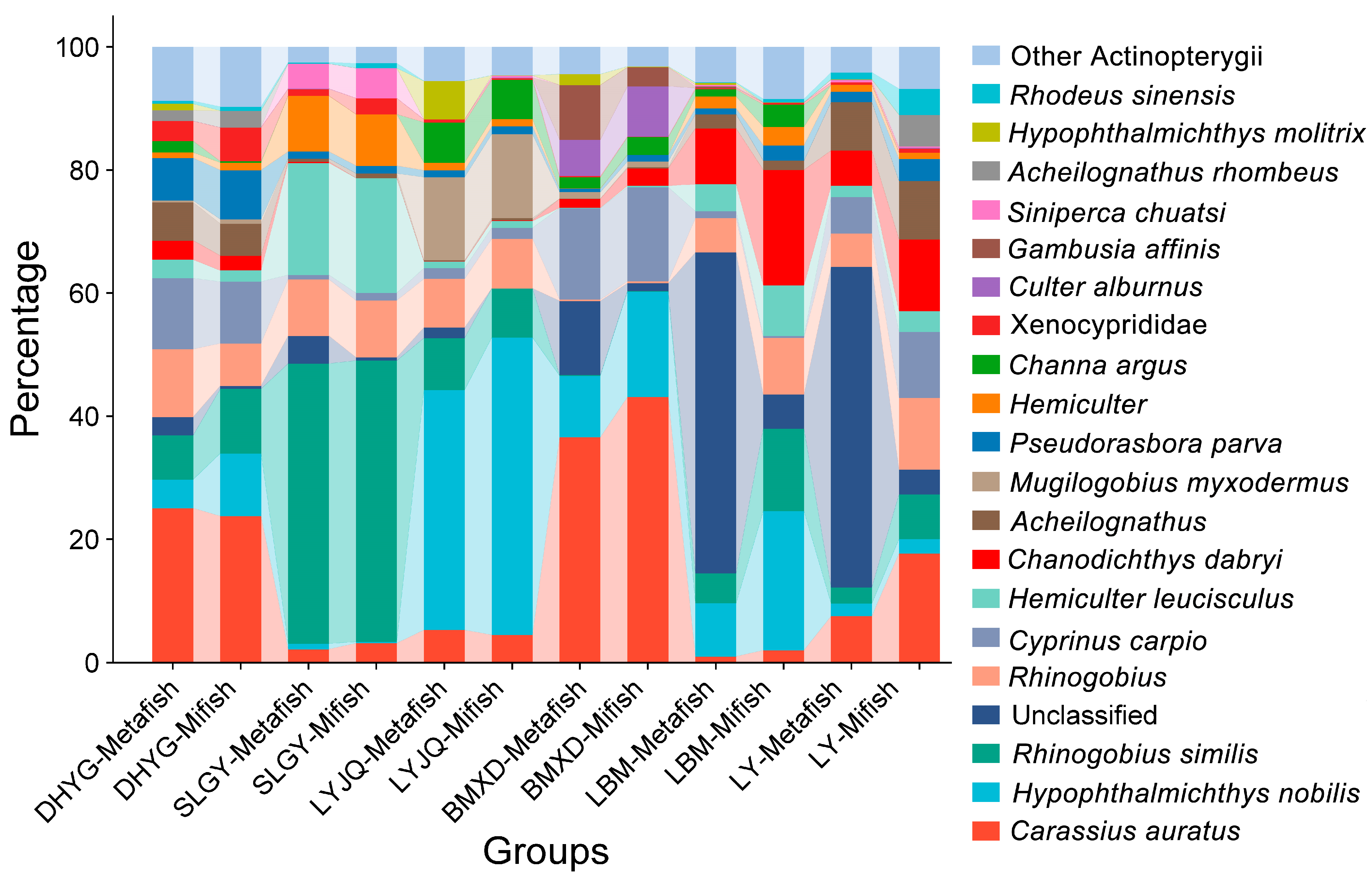
3.1. Species Composition and Diversity
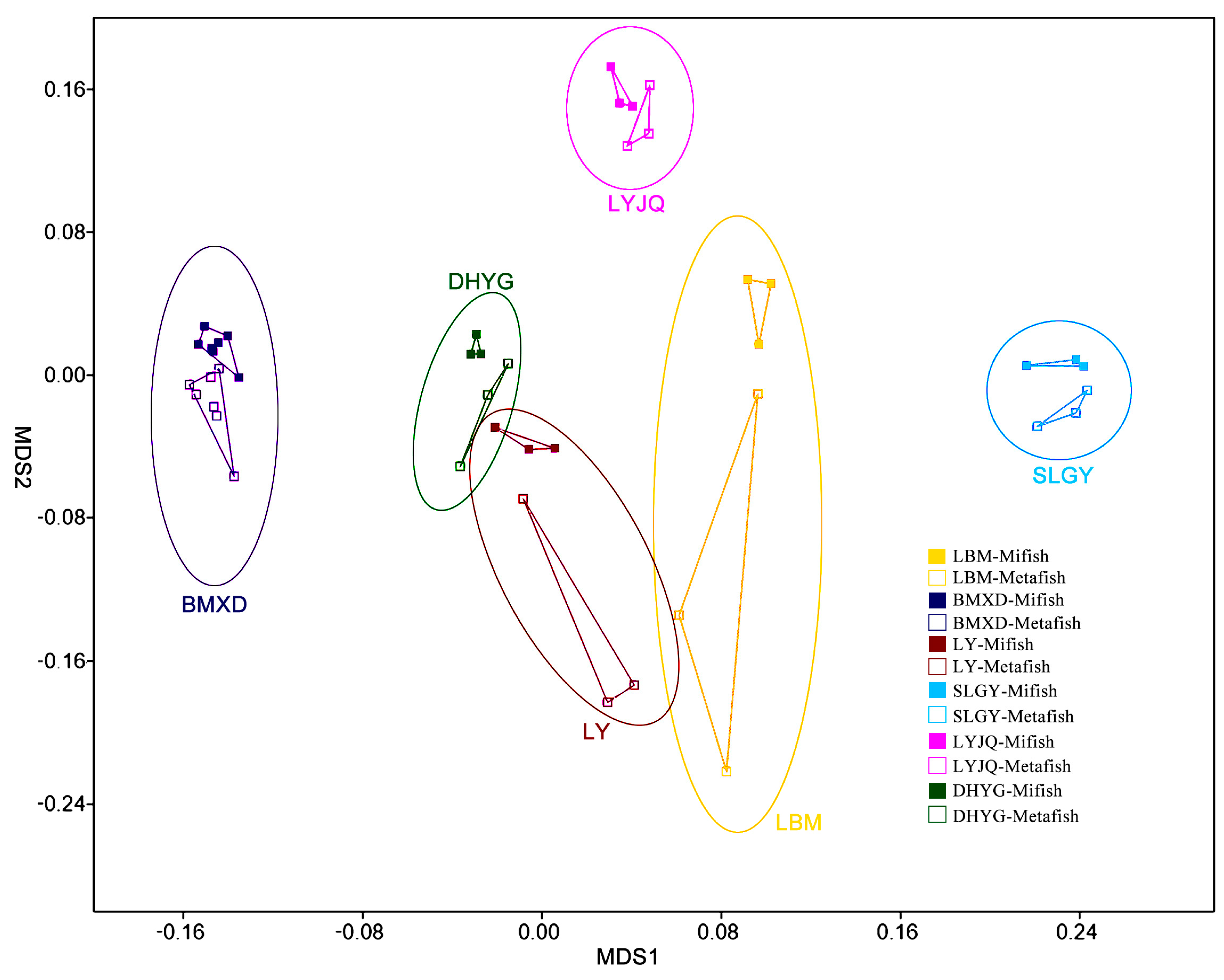
3.2. Community Diversity
3.3. Species Distribution by Primers
3.4. Species Distribution by Location
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Ge, J.; Liu, M.; Yun, X.; Yang, Y.; Zhang, M.; Li, Q.; Wang, J. Occurrence, distribution and seasonal variations of polychlorinated biphenyls and polybrominated diphenyl ethers in surface waters of the East Lake, China. Chemosphere 2014, 103, 256–262. [Google Scholar] [CrossRef]
- Liu, J.; Chen, S. An assessment of the impact of fish stocking on lake eutrophication in China. SIL Commun. 1994, 24, 231–235. [Google Scholar] [CrossRef]
- Yao, M.; Zhang, S.; Lu, Q.; Chen, X.; Zhang, S.Y.; Kong, Y.; Zhao, J. Fishing for Fish Environmental DNA: Ecological Applications, Methodological Considerations, Surveying Designs, and Ways forward. Mol. Ecol. 2022, 31, 5132–5164. [Google Scholar] [CrossRef]
- Gan, Y.; Guo, Y. Evaluation analysis and remedy strategy for eutrophication in Wuhan lake Donghu. Resour. Environ. Yangtze 2004, 13, 277–281. [Google Scholar]
- Du, Y.; Xue, H.P.; Wu, S.J.; Ling, F.; Xiao, F.; Wei, X. Lake area changes in the middle Yangtze region of China over the 20th century. J. Environ. Manag. 2011, 92, 1248–1255. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yun, X.; Liu, M.; Jiang, Y.; Li, Q.X.; Wang, J. Concentrations, distributions, sources, and risk assessment of organochlorine pesticides in surface water of the East Lake, China. Environ. Sci. Pollut. Res. 2014, 21, 3041–3050. [Google Scholar] [CrossRef]
- Rees, H.C.; Maddison, B.C.; Middleditch, D.J.; Patmore, J.R.M.; Gough, K.C. The detection of aquatic animal species using environmental DNA—A review of eDNA as a survey tool in ecology. J. Appl. Ecol. 2014, 51, 1450–1459. [Google Scholar] [CrossRef]
- Clements, C.F.; Blanchard, J.L.; Nash, K.L.; Hindell, M.A.; Ozgul, A. Body size shifts and early warning signals precede the historic collapse of whale stocks. Nat. Ecol. Evol. 2017, 1, 188. [Google Scholar] [CrossRef]
- Hopkins, G.; Freckleton, R. Declines in the numbers of amateur and professional taxonomists: Implications for conservation. Anim. Conserv. 2002, 5, 245–249. [Google Scholar] [CrossRef]
- Hering, D.; Borja, A.; Jones, J.I.; Pont, D.; Boets, P.; Bouchez, A.; Bruce, K.; Drakare, S.; Hänfling, B.; Kahlert, M.; et al. Implementation options for DNA-based identification into ecological status assessment under the European Water Framework Directive. Water Resour. 2018, 138, 192–205. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhao, J.; Yao, M. A comprehensive and comparative evaluation of primers for metabarcoding eDNA from fish. Methods Ecol. Evol. 2020, 11, 1609–1625. [Google Scholar] [CrossRef]
- Yamamoto, S.; Minami, K.; Fukaya, K.; Takahashi, K.; Sawada, H.; Murakami, H.; Tsuji, S.; Hashizume, H.; Kubonaga, S.; Horiuchi, T.; et al. Environmental DNA as a ‘snapshot’ of fish distribution: A case study of Japanese Jack Mackerel in Maizuru Bay, sea of Japan. PLoS ONE 2016, 11, e0149786. [Google Scholar]
- Wang, S.; Yan, Z.; Hänfling, B.; Zheng, X.; Wang, P.; Fan, J.; Li, J. Methodology of fish eDNA and its applications in ecology and environment. Sci. Total Environ. 2021, 755, 142622. [Google Scholar] [CrossRef]
- Burian, A.; Bruce, K.; Tovela, E.; Bakker, J.; Balcells, L.; Bennett, R.; Chordekar, S.; Costa, H.M.; Crampton-Platt, A.; Boer, H.; et al. Merging two eDNA metabarcoding approaches and citizen-science-based sampling to facilitate fish community monitoring along vast Sub-Saharan coastlines. Mol. Ecol. Resour. 2023, 23, 1641–1655. [Google Scholar] [CrossRef] [PubMed]
- Osathanunkul, M.; Minamoto, T. A molecular survey based on eDNA to assess the presence of a clown featherback (Chitala ornata) in a confined environment. PeerJ 2020, 8, e10338. [Google Scholar] [CrossRef] [PubMed]
- Bohmann, K.; Evans, A.; Gilbert, M.T.P.; Carvalho, G.R.; Creer, S.; Knapp, M.; Yu, D.W.; Bruyn, M. Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol. Evol. 2014, 29, 358–367. [Google Scholar] [CrossRef] [PubMed]
- Port, J.A.; O’Donnell, J.L.; Romero-Maraccini, O.C.; Leary, P.R.; Litvin, S.Y.; Nickols, K.J.; Yamahara, K.M.; Kelly, R.P. Assessing vertebrate biodiversity in a kelp forest ecosystem using environmental DNA. Mol. Ecol. 2016, 25, 527–541. [Google Scholar] [CrossRef]
- Shaw, J.L.A.; Clarke, L.J.; Wedderburn, S.D.; Barnes, T.C.; Weyrich, L.S.; Cooper, A. Comparison of environmental DNA metabarcoding and conventional fish survey methods in a river system. Biol. Conserv. 2016, 197, 131–138. [Google Scholar] [CrossRef]
- Valentini, A.; Taberlet, P.; Miaud, C.; Civade, R.; Herder, J.; Thomsen, P.F.; Bellemain, E.; Besnard, A.; Coissac, E.; Boyer, F.; et al. Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Mol. Ecol. 2016, 25, 929–942. [Google Scholar] [CrossRef]
- Collins, R.A.; Bakker, J.; Wangensteen, O.S.; Soto, A.Z.; Corrigan, L.; Sims, D.W.; Genner, M.J.; Mariani, S. Non-specific amplification compromises environmental DNA metabarcoding with COI. Methods Ecol. Evol. 2019, 10, 1985–2001. [Google Scholar] [CrossRef]
- Polanco, A.; Richards, E.; Flück, B.; Valentini, A.; Altermatt, F.; Brosse, S.; Walser, J.C.; Eme, D.; Marques, V.; Manel, S.; et al. Comparing the performance of 12S mitochondrial primers for fish environmental DNA across ecosystems. Environ. DNA 2006, 3, 1113–1127. [Google Scholar] [CrossRef]
- Clarke, L.J.; Soubrier, J.; Weyrich, L.S.; Cooper, A. Environmental metabarcodes for insects: In silico PCR reveals potential for taxonomic bias. Mol. Ecol. Resour. 2014, 14, 1160–1170. [Google Scholar] [CrossRef] [PubMed]
- Miya, M.; Gotoh, R.O.; Sado, T. Mifish metabarcoding: A high-throughput approach for simultaneous detection of multiple fish species from environmental DNA and other samples. Fish. Sci. 2020, 86, 939–970. [Google Scholar] [CrossRef]
- Bylemans, J.; Gleeson, D.M.; Hardy, C.M.; Furlan, E. Toward an ecoregion scale evaluation of eDNA metabarcoding primers: A case study for the freshwater fish biodiversity of the Murray-Darling Basin (Australia). Ecol. Evol. 2018, 8, 8697–8712. [Google Scholar] [CrossRef] [PubMed]
- Wilcox, T.M.; McKelvey, K.S.; Young, M.K.; Jane, S.F.; Lowe, W.H.; Whiteley, A.R.; Schwartz, M.K. Robust detection of rare species using environmental DNA: The importance of primer specificity. PLoS ONE 2013, 8, e59520. [Google Scholar] [CrossRef] [PubMed]
- Freeland, J.R. The importance of molecular markers and primer design when characterizing biodiversity from environmental DNA. Genome 2017, 60, 358–374. [Google Scholar] [CrossRef] [PubMed]
- Marques, V.; Guérin, P.-É.; Rocle, M.; Valentini, A.; Manel, S.; Mouillot, D.; Dejean, T. Blind assessment of vertebrate taxonomic diversity across spatial scales by clustering environmental DNA metabarcoding sequences. Ecography 2020, 43, 1779–1790. [Google Scholar] [CrossRef]
- Jo, T.S.; Tsuri, K.; Yamanaka, H. Can nuclear aquatic environmental DNA be a genetic marker for the accurate estimation of a species abundance? Sci. Nat. 2022, 109, 38. [Google Scholar] [CrossRef]
- Shu, L.; Ludwig, A.; Peng, Z. Environmental DNA metabarcoding primers for freshwater fish detection and quantification: In silico and in tanks. Ecol. Evol. 2021, 11, 8281–8294. [Google Scholar] [CrossRef]
- Riaz, T.; Shehzad, W.; Viari, A.; Pompanon, F.; Taberlet, P.; Coissac, E. ecoPrimers: Inference of new DNA barcode markers from whole genome sequence analysis. Nucleic Acids Res. 2011, 39, e145. [Google Scholar] [CrossRef]
- Kelly, R.P.; Port, J.A.; Yamahara, K.M.; Crowder, L.B. Using environmental DNA to census marine fishes in a large mesocosm. PLoS ONE 2014, 9, e86175. [Google Scholar] [CrossRef]
- Evans, N.T.; Olds, B.P.; Renshaw, M.A.; Turner, C.R.; Li, Y.; Jerde, C.L.; Mahon, A.R.; Pfrender, M.E.; Lamberti, G.A.; Lodge, D.M. Quantification of mesocosm fish and amphibian species diversity via environmental DNA metabarcoding. Mol. Ecol. Resour. 2016, 16, 29–41. [Google Scholar] [CrossRef]
- Bellemain, E.; Carlsen, T.; Brochmann, C.; Coissac, E.; Taberlet, P.; Kauserud, H. ITS as an environmental DNA barcode for fungi: An in silico approach reveals potential PCR biases. BMC Microbiol. 2010, 10, 189. [Google Scholar] [CrossRef]
- García-Machado, E.; Normandeau, E.; Côté, G.; Bernatchez, L. How eDNA data filtration, sequence coverage, and primer selection influence assessment of fish communities in northern temperate lakes. Environ. DNA 2023, 5, 1216–1233. [Google Scholar] [CrossRef]
- Xiong, F.; Shu, L.; Zeng, H.; Gan, X.; He, S.; Peng, Z. Methodology for fish biodiversity monitoring with environmental DNA metabarcoding: The primers, databases and bioinformatic pipelines. Water Biol. Secur. 2022, 1, 100007. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, X.W. Universal Metabarcoding Amplification Primers for Freshwater Fish Mitochondria 12S and Application Method Thereof. China Patent CN109943645B, 14 April 2023. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Steven, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package, R package version 2.4-3; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Ricker, W.E. Linear Regressions in Fishery Research. J. Fish. Res. Board Can. 1973, 30, 409–434. [Google Scholar] [CrossRef]
- Goslee, S.C.; Urban, D.L. The ecodist package for dissimilarity-based analysis of ecological data. J. Stat. Softw. 2007, 22, 7. [Google Scholar] [CrossRef]
- Kelly, R.P.; Gallego, R.; Jacobs-Palmer, E. The effect of tides on nearshore environmental DNA. PeerJ 2018, 6, e4521. [Google Scholar] [CrossRef] [PubMed]
- Evans, N.T.; Lamberti, G.A. Freshwater fisheries assessment using environmental DNA: A primer on the method, its potential, and shortcomings as a conservation tool. Fish. Res. 2018, 197, 60–66. [Google Scholar] [CrossRef]
- Klobucar, S.L.; Rodgers, T.W.; Budy, P. At the forefront: Evidence of the applicability of using environmental DNA to quantify the abundance of fish populations in natural lentic waters with additional sampling considerations. Can. J. Fish. Aquat. Sci. 2017, 74, 2030–2034. [Google Scholar] [CrossRef]
- Ushio, M.; Murakami, H.; Masuda, R.; Sado, T.; Miya, M.; Sakurai, S.; Yamanaka, H.; Minamoto, T.; Kondoh, M. Quantitative monitoring of multispecies fish environmental DNA using high-throughput sequencing. Metabarcoding Metagenom. 2018, 2, e23297. [Google Scholar]
- Lacoursiere-Roussel, A.; Deiner, K. Environmental DNA is not the tool by itself. J. Fish Biol. 2019, 98, 383–386. [Google Scholar] [CrossRef]
- Stoeckle, M.Y.; Ausubel, J.H.; Coogan, M. 12S gene metabarcoding with DNA standard quantifies marine bony fish environmental DNA, identifies threshold for reproducible detection, and overcomes distortion due to amplification of non-fish DNA. Environ. DNA 2022, 6, e376. [Google Scholar] [CrossRef]
- Allan, E.A.; Zhang, W.G.; Lavery, A.C.; Govindarajan, A.F. Environmental DNA shedding and decay rates from diverse animal forms and thermal regimes. Environ. DNA 2020, 3, 492–514. [Google Scholar] [CrossRef]

| Location-Primer | Species | Genus | Family |
|---|---|---|---|
| DHYG-Metafish | 29 | 8 | 4 |
| SLGY-Metafish | 22 | 6 | 2 |
| LYJQ-Metafish | 30 | 7 | 2 |
| BMXD-Metafish | 20 | 5 | 1 |
| LBM-Metafish | 23 | 9 | 3 |
| LY-Metafish | 25 | 10 | 1 |
| DHYG-Mifish | 30 | 11 | 2 |
| SLGY-Mifish | 18 | 7 | 2 |
| LYJQ-Mifish | 28 | 7 | 2 |
| BMXD-Mifish | 22 | 7 | 1 |
| LBM-Mifish | 21 | 10 | 2 |
| LY-Mifish | 24 | 10 | 2 |
| Total-Metafish | 44 | 32 | 14 |
| Total-Mifish | 45 | 34 | 15 |
| Total-Metafish and Mifish | 51 | 36 | 16 |
| Species | DHYG-Metafish | SLGY-Metafish | LYJQ-Metafish | BMXD-Metafish | LBM-Metafish | LY-Metafish | DHYG-Mifish | SLGY-Mifish | LYJQ-Mifish | BMXD-Mifish | LBM-Mifish | LY-Mifish |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unclassified | 3.0 | 4.5 | 1.7 | 12.1 | 52.1 | 52.1 | 0.5 | 0.5 | <0.1 | 1.3 | 5.6 | 4.1 |
| Acheilognathus rhombeus | 1.7 | <0.1 | <0.1 | 0 | 0 | <0.1 | 2.7 | 0 | <0.1 | <0.1 | <0.1 | 5.1 |
| Acheilognathus tonkinensis | <0.1 | 0 | 0 | 0 | 0 | <0.1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Culter alburnus | 0 | <0.1 | 0 | 5.9 | 0 | 0 | 0 | 0 | <0.1 | 8.2 | 0 | <0.1 |
| Carassius auratus | 25.1 | 2.2 | 5.3 | 36.6 | 1.0 | 7.6 | 23.8 | 3.2 | 4.5 | 43.1 | 2.0 | 17.7 |
| Carassius carassius | 0 | 0 | 0 | 0 | <0.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Channa argus | 1.6 | 0 | 6.6 | 1.8 | 1.2 | 0 | 0.2 | 0 | 6.4 | 1.8 | 3.6 | 0 |
| Chanodichthys dabryi | 3.1 | 0.2 | <0.1 | 1.5 | 9.1 | 5.7 | 2.4 | 0 | 0.13 | 2.8 | 18.8 | 11.7 |
| Coptodon zillii | 0 | 0 | 0 | 0 | 0 | 0 | <0.1 | <0.1 | 0 | <0.1 | <0.1 | <0.1 |
| Gambusia affinis | 0 | 0 | 0 | 9.0 | 0.3 | 0 | 0 | 0 | 0 | 3.0 | 0 | 0 |
| Gambusia holbrooki | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | <0.1 | 0 | 0 |
| Hemibarbus barbus | 0 | 0 | <0.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Hypophthalmichthys molitrix | 1.1 | 0 | 6.3 | 1.9 | 0.4 | <0.1 | <0.1 | 0 | <0.1 | 0.2 | <0.1 | <0.1 |
| Hypophthalmichthys nobilis | 5.1 | 0.9 | 38.7 | 9.6 | 8.1 | 1.9 | 10.2 | 0.24 | 48.2 | 16.9 | 22.1 | 2.4 |
| Hyporhamphus intermedius | 0 | 0 | <0.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Misgurnus anguillicaudatus | 0 | 0 | <0.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Misgurnus bipartitus | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | <0.1 | 0 | 0 | 0 |
| Mystacoleucus marginatus | <0.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Pagellus bellottii | 0 | 0 | 0 | 0 | 0 | 0 | <0.1 | 0 | 0 | 0 | 0 | 0 |
| Paramisgurnus dabryanus | 0 | 0 | 0 | 0 | 0 | 0 | <0.1 | 0 | <0.1 | 0 | 0 | 0 |
| Rhinogobius similis | 7.6 | 45.6 | 8.2 | <0.1 | 4.7 | 2.5 | 10.5 | 45.5 | 7.9 | <0.1 | 13.1 | 7.2 |
| Sarcocheilichthys sinensis | 0 | 0 | <0.1 | 0 | 0 | 0 | 0.3 | 0 | 0 | 0 | 0 | 0 |
| Saurogobio lissilabris | 0 | 0 | 0 | 0 | 0 | 0 | <0.1 | 0 | 0 | 0 | 0 | 0 |
| Saurogobio xiangjiangensis | 0 | 0 | 0 | 0 | 0 | 0 | <0.1 | 0 | 0 | 0 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Tang, M.; Lu, S.; Zhang, X.; Fang, C.; Tan, L.; Xiong, F.; Zeng, H.; He, S. A Comparative Evaluation of eDNA Metabarcoding Primers in Fish Community Monitoring in the East Lake. Water 2024, 16, 631. https://doi.org/10.3390/w16050631
Li Y, Tang M, Lu S, Zhang X, Fang C, Tan L, Xiong F, Zeng H, He S. A Comparative Evaluation of eDNA Metabarcoding Primers in Fish Community Monitoring in the East Lake. Water. 2024; 16(5):631. https://doi.org/10.3390/w16050631
Chicago/Turabian StyleLi, Yiwen, Minzhe Tang, Suxiang Lu, Xiaochun Zhang, Chengchi Fang, Li Tan, Fan Xiong, Honghui Zeng, and Shunping He. 2024. "A Comparative Evaluation of eDNA Metabarcoding Primers in Fish Community Monitoring in the East Lake" Water 16, no. 5: 631. https://doi.org/10.3390/w16050631
APA StyleLi, Y., Tang, M., Lu, S., Zhang, X., Fang, C., Tan, L., Xiong, F., Zeng, H., & He, S. (2024). A Comparative Evaluation of eDNA Metabarcoding Primers in Fish Community Monitoring in the East Lake. Water, 16(5), 631. https://doi.org/10.3390/w16050631








