Ultraviolet (Spot)light on Water Treatment: Targeting Inactivation Efficiency and Stress Responses of Gram-Positive and Gram-Negative Bacteria Using UV-B and UV-C LEDs
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Cultivation
2.2. UV-LED Device
2.3. Survival Experiments
2.4. Inactivation Kinetics, Model Fitting and Statistical Analysis
2.5. Extraction of RNA, Synthesis of Complementary DNA (cDNA) and RT-qPCR
3. Results
3.1. Wavelength Dependent Inactivation Kinetics
3.1.1. Disinfection Performance
3.1.2. Inactivation Rates
3.2. Wavelength Dependent Stress Mechanisms
4. Discussion
4.1. Wavelength Dependent Inactivation Kinetics
4.2. Wavelength-Dependent Stress Mechanisms
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kebbi, Y.; Muhammad, A.I.; Sant’Ana, A.S.; do Prado-Silva, L.; Liu, D.; Ding, T. Recent advances on the application of UV-LED technology for microbial inactivation: Progress and mechanism. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3501–3527. [Google Scholar] [CrossRef]
- Soro, A.B.; Shokri, S.; Nicolau-Lapeña, I.; Ekhlas, D.; Burgess, C.M.; Whyte, P.; Bolton, D.J.; Bourke, P.; Tiwari, B.K. Current challenges in the application of the UV-LED technology for food decontamination. Trends Food Sci. Technol. 2023, 131, 264–276. [Google Scholar] [CrossRef]
- Song, K.; Mohseni, M.; Taghipour, F. Application of ultraviolet light-emitting diodes (UV-LEDs) for water disinfection: A review. Water Res. 2016, 94, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Rattanakul, S.; Oguma, K. Inactivation kinetics and efficiencies of UV-LEDs against Pseudomonas aeruginosa, Legionella pneumophila, and surrogate microorganisms. Water Res. 2018, 130, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, P.; Autin, O.; Goslan, E.H.; Hassard, F. Application of ultraviolet light-emitting diodes (UV-LED) to full-scale drinking-water disinfection. Water 2019, 11, 1894. [Google Scholar] [CrossRef]
- Ghosh, S.; Chen, Y.; Hu, J. Application of UVC and UVC based advanced disinfection technologies for the inactivation of antibiotic resistance genes and elimination of horizontal gene transfer activities: Opportunities and challenges. Chem. Eng. J. 2022, 450, 138234. [Google Scholar] [CrossRef]
- Gayán, E.; Serrano, M.J.; Pagán, R.; Álvarez, I.; Condón, S. Environmental and biological factors influencing the UV-C resistance of Listeria monocytogenes. Food Microbiol. 2015, 46, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.Z.; Craik, S.A.; Bolton, J.R. Comparison of the action spectra and relative DNA absorbance spectra of microorganisms: Information important for the determination of germicidal fluence (UV dose) in an ultraviolet disinfection of water. Water Res. 2009, 43, 5087–5096. [Google Scholar] [CrossRef]
- Mamane-Gravetz, H.; Linden, K.G.; Cabaj, A.; Sommer, R. Spectral sensitivity of Bacillus subtilis spores and MS2 coliphage for validation testing of ultraviolet reactors for water disinfection. Environ. Sci. Technol. 2005, 39, 7845–7852. [Google Scholar] [CrossRef] [PubMed]
- Guglielmetti, P.; Bartoloni, A. Escherichia coli-Occurrence and Epidemiology of Species other than Escherichia coli. In Encyclopedia of Food Sciences and Nutrition; Academic Press: London, UK, 2003; pp. 2162–2166. [Google Scholar]
- Liang, R.; Li, J.; Tang, H.; Sima, W.; Tang, J.; Wu, L.; Li, Z.; Liao, Y.; Lin, C. Influence of extracellular polymeric substances on electrochemical behaviours of stainless steels in circulating cooling water. Mater. Chem. Phys. 2023, 293, 126892. [Google Scholar] [CrossRef]
- Zhang, J.; Li, J.; Wu, C.; Liang, X. Citrobacter Freundii on Corrosion Behavior of HSn701-A in Reclaimed Water. In Proceedings of the 2010 International Conference on E-Product E-Service and E-Entertainment, Henan, China, 7–9 November 2010; pp. 1–6. [Google Scholar]
- Chaudhry, W.N.; Haq, I.U.; Andleeb, S.; Qadri, I. Characterization of a virulent bacteriophage LK1 specific for Citrobacter freundii isolated from sewage water. J. Basic Microbiol. 2014, 54, 531–541. [Google Scholar] [CrossRef]
- Yang, L.; Li, P.; Liang, B.; Hu, X.; Li, J.; Xie, J.; Yang, C.; Hao, R.; Wang, L.; Jia, L.; et al. Multidrug-resistant Citrobacter freundii ST139 co-producing NDM-1 and CMY-152 from China. Sci. Rep. 2018, 8, 10653. [Google Scholar] [CrossRef] [PubMed]
- Haase, J.K.; Didelot, X.; Lecuit, M.; Korkeala, H.L.; Monocytogenes MLST Study Group; Achtman, M. The ubiquitous nature of L. isteria monocytogenes clones: A large-scale Multilocus Sequence Typing study. Environ. Microbiol. 2014, 16, 405–416. [Google Scholar] [CrossRef] [PubMed]
- El Zowalaty, M.E.; Hickman, R.A.; Moura, A.; Lecuit, M.; Zishiri, O.T.; Noyes, N.; Järhult, J.D. Genome sequence of Listeria innocua strain MEZLIS26, isolated from a goat in South Africa. Microbiol. Resour. Announc. 2019, 8, 10–1128. [Google Scholar] [CrossRef]
- Hassan, A.N.; Frank, J.F. Microorganisms associated with milk. In Encyclopedia of Dairy Science; Fuquay, J.W., Ed.; Academic Press: San Diego, CA, USA, 2011; Volume 3, pp. 447–457. [Google Scholar]
- Luque-Sastre, L.; Fox, E.M.; Jordan, K.; Fanning, S. A comparative study of the susceptibility of Listeria species to sanitizer treatments when grown under planktonic and biofilm conditions. J. Food Prot. 2018, 81, 1481–1490. [Google Scholar] [CrossRef]
- de Carvalho, C.C. Biofilms: Microbial strategies for surviving UV exposure. Adv. Exp. Med. Biol. 2017, 996, 233–239. [Google Scholar]
- Bortolussi, R. Listeriosis: A primer. Can. Med. Assoc. J. CMAJ 2008, 179, 795–797. [Google Scholar] [CrossRef]
- Berlec, A.; Janež, N.; Sterniša, M.; Klančnik, A.; Sabotič, J. Listeria innocua Biofilm Assay Using NanoLuc Luciferase. Bio Protoc. 2022, 12, e4308. [Google Scholar] [CrossRef] [PubMed]
- Mafuna, T.; Matle, I.; Magwedere, K.; Pierneef, R.E.; Reva, O.N. Comparative Genomics of Listeria Species Recovered from Meat and Food Processing Facilities. Microbiol. Spectr. 2022, 10, e01189-22. [Google Scholar] [CrossRef]
- Schöbel, H.; Diem, G.; Kiechl, J.; Chistè, D.; Bertacchi, G.; Mayr, A.; Wilflingseder, D.; Lass-Flörl, C.; Posch, W. Antimicrobial efficacy and inactivation kinetics of a novel LED-based UV-irradiation technology. J. Hosp. Infect. 2023, 135, 11–17. [Google Scholar] [CrossRef]
- Andrés, C.M.C.; Pérez de la Lastra, J.M.; Andrés Juan, C.; Plou, F.J.; Pérez-Lebeña, E. Impact of reactive species on amino acids—Biological relevance in proteins and induced pathologies. Int. J. Mol. Sci. 2022, 23, 14049. [Google Scholar] [CrossRef] [PubMed]
- ISO 20776-1:2019; Susceptibility Testing of Infectious Agents and Evaluation of Performance of Antimicrobial Susceptibility Test Devices. ISO: Geneva, Switzerland, 2019. Available online: https://www.iso.org/standard/70464.html (accessed on 20 June 2024).
- Xu, L.; Zhang, C.; Xu, P.; Wang, X.C. Mechanisms of ultraviolet disinfection and chlorination of Escherichia coli: Culturability, membrane permeability, metabolism, and genetic damage. J. Environ. Sci. 2018, 65, 356–366. [Google Scholar] [CrossRef] [PubMed]
- Hiatt, C.W. Kinetics of the inactivation of viruses. Bacteriol. Rev. 1964, 28, 150–163. [Google Scholar] [CrossRef] [PubMed]
- Cerf, O. A review tailing of survival curves of bacterial spores. J. Appl. Microbiol. 1977, 42, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, W. Ultraviolet Germicidal Irradiation Handbook: UVGI for Air and Surface Disinfection; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- The MathWorks Inc. MATLAB Version: 9.13.0.2080170 (R2022b); The MathWorks Inc.: Natick, MA, USA, 2022; Available online: https://www.mathworks.com (accessed on 20 June 2024).
- Gomes, A.É.I.; Stuchi, L.P.; Siqueira, N.M.G.; Henrique, J.B.; Vicentini, R.; Ribeiro, M.L.; Darrieux, M.; Ferraz, L.F.C. Selection and validation of reference genes for gene expression studies in Klebsiella pneumoniae using Reverse Transcription Quantitative real-time PCR. Sci. Rep. 2018, 8, 9001. [Google Scholar] [CrossRef]
- Nicot, N.; Hausman, J.F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef]
- Linardić, M.; Braybrook, S.A. Identification and selection of optimal reference genes for qPCR-based gene expression analysis in Fucus distichus under various abiotic stresses. PLoS ONE 2021, 16, e0233249. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Malayeri, A.H.; Mohseni, M.; Cairns, B.; Bolton, J.R.; Chevrefils, G.; Caron, E.; Barbeau, B.; Wright, H.; Linden, K.G. Fluence (UV dose) required to achieve incremental log inactivation of bacteria, protozoa, viruses and algae. IUVA News 2016, 18, 4–6. [Google Scholar]
- Gause, S.; Chauhan, A. UV-blocking potential of oils and juices. Int. J. Cosmet. Sci. 2016, 38, 354–363. [Google Scholar] [CrossRef]
- Ibarz, A.; Pagán, J.; Panadés, R.; Garza, S. Photochemical destruction of color compounds in fruit juices. J. Food Eng. 2005, 69, 155–160. [Google Scholar] [CrossRef]
- Usaga, J.; Manns, D.C.; Moraru, C.I.; Worobo, R.W.; Padilla-Zakour, O.I. Ascorbic acid and selected preservatives influence effectiveness of UV treatment of apple juice. LWT 2017, 75, 9–16. [Google Scholar] [CrossRef]
- Bernbom, N.; Vogel, B.F.; Gram, L. Listeria monocytogenes survival of UV-C radiation is enhanced by presence of sodium chloride, organic food material and by bacterial biofilm formation. Int. J. Food Microbiol. 2011, 147, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Tikekar, R.V.; Anantheswaran, R.C.; Elias, R.J.; LaBorde, L.F. Ultraviolet-induced oxidation of ascorbic acid in a model juice system: Identification of degradation products. J. Agric. Food Chem. 2011, 59, 8244–8248. [Google Scholar] [CrossRef] [PubMed]
- Sholtes, K.; Linden, K.G. Pulsed and continuous light UV LED: Microbial inactivation, electrical, and time efficiency. Water Res. 2019, 165, 114965. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, I.A.; Niedziela, J.C.; Ogden, I.D. The synergistic effect of excimer and low-pressure mercury lamps on the disinfection of flowing water. J. Food Prot. 2000, 63, 1529–1533. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Chang, J.; Singh, M. Peptidoglycan architecture of Gram-positive bacteria by solid-state NMR. Biochim. Biophys. Acta (BBA) Biomembr. 2015, 1848, 350–362. [Google Scholar] [CrossRef] [PubMed]
- Orlandi, V.T.; Martegani, E.; Giaroni, C.; Baj, A.; Bolognese, F. Bacterial pigments: A colorful palette reservoir for biotechnological applications. Biotechnol. Appl. Biochem. 2022, 69, 981–1001. [Google Scholar] [CrossRef] [PubMed]
- McSharry, S.; Koolman, L.; Whyte, P.; Bolton, D. Inactivation of Listeria monocytogenes and Salmonella typhimurium in beef broth and on diced beef using an ultraviolet light emitting diode (UV-LED) system. LWT 2022, 158, 113150. [Google Scholar] [CrossRef]
- Endarko, E.; Maclean, M.; Timoshkin, I.V.; MacGregor, S.J.; Anderson, J.G. High-intensity 405 nm light inactivation of Listeria monocytogenes. Photochem. Photobiol. 2012, 88, 1280–1286. [Google Scholar] [CrossRef]
- Li, Y.; Huang, T.-Y.; Mao, Y.; Chen, Y.; Shi, F.; Peng, R.; Chen, J.; Yuan, L.; Bai, C.; Chen, L.; et al. Study on the viable but non-culturable (VBNC) state formation of Staphylococcus aureus and its control in food system. Front. Microbiol. 2020, 11, 599739. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ye, C.; Lin, H.; Lv, L.; Yu, X. UV disinfection induces a VBNC state in Escherichia coli and Pseudomonas aeruginosa. Environ. Sci. Technol. 2015, 49, 1721–1728. [Google Scholar] [CrossRef]
- Trevors, J.T. Viable but non-culturable (VBNC) bacteria: Gene expression in planktonic and biofilm cells. J. Microbiol. Methods 2011, 86, 266–273. [Google Scholar] [CrossRef]
- Trevors, J.T. Can dead bacterial cells be defined and are genes expressed after cell death? J. Microbiol. Methods 2012, 90, 25–28. [Google Scholar] [CrossRef] [PubMed]
- Ramamurthy, T.; Ghosh, A.; Pazhani, G.P.; Shinoda, S. Current perspectives on viable but non-culturable (VBNC) pathogenic bacteria. Front. Public Health 2014, 2, 91118. [Google Scholar] [CrossRef] [PubMed]
- Ojha, D.; Patil, K.N. Molecular and functional characterization of the Listeria monocytogenes RecA protein: Insights into the homologous recombination process. Int. J. Biochem. Cell Biol. 2020, 119, 105642. [Google Scholar] [CrossRef] [PubMed]
- Kidambi, S.P.; Booth, M.G.; Kokjohn, T.A.; Miller, R.V. recA-dependence of the response of Pseudomonas aeruginosa to UVA and UVB irradiation. Microbiology 1996, 142, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Pousty, D.; Hofmann, R.; Gerchman, Y.; Mamane, H. Wavelength-dependent time–dose reciprocity and stress mechanism for UV-LED disinfection of Escherichia coli. J. Photochem. Photobiol. B 2021, 217, 112129. [Google Scholar] [CrossRef] [PubMed]
- Matallana-Surget, S.; Meador, J.A.; Joux, F.; Douki, T. Effect of the GC content of DNA on the distribution of UVB-induced bipyrimidine photoproducts. Photochem. Photobiol. Sci. 2008, 7, 794–801. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Kuru, E.; Sachdeva, A.; Vendrell, M. Fluorescent amino acids as versatile building blocks for chemical biology. Nat. Rev. Chem. 2020, 4, 275–290. [Google Scholar] [CrossRef]
- González, Y.; Gómez, G.; Moeller-Chávez, G.E.; Vidal, G. UV Disinfection Systems for wastewater treatment: Emphasis on reactivation of microorganisms. Sustainability 2023, 15, 11262. [Google Scholar] [CrossRef]
- Popovic, V.; Reman, J.; Koutchma, T. Comparison of UV-LED treatments at 255 and 281 nm in continuous-flow on bacterial inactivation, quality, nutrients and electrical efficiency in beverages. J. Food. Eng. 2023, 357, 111596. [Google Scholar] [CrossRef]
- Martino, V.; Ochsner, K.; Peters, P.; Zitomer, D.H.; Mayer, B.K. Virus and Bacteria Inactivation Using Ultraviolet Light-Emitting Diodes. Environ. Eng. Sci. 2021, 38, 458–468. [Google Scholar] [CrossRef]
- Amano, H.; Collazo, R.; De Santi, C.; Einfeldt, S.; Funato, M.; Glaab, J.; Hagedorn, S.; Hirano, A.; Hirayama, H.; Ishii, R.; et al. The 2020 UV emitter roadmap. J. Phys. D Appl. Phys. 2020, 53, 503001. [Google Scholar] [CrossRef]
- Martín-Sómer, M.; Pablos, C.; Adán, C.; van Grieken, R.; Marugán, J. A review on led technology in water photodisinfection. Sci. Total Environ. 2023, 885, 163963. [Google Scholar] [CrossRef] [PubMed]
- Li, G.Q.; Wang, W.L.; Huo, Z.Y.; Lu, Y.; Hu, H.Y. Comparison of UV-LED and low pressure UV for water disinfection: Photoreactivation and dark repair of Escherichia coli. Water Res. 2017, 126, 134–143. [Google Scholar] [CrossRef]
- Itani, N.; El Fadel, M. Microbial inactivation kinetics of UV LEDs and effect of operating conditions: A methodological critical analysis. Sci. Total Environ. 2023, 885, 163727. [Google Scholar] [CrossRef]
- MacIsaac, S.A.; Rauch, K.D.; Prest, T.; Simons, R.M.; Gagnon, G.A.; Stoddart, A.K. Improved disinfection performance for 280 nm LEDs over 254 nm low-pressure UV lamps in community wastewater. Sci. Rep. 2023, 13, 7576. [Google Scholar] [CrossRef]
- Pizzichetti, R.; Martín-Gamboa, M.; Pablos, C.; Reynolds, K.; Stanley, S.; Dufour, J.; Marugán, J. Environmental life cycle assessment of UV-C LEDs vs. mercury lamps and oxidant selection for diclofenac degradation. Sustain. Mater. Technol. 2024, 41, e01002. [Google Scholar] [CrossRef]
- Santos, A.L.; Gomes, N.C.; Henriques, I.; Almeida, A.; Correia, A.; Cunha, Â. Contribution of reactive oxygen species to UV-B-induced damage in bacteria. J. Photochem. Photobiol. B 2012, 117, 40–46. [Google Scholar] [CrossRef]
- Xiao, Y.; Chu, X.N.; He, M.; Liu, X.C.; Hu, J.Y. Impact of UVA pre-radiation on UVC disinfection performance: Inactivation, repair and mechanism study. Water Res. 2018, 141, 279–288. [Google Scholar] [CrossRef] [PubMed]
 ; the bacterial concentration, adjusting the initial bacterial concentration using a McFarland standard
; the bacterial concentration, adjusting the initial bacterial concentration using a McFarland standard  ; irradiation of the different bacteria with ultraviolet light at varying wavelengths (255 vs. 285 nm) and doses
; irradiation of the different bacteria with ultraviolet light at varying wavelengths (255 vs. 285 nm) and doses  ; and spreading of the irradiated bacteria on agar plates to reveal the inactivation kinetics
; and spreading of the irradiated bacteria on agar plates to reveal the inactivation kinetics  ; and performing RT-qPCR to investigate the expression of selected genes involved in SOS repair
; and performing RT-qPCR to investigate the expression of selected genes involved in SOS repair  . Created with BioRender.com.
. Created with BioRender.com.
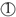 ; the bacterial concentration, adjusting the initial bacterial concentration using a McFarland standard
; the bacterial concentration, adjusting the initial bacterial concentration using a McFarland standard  ; irradiation of the different bacteria with ultraviolet light at varying wavelengths (255 vs. 285 nm) and doses
; irradiation of the different bacteria with ultraviolet light at varying wavelengths (255 vs. 285 nm) and doses  ; and spreading of the irradiated bacteria on agar plates to reveal the inactivation kinetics
; and spreading of the irradiated bacteria on agar plates to reveal the inactivation kinetics  ; and performing RT-qPCR to investigate the expression of selected genes involved in SOS repair
; and performing RT-qPCR to investigate the expression of selected genes involved in SOS repair  . Created with BioRender.com.
. Created with BioRender.com.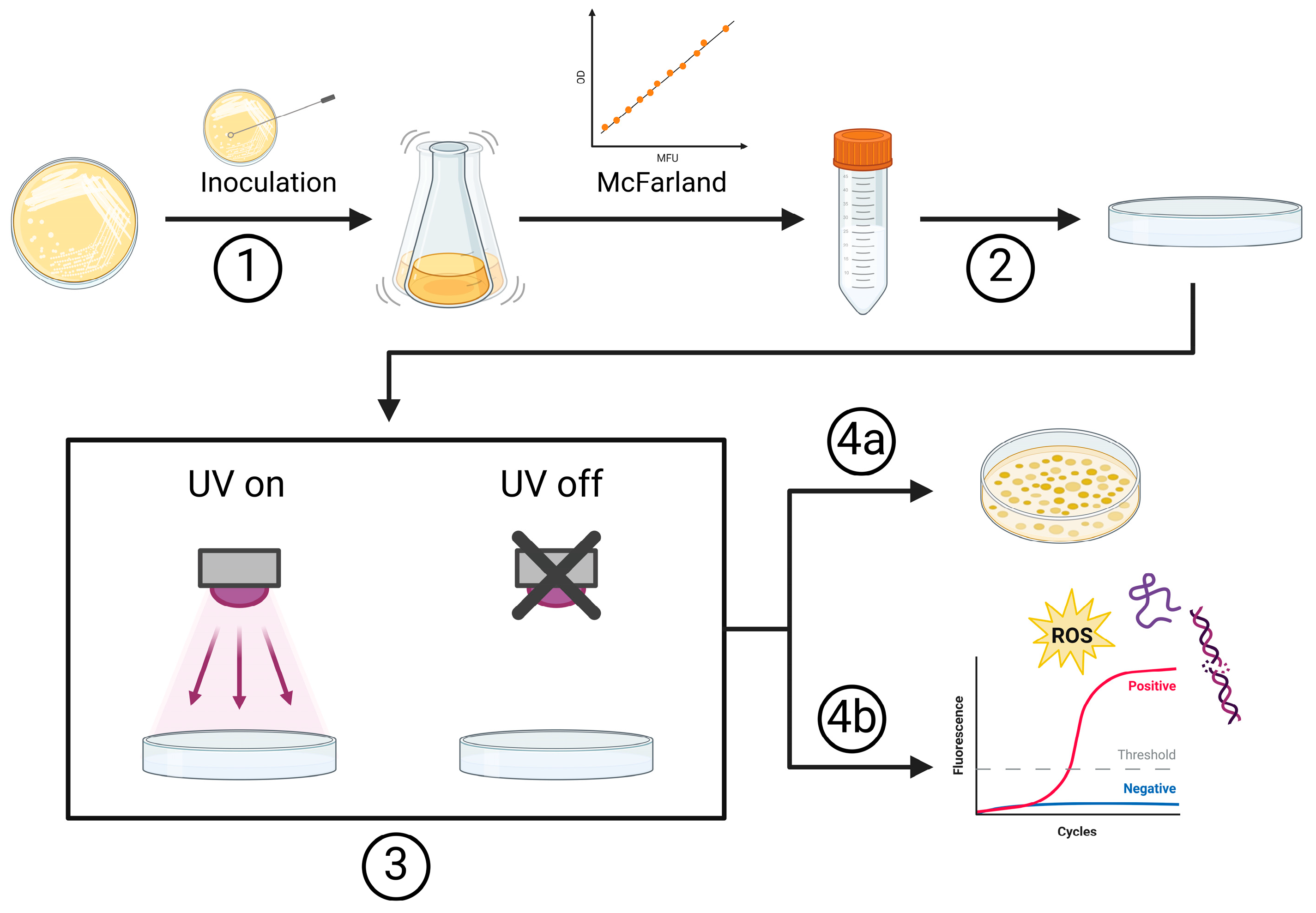


| Microorganism | |||
|---|---|---|---|
| C. freundii | |||
| E. coli | |||
| L. innocua | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mutschlechner, M.; Chisté, D.; Hauptmann, D.; Schöbel, H. Ultraviolet (Spot)light on Water Treatment: Targeting Inactivation Efficiency and Stress Responses of Gram-Positive and Gram-Negative Bacteria Using UV-B and UV-C LEDs. Water 2024, 16, 2028. https://doi.org/10.3390/w16142028
Mutschlechner M, Chisté D, Hauptmann D, Schöbel H. Ultraviolet (Spot)light on Water Treatment: Targeting Inactivation Efficiency and Stress Responses of Gram-Positive and Gram-Negative Bacteria Using UV-B and UV-C LEDs. Water. 2024; 16(14):2028. https://doi.org/10.3390/w16142028
Chicago/Turabian StyleMutschlechner, Mira, Daniela Chisté, Daniel Hauptmann, and Harald Schöbel. 2024. "Ultraviolet (Spot)light on Water Treatment: Targeting Inactivation Efficiency and Stress Responses of Gram-Positive and Gram-Negative Bacteria Using UV-B and UV-C LEDs" Water 16, no. 14: 2028. https://doi.org/10.3390/w16142028
APA StyleMutschlechner, M., Chisté, D., Hauptmann, D., & Schöbel, H. (2024). Ultraviolet (Spot)light on Water Treatment: Targeting Inactivation Efficiency and Stress Responses of Gram-Positive and Gram-Negative Bacteria Using UV-B and UV-C LEDs. Water, 16(14), 2028. https://doi.org/10.3390/w16142028






