Mechanism of Crude Oil Biodegradation in Bioreactors: A Model Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Oil-Degrading Microorganisms
2.2. Culture Media
2.3. Bioreactors
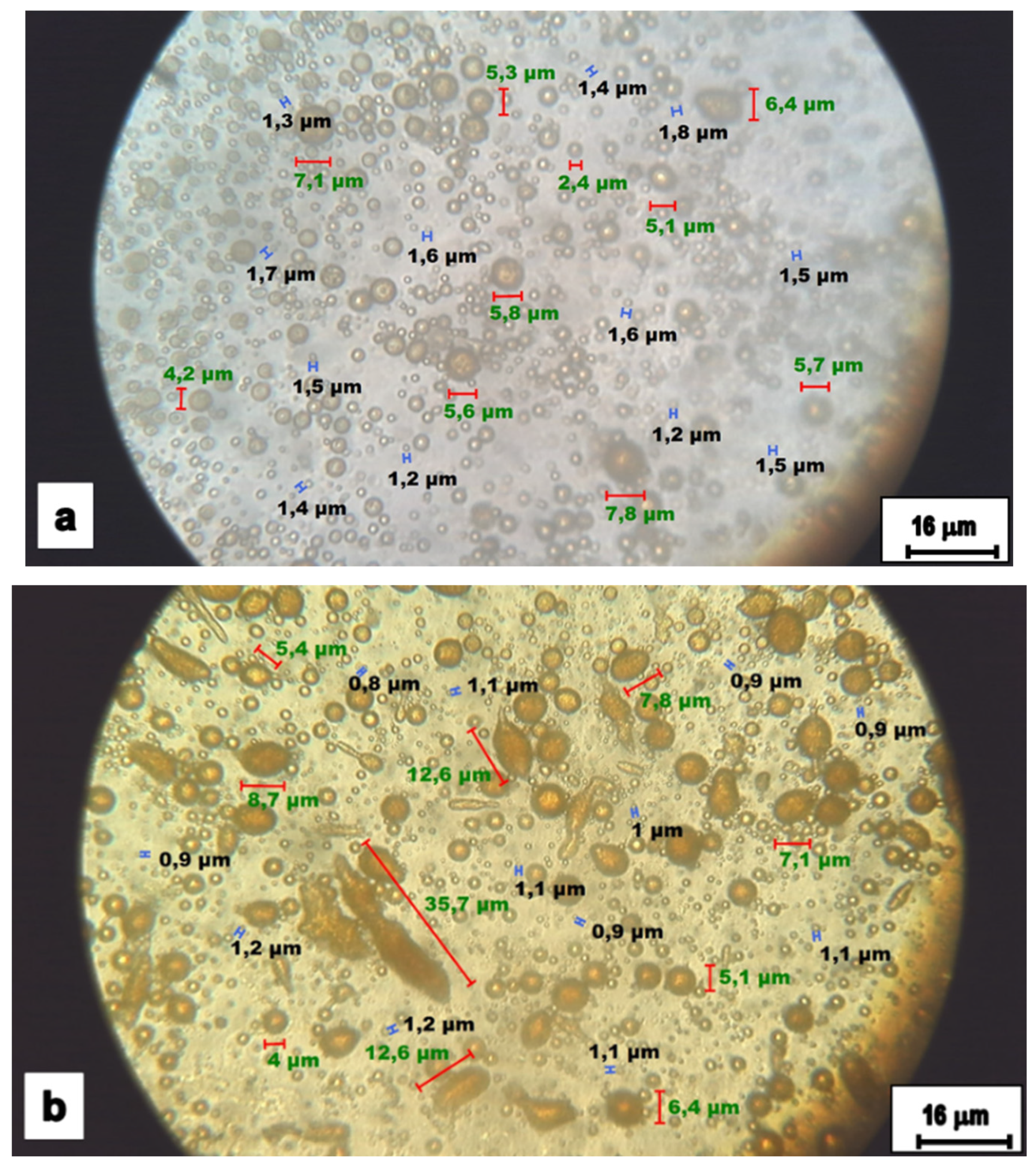
2.4. Microphotographs
2.5. Crude Oil Analysis by GC-FID
3. Proposed Mechanism
3.1. Kinetics
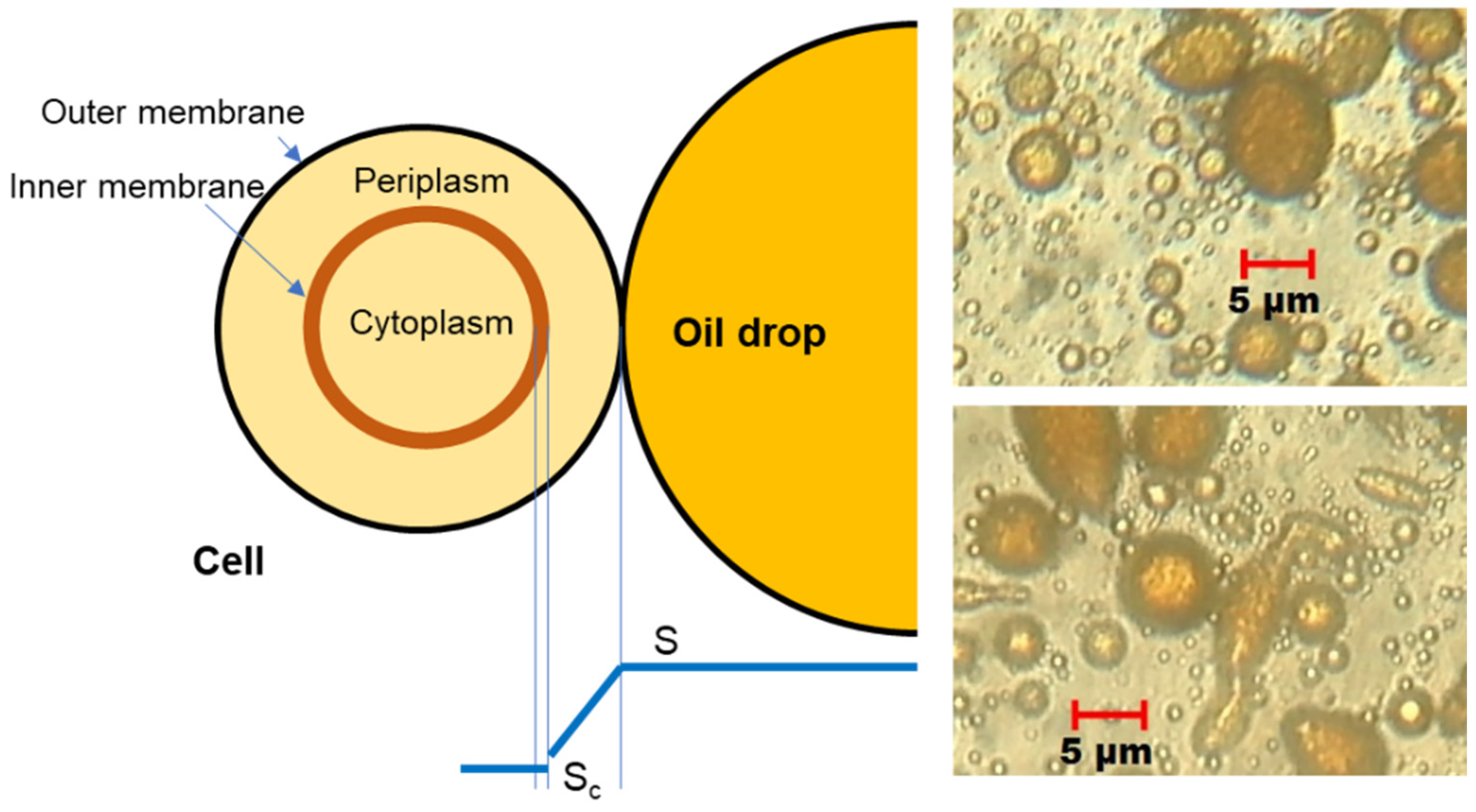
3.2. Mass Transport
4. Results and Discussion
4.1. Critical Time
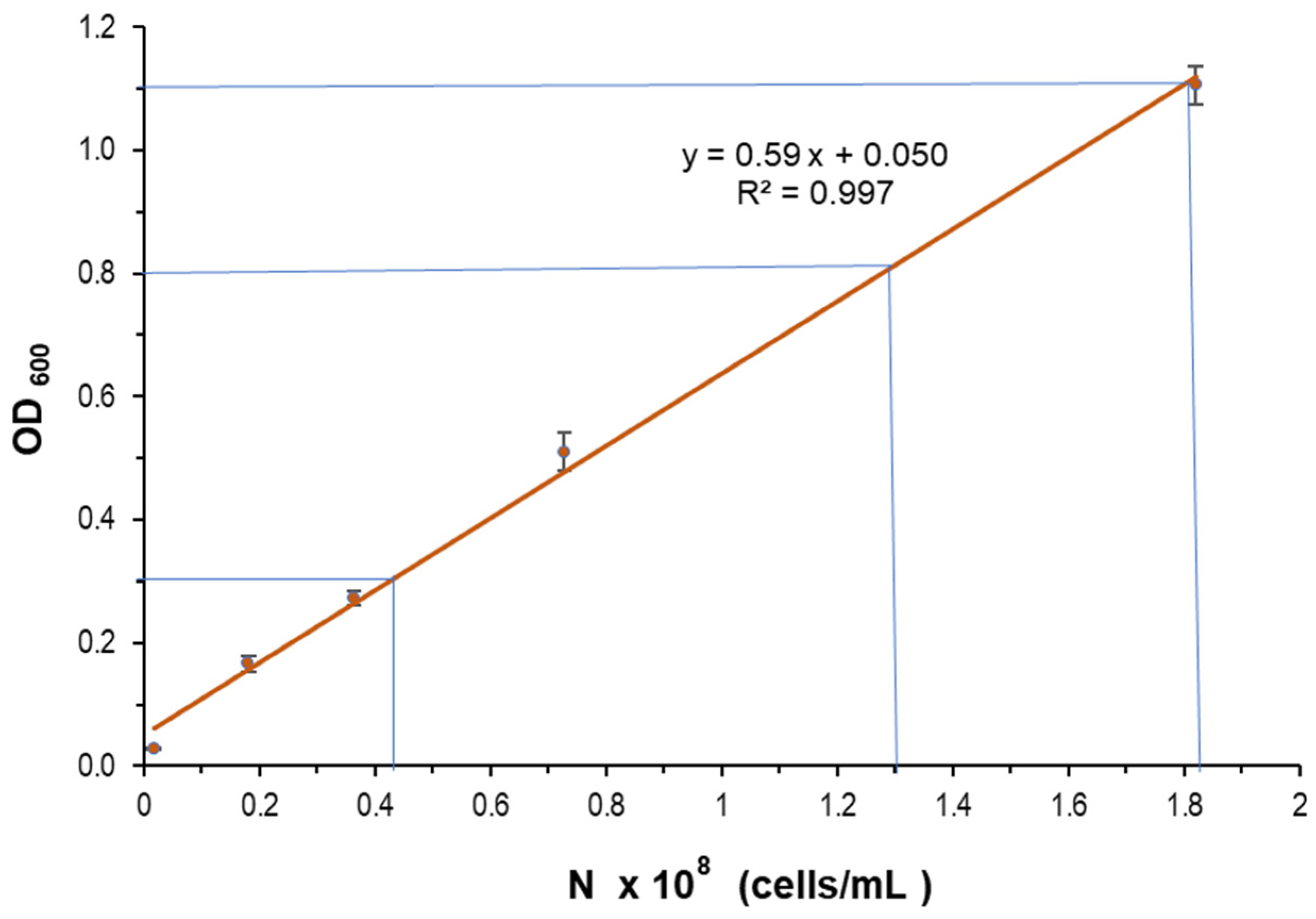
4.2. Number of Cells
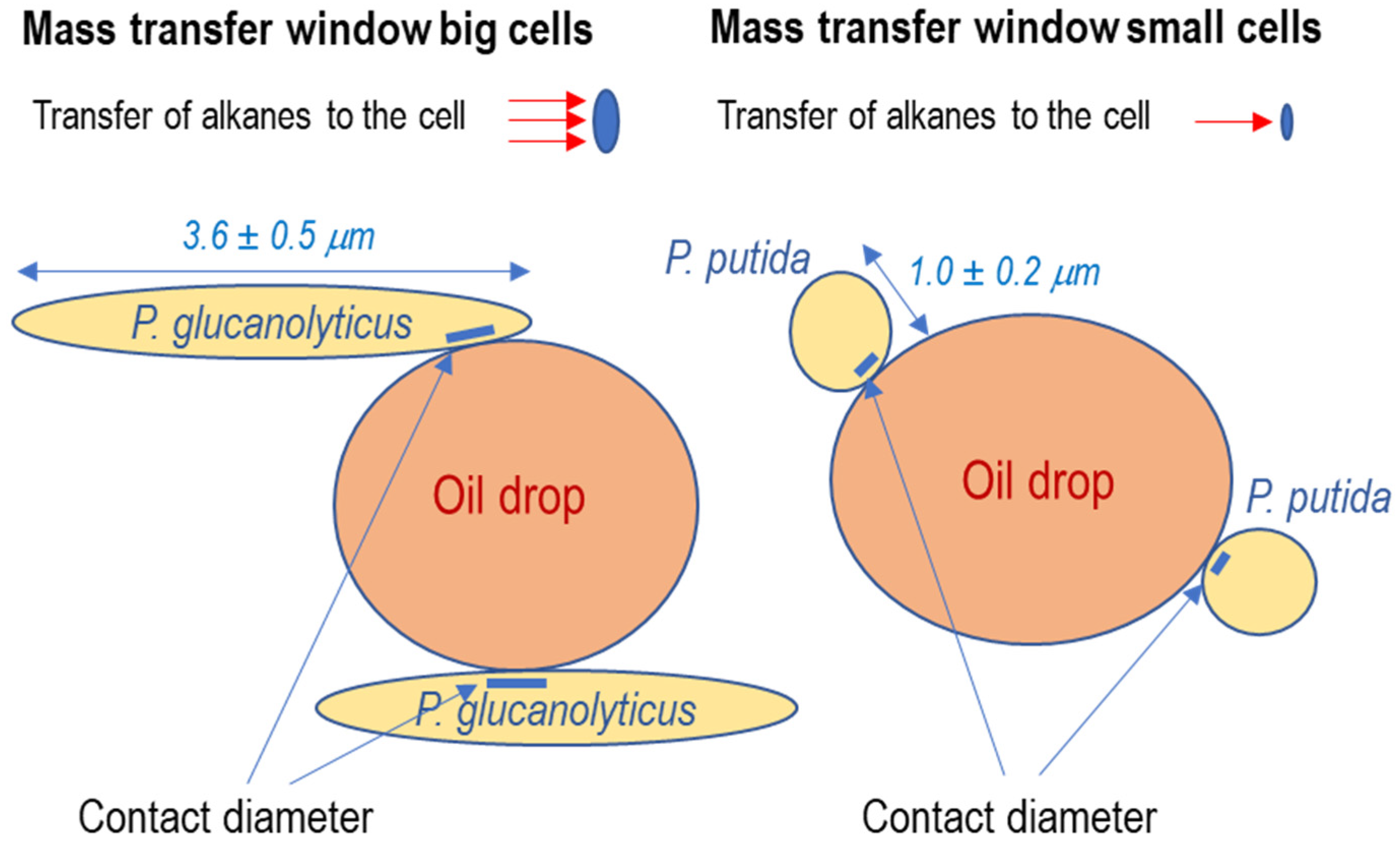
4.3. Projected Area of the Cells
4.4. Mass Transfer Coefficients
4.5. Future Research
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Acronyms | |
| AlkH | alkane hydroxylase |
| DCM | dichloromethane |
| MSM | mineral salt medium |
| TPH | total petroleum hydrocarbons |
| Variables | |
| Ac | projected area of the cell (m2) |
| d | diameter of the cell (μm) |
| Js | mass flux (g/m2d) |
| k | kinetic coefficient (d−1) |
| kL | permeability coefficient (m/d) |
| N | number of cells for a unit volume of the bioreactor (cell/m3) |
| P | polystyrene microspheres density (g/mL) |
| r | degradation rate (g/Ld) |
| rTPH | degradation rate of TPH (g/Ld) |
| S | substrate concentration in the oil phase (g/L) |
| Stc | substrate concentration in the oil phase at the critical time (g/L) |
| Sc | substrate concentration inside the cell (outer side of the inner membrane) (g/L) |
| So | initial substrate concentration in the oil phase (g/L) |
| t | time (d) |
| tc | critical time, at which the kinetics and mass transport are in balance (d) |
| V | bioreactor volume (m3) |
| W | polystyrene microspheres mass (g) |
| Greek letters | |
| α | fraction of the contact area related to the projected area of the cell |
References
- Atlas, R.M. Microbial degradation of petroleum hydrocarbons: An environmental perspective. Microbiol. Rev. 1981, 45, 180–209. [Google Scholar] [CrossRef] [PubMed]
- Atlas, R.M.; Hazen, T.C. Oil biodegradation and bioremediation: A tale of the two worst spills in U.S. history. Environ. Sci. Technol. 2011, 45, 6709–6715. [Google Scholar] [CrossRef] [PubMed]
- Abbasian, F.; Lockington, R.; Mallavarapu, M.; Naidu, R. A comprehensive review of aliphatic hydrocarbon biodegradation by bacteria. Appl. Biochem. Biotechnol. 2015, 176, 670–699. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, M. Dioxygen activation responsible for oxidation of aliphatic and aromatic hydrocarbon compounds: Current state and variants. Appl. Microbiol. Biotechnol. 2010, 87, 1595–1603. [Google Scholar] [CrossRef]
- Urlacher, V.B.; Girhard, M. Cytochrome P450 monooxygenases: An update on perspectives for synthetic application. Trends Biotechnol. 2012, 30, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Van Beilen, J.B.; Funhoff, E.G. Alkane hydroxylases involved in microbial alkane degradation. Appl. Microbiol. Biotechnol. 2007, 74, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Al-Hawash, A.B.; Dragh, M.A.; Li, S.; Alhujaily, A.; Abbood, H.A.; Zhang, X.; Ma, F. Principles of microbial degradation of petroleum hydrocarbons in the environment. Egypt. J. Aquat. Res. 2018, 44, 71–76. [Google Scholar] [CrossRef]
- Gricman, Ł.; Vogel, C.; Pleiss, J. Conservation analysis of class-specific positions in cytochrome P450 monooxygenases: Functional and structural relevance. Proteins 2014, 82, 491–504. [Google Scholar] [CrossRef]
- Viggor, S.; Jöesaar, M.; Vedler, E.; Kiiker, R.; Pärnpuu, L.; Heinaru, A. Occurrence of diverse alkane hydroxylase alkB genes in indigenous oil-degrading bacteria of Baltic Sea surface water. Marine Pollut. Bull. 2015, 101, 507–516. [Google Scholar] [CrossRef]
- Xu, N.; Bao, M.; Sun, P.; Li, Y. Study on bioadsorption and biodegradation of petroleum hydrocarbons by a microbial consortium. Bioresour. Technol. 2013, 149, 22–30. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Bao, M.; Sun, P. Metabolic pathway for a new strain Pseudomonas synxantha LSH-7’: From chemotaxis to uptake of n-hexadecane. Sci. Rep. 2017, 7, 39068. [Google Scholar] [CrossRef] [PubMed]
- Ladygina, N.; Dedyukhina, E.; Vainshtein, M. A review on microbial synthesis of hydrocarbons. Proc. Biochem. 2006, 41, 1001–1014. [Google Scholar] [CrossRef]
- Rojo, F. Degradation of alkanes by bacteria. Environ. Microbiol. 2009, 11, 2477–2490. [Google Scholar] [CrossRef]
- Sajna, K.V.; Sukumaran, R.K.; Gottumukkala, L.D.; Pandey, A. Crude oil biodegradation aided by biosurfactants from Pseudozyma sp. NII 08165 or its culture broth. Bioresour. Technol. 2015, 191, 133–139. [Google Scholar] [CrossRef]
- Zhang, Z.; Hou, Z.; Yang, C.; Ma, C.; Tao, F.; Xu, P. Degradation of n-alkanes and polycyclic aromatic hydrocarbons in petroleum by a newly isolated Pseudomonas aeruginosa DQ8. Bioresour. Technol. 2011, 102, 4111–4116. [Google Scholar] [CrossRef] [PubMed]
- Varjani, S.J. Microbial degradation of petroleum hydrocarbons. Bioresour. Technol. 2017, 223, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Das, N.; Chandran, P. Microbial degradation of petroleum hydrocarbon contaminants: An overview. Biotechnol. Res. Int. 2011, 2011, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Paisse, S.; Duran, R.; Coulon, F.; Goñi-Urriza, M. Are alkane hydroxylase genes (alkB) relevant to assess petroleum bioremediation processes in chronically polluted coastal sediments? Appl. Microbiol. Biotechnol. 2011, 92, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Pi, Y.-R.; Bao, M.-T. Investigation of kinetics in bioaugmentation of crude oil via high-throughput sequencing: Enzymatic activities, bacterial community composition and functions. Pet. Sci. 2022, 19, 1905–1914. [Google Scholar] [CrossRef]
- Van Beilen, J.B.; Wubbolts, M.G.; Witholt, B. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation 1994, 5, 161–174. [Google Scholar] [CrossRef]
- Chandra, S.; Sharma, R.; Singh, K.; Sharma, A. Application of bioremediation technology in the environment contaminated with petroleum hydrocarbon. Ann. Microbiol. 2013, 63, 417–431. [Google Scholar] [CrossRef]
- Kumari, B.; Singh, S.N.; Singh, D.P. Characterization of two biosurfactant producing strains in crude oil degradation. Proc. Biochem. 2012, 47, 2463–2471. [Google Scholar] [CrossRef]
- Liu, Y.; Wan, Y.Y.; Wang, C.; Ma, Z.; Liu, X.; Li, S. Biodegradation of n-alkanes in crude oil by three identified bacterial strains. Fuel 2020, 275, 117897. [Google Scholar] [CrossRef]
- Ron, E.Z.; Rosenberg, E. Enhanced bioremediation of oil spills in the sea. Curr. Opin. Biotechnol. 2014, 27, 191–194. [Google Scholar] [CrossRef]
- Xia, M.; Fu, D.; Chakraborty, R.; Singh, R.P.; Terry, N. Enhanced crude oil depletion by constructed bacterial consortium comprising bioemulsifier producer and petroleum hydrocarbon degraders. Bioresour. Technol. 2019, 282, 456–463. [Google Scholar] [CrossRef]
- Varjani, S.J.; Upasani, V.N. Carbon spectrum utilization by an indigenous strain of Pseudomonas aeruginosa NCIM 5514: Production, characterization and surface active properties of biosurfactant. Bioresour. Technol. 2016, 221, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Zhou, J.-D.; Ma, F.; Shi, R.-J.; Han, S.-Q.; Zhang, J.; Zhang, Y. Simultaneous inhibition of sulfate-reducing bacteria, removal of H2S and production of rhamnolipid by recombinant Pseudomonas stutzeri Rhl: Applications for microbial enhanced oil recovery. Bioresour. Technol. 2016, 207, 24–30. [Google Scholar] [CrossRef]
- Abdullah, S.R.S.; Al-Baldawi, I.A.; Almansoory, A.F.; Purwanti, I.F.; Al-Sbani, N.H.; Sharuddin, S.S.N. Plant-assisted remediation of hydrocarbons in water and soil: Application, mechanisms, challenges and opportunities. Chemosphere 2020, 247, 125932. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, V.S.; Hollenbach, E.B.; Maboni, F.; Vainstein, M.; Camargo, F.; Do-Carmo, R.P.M.; Bento, F.M. Biodegradation potential of oily sludge by pure and mixed bacterial cultures. Bioresour. Technol. 2011, 102, 11003–11010. [Google Scholar] [CrossRef]
- Kavitha, V.; Mandal, A.B.; Gnanamani, A. Microbial biosurfactant mediated removal and/or solubilization of crude oil contamination from soil and aqueous phase: An approach with Bacillus licheniformis MTCC 5514. Int. Biodeterior. Biodegrad. 2014, 94, 24–30. [Google Scholar] [CrossRef]
- Nievas, M.L.; Commendatore, M.G.; Esteves, J.L.; Bucalá, V. Biodegradation pattern of hydrocarbons from a fuel oil-type complex residue by an emulsifier-producing microbial consortium. J. Hazard. Mat. 2008, 154, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Xue, J.; Xiao, X.; Qiao, Y.; Wu, Y.; Gao, Y. Mechanism of Degrading Petroleum Hydrocarbons by Compound Marine Petroleum-Degrading Bacteria: Surface Adsorption, Cell Uptake, and Biodegradation. Energy Fuels 2019, 33, 11373–11379. [Google Scholar] [CrossRef]
- Wang, X.B.; Chi, C.Q.; Nie, Y.; Tang, Y.Q.; Tan, Y.; Wu, G.; Wu, X.L. Degradation of petroleum hydrocarbons (C6–C40) and crude oil by a novel Dietzia strain. Bioresour. Technol. 2011, 102, 7755–7761. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.K.; Bae, H. Biotransformation and chemotaxis of 4-chloro-2-nitrophenol by Pseudomonas sp. JHN. Microbial Cell Factories 2014, 13, 110. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Costa, C.; Santos, A.; Vega, M.A. Kinetics of Arab Light crude oil degradation by Pseudomonas and Bacillus strains. Water 2022, 14, 3802. [Google Scholar] [CrossRef]
- Yudono, B.; Said, M.; Sabaruddin; Napoleon, A.; Fanani, Z. Kinetics Approach of Biodegradation of Petroleum Contaminated Soil by using Indigenous Isolated Bacteria. J. Trop. Soils 2011, 16, 33–38. [Google Scholar] [CrossRef][Green Version]
- Roy, A.S.; Baruah, R.; Borah, M.; Singh, A.K.; Deka Boruah, H.P.; Saikia, N.; Deka, M.; Dutta, N.; Bora, T.C. Bioremediation potential of native hydrocarbon degrading bacterial strains in crude oil contaminated soil under microcosm study. Int. Biodeter. Biodegr. 2014, 94, 79–89. [Google Scholar] [CrossRef]
- Varjani, S.J.; Rana, D.P.; Jain, A.K.; Bateja, S.; Upasani, V.N. Synergistic ex-situ biodegradation of crude oil by halotolerant bacterial consortium of indigenous strains isolated from on shore sites of Gujarat, India. Int. Biodeter. Biodegr. 2015, 103, 116–124. [Google Scholar] [CrossRef]
- Choi, D.H.; Hori, K.; Tanji, Y.; Unno, H. Microbial degradation kinetics of solid alkane dissolved in nondegradable oil phase. Biochem. Eng. J. 1999, 3, 71–78. [Google Scholar] [CrossRef][Green Version]
- Xu, X.; Liu, W.; Tian, S.; Wang, W.; Qi, Q.; Jiang, P.; Gao, X.; Li, H.; Yu, H. Petroleum hydrocarbon-degrading bacteria for the remediation of oil pollution under aerobic conditions: A perspective analysis. Front. Microbiol. 2015, 9, 2885. [Google Scholar] [CrossRef]
- Zheng, M.; Wang, W.; Hayes, M.; Nydell, A.; Tarr, M.A.; Van Bael, S.A.; Papadopoulos, K. Degradation of Macondo 252 oil by endophytic Pseudomonas putida. J. Environ. Chem. Eng. 2018, 6, 643–648. [Google Scholar] [CrossRef]
- Chettri, B.; Mukherjee, A.; Langpoklakpam, J.S.; Chattopadhyay, D.; Singh, A.K. Kinetics of nutrient enhanced crude oil degradation by Pseudomonas aeruginosa AKS1 and Bacillus sp. AKS2 isolated from Guwahati refinery, India. Environ. Pollut. 2016, 216, 548–558. [Google Scholar] [CrossRef]
- Beal, J.; Farny, N.G.; Haddock-Angelli, T.; Selvarajah, V.; Baldwin, G.S.; Buckley-Taylor, R.; Gershater, M.; Kiga, D.; Marken, J.; Sanchania, V.; et al. Robust estimation of bacterial cell count from optical density. Commun. Biol. 2020, 3, 512. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, K.; McVey, A.F.; Clark, I.B.N.; Swain, P.S.; Pilizota, T. General calibration of microbial growth in microplate readers. Sci. Rep. 2016, 6, 38828. [Google Scholar] [CrossRef] [PubMed]
- Hajieghrari, M.; Hejazi, P. Enhanced biodegradation of n-Hexadecane in solid-phase of soil by employing immobilized Pseudomonas aeruginosa on size-optimized coconut fibers. J. Hazard. Mat. 2020, 389, 122134. [Google Scholar] [CrossRef]
- Sharma, S.; Pandey., L.M. Biodegradation kinetics of binary mixture of hexadecane and phenanthrene by the bacterial microconsortium. Bioresour. Technol. 2022, 358, 127408. [Google Scholar] [CrossRef]
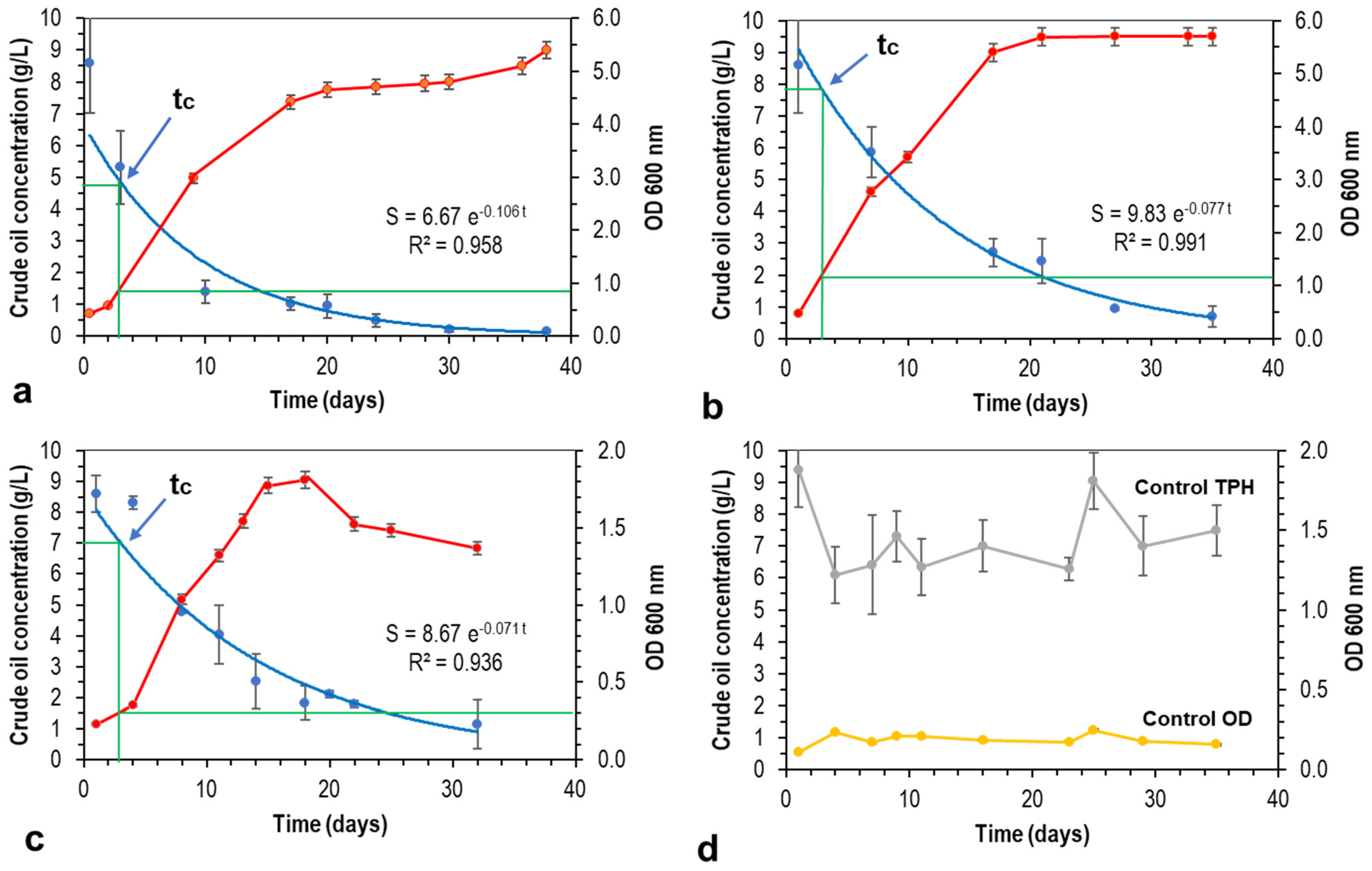


| Oil-Degrader | k (d−1) | Stc (g/L) | Sc (g/L) |
|---|---|---|---|
| B. licheniformis | 0.106 | 4.7 | 0.2 |
| P. putida | 0.077 | 7.8 | 0.7 |
| P. glucanolyticus | 0.071 | 7.0 | 1.1 |
| Oil-Degrader | d (μm) * | Ac × 10−12 (m2) ** | N × 1014 (cells/m3) | kL α (m/d) |
|---|---|---|---|---|
| B. licheniformis | 1.5 ± 0.3 (0.4) | 0.71 ± 0.09 | 1.30 (ABS600 = 0.8) | 1.60 × 10−3 |
| P. putida | 1.0 ± 0.2 | 0.77 ± 0.04 | 1.83 (ABS600 = 1.1) | 5.25 × 10−4 |
| P. glucanolyticus | 3.6 ± 0.5 (0.4) | 3.14 ± 0.25 | 0.43 (ABS600 = 0.3) | 6.19 × 10−4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa, C.; Millán, N. Mechanism of Crude Oil Biodegradation in Bioreactors: A Model Approach. Water 2024, 16, 1653. https://doi.org/10.3390/w16121653
Costa C, Millán N. Mechanism of Crude Oil Biodegradation in Bioreactors: A Model Approach. Water. 2024; 16(12):1653. https://doi.org/10.3390/w16121653
Chicago/Turabian StyleCosta, Carlos, and Nicolás Millán. 2024. "Mechanism of Crude Oil Biodegradation in Bioreactors: A Model Approach" Water 16, no. 12: 1653. https://doi.org/10.3390/w16121653
APA StyleCosta, C., & Millán, N. (2024). Mechanism of Crude Oil Biodegradation in Bioreactors: A Model Approach. Water, 16(12), 1653. https://doi.org/10.3390/w16121653








