Spatial and Temporal Variation in the Fish Diversity in Dianchi Lake and the Influencing Factors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Determination of the Sampling Points
2.2. Sample Collection and Storage
2.3. The Extraction of Total Aqueous DNA
2.4. Evaluation of Universal Detection Primers for Fish Screening
2.5. Analysis of the Environmental DNA Metabarcoding Sequencing Data
2.6. Identification of the Environmental Factors
3. Results
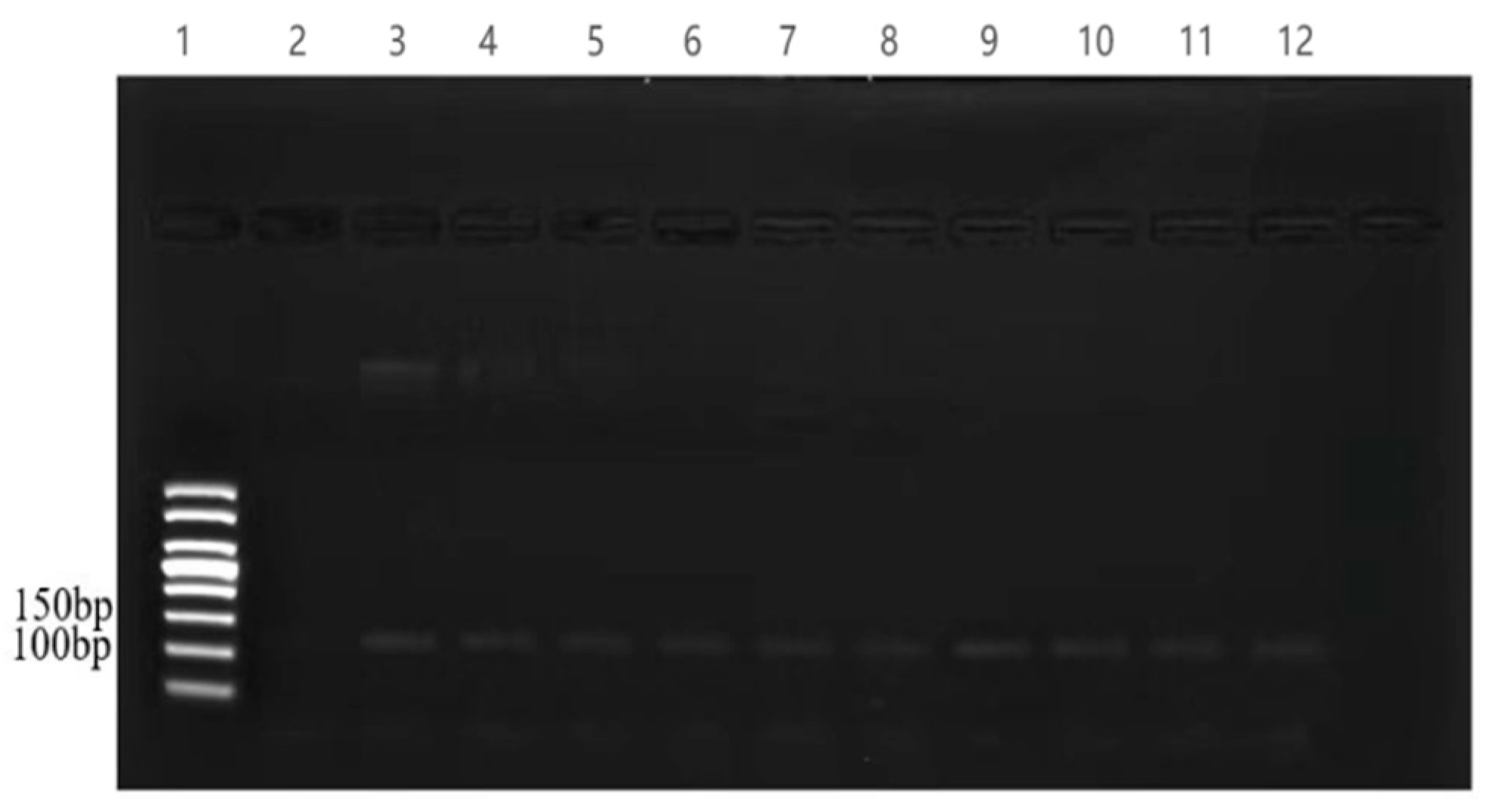
3.1. Screening of the Universal Primer for the Detection of Fish
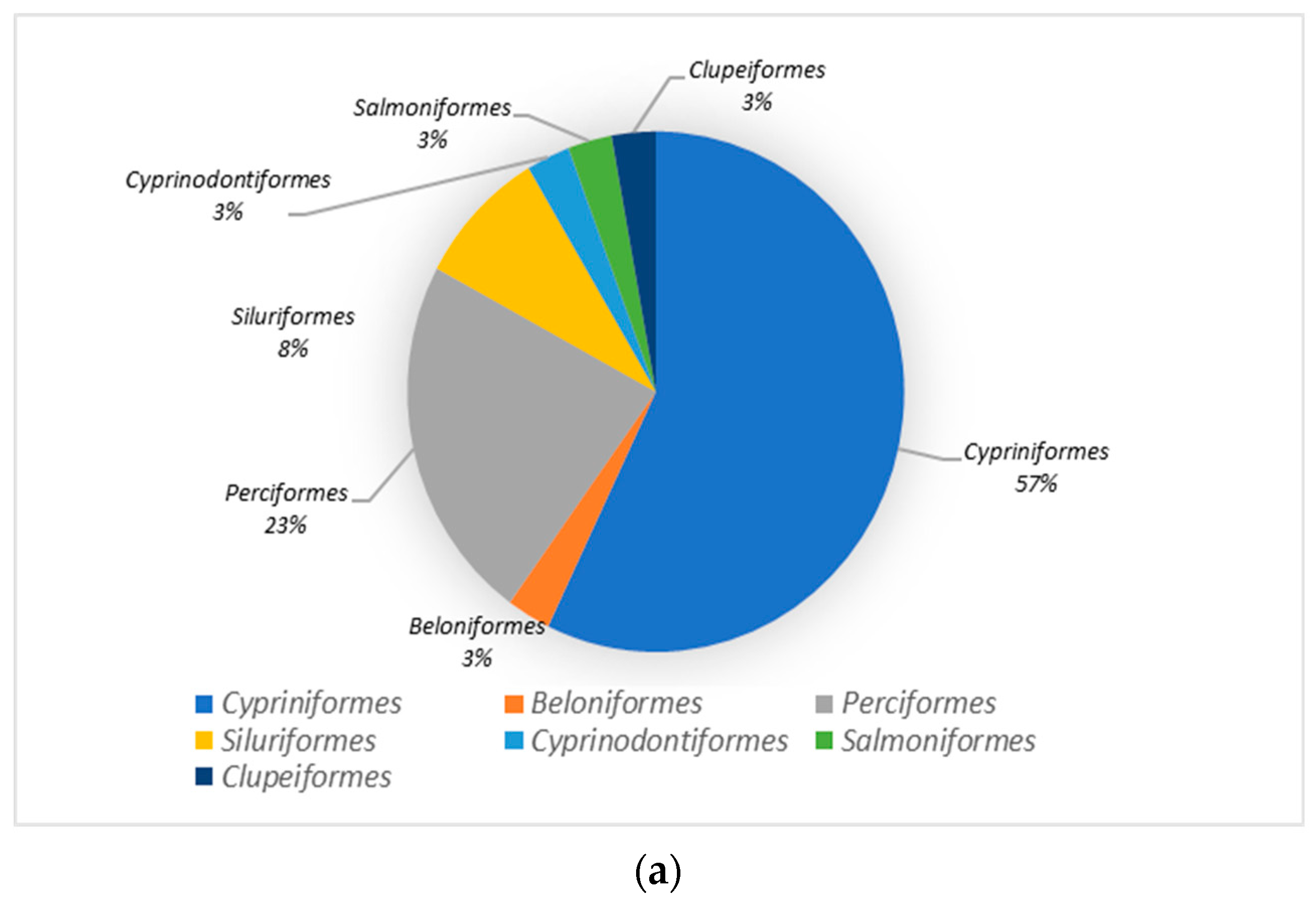
3.2. Species Analysis of the Dianchi Basin’s Fish Communities
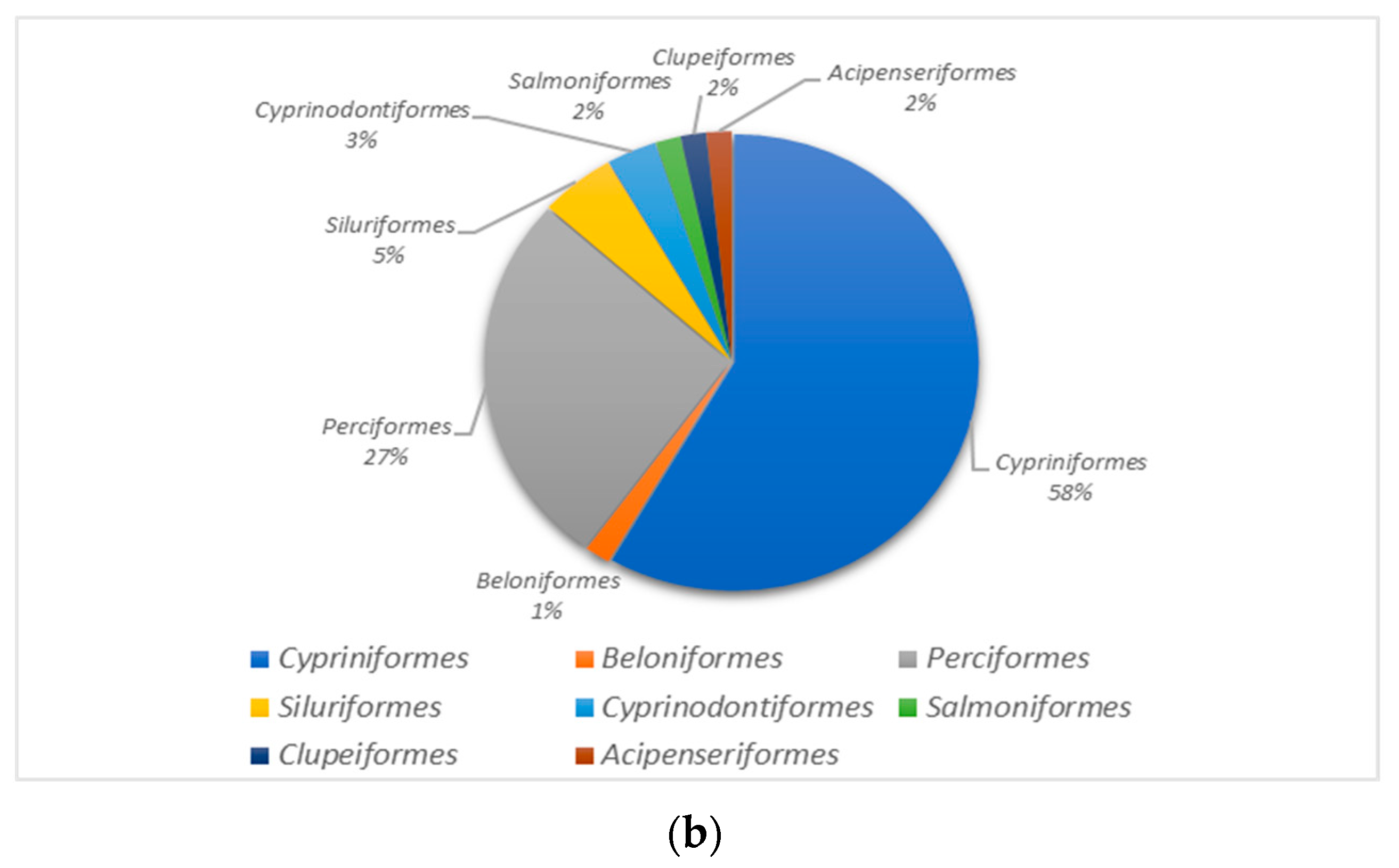
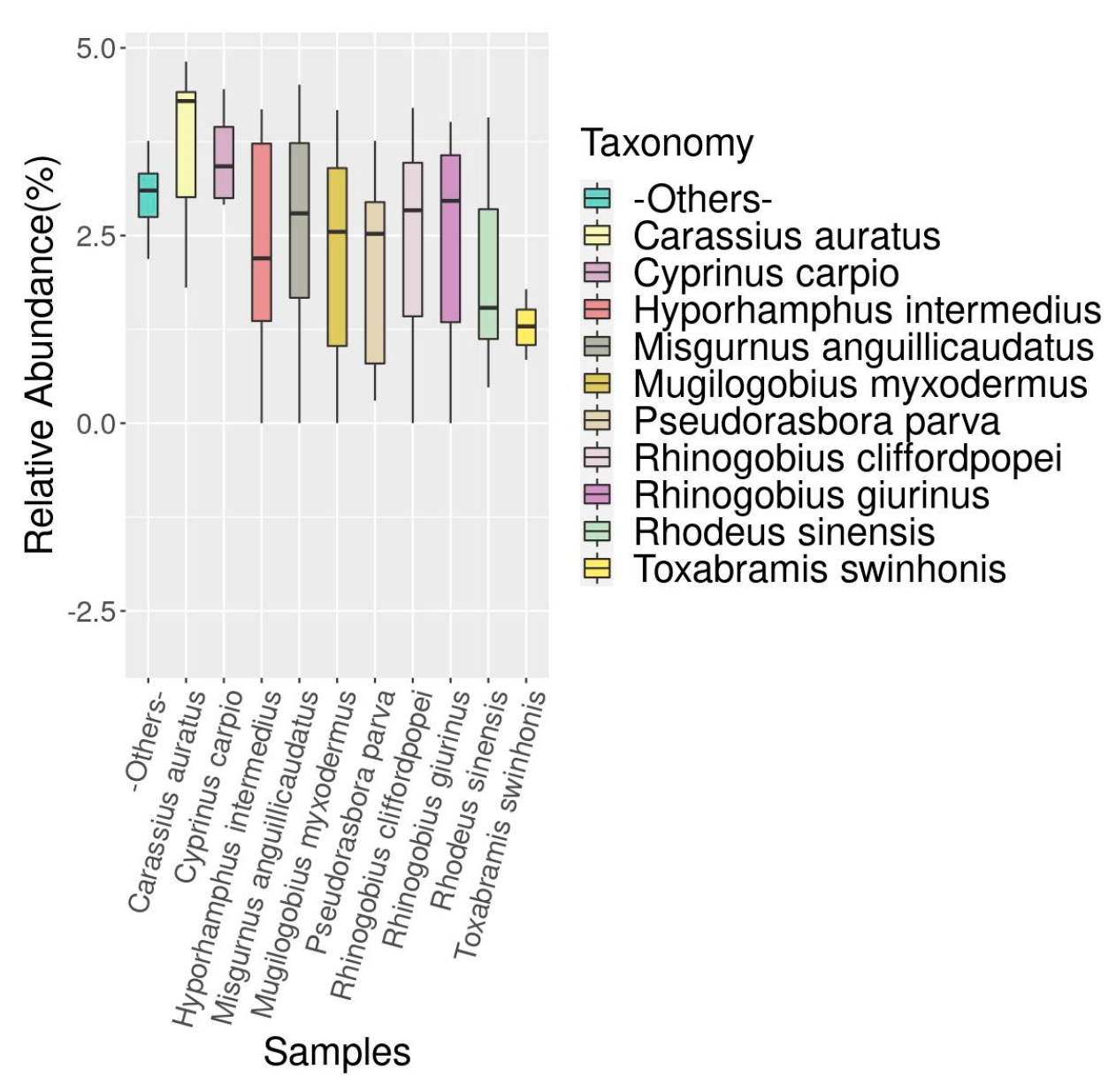
3.3. Analysis of Fish Species Abundance in the Dianchi Basin during Dry and Rich Seasons
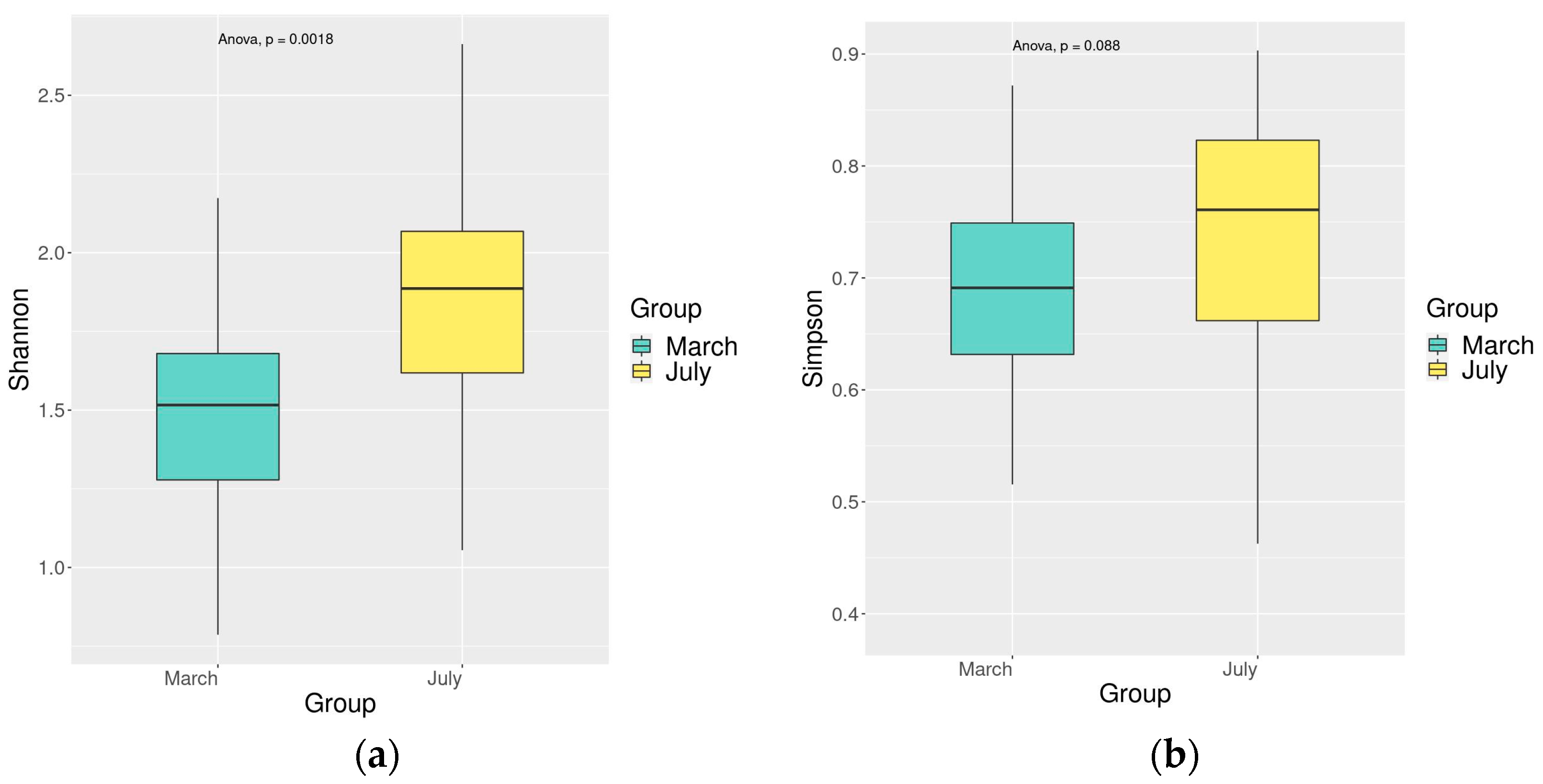
3.4. Comparison of Fish Diversity in the Dianchi Basin during Dry and Rich Periods
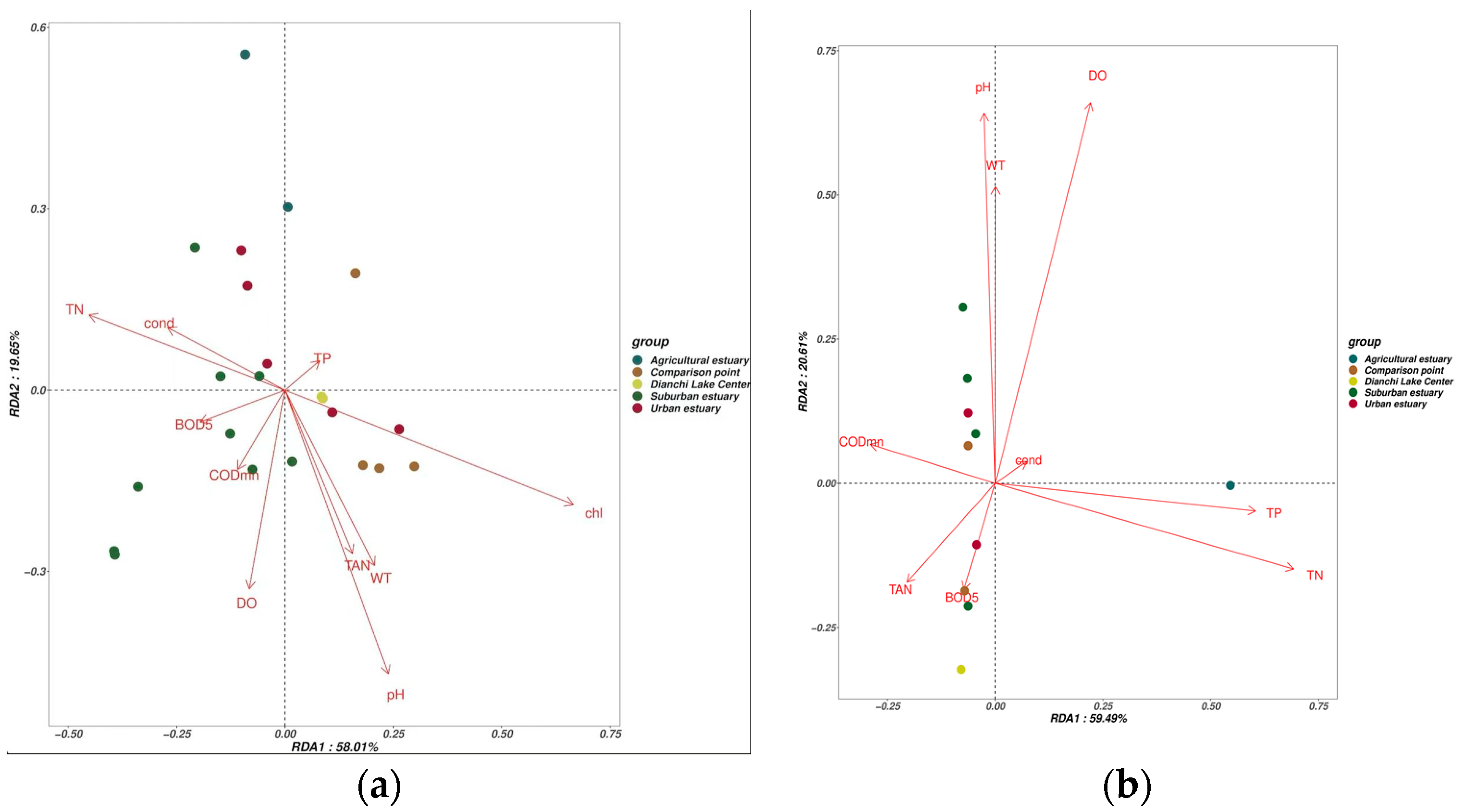
3.5. The Impact of Environmental Conditions on Fish Communities
3.6. The Spatial Heterogeneity of Numerous Kinds of Estuarine Fishes
3.6.1. Differences in Fish Abundance Amongst Various Kinds of Estuaries
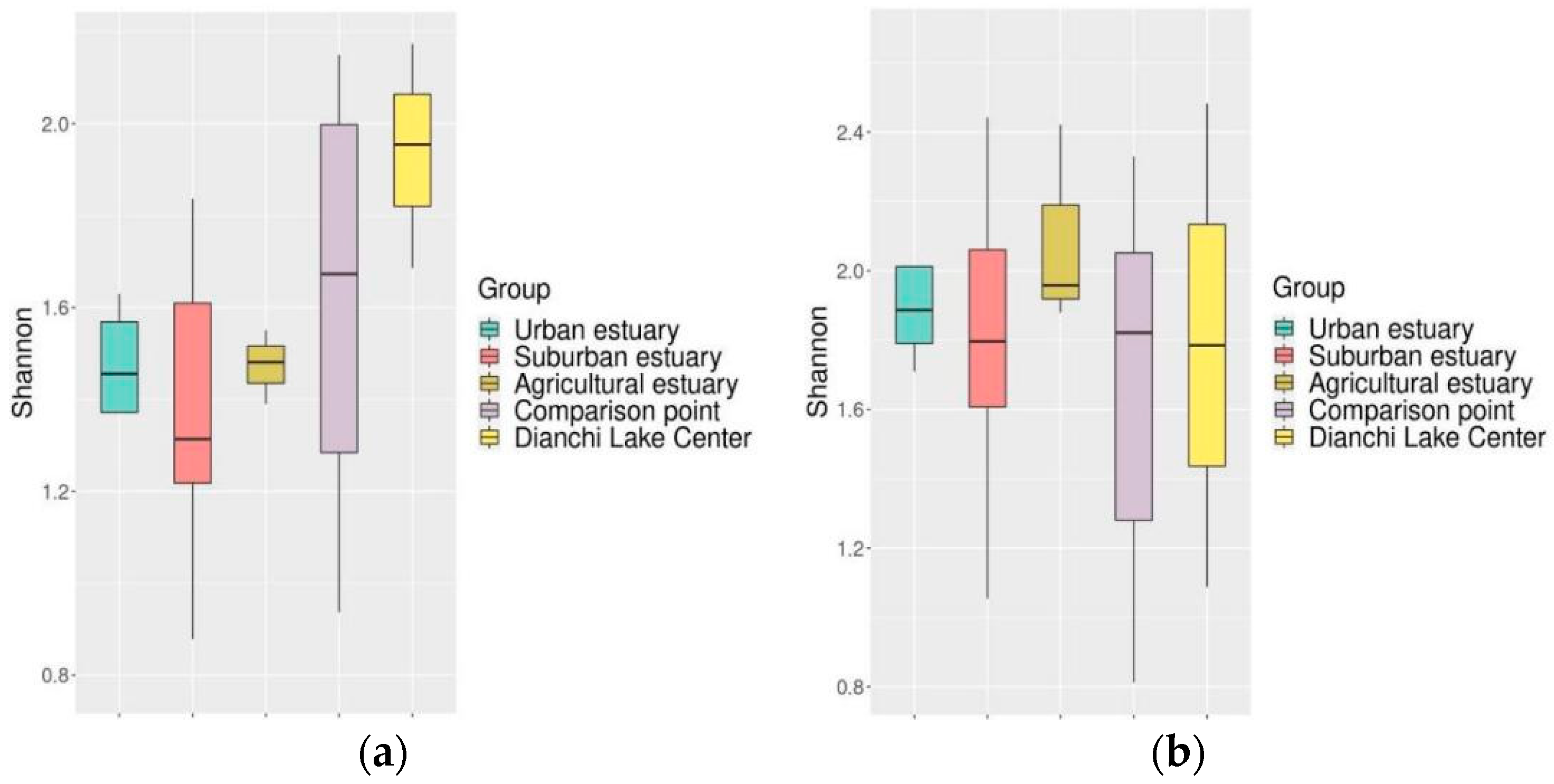
3.6.2. Of Fish Diversity among the Estuaries within the Lake
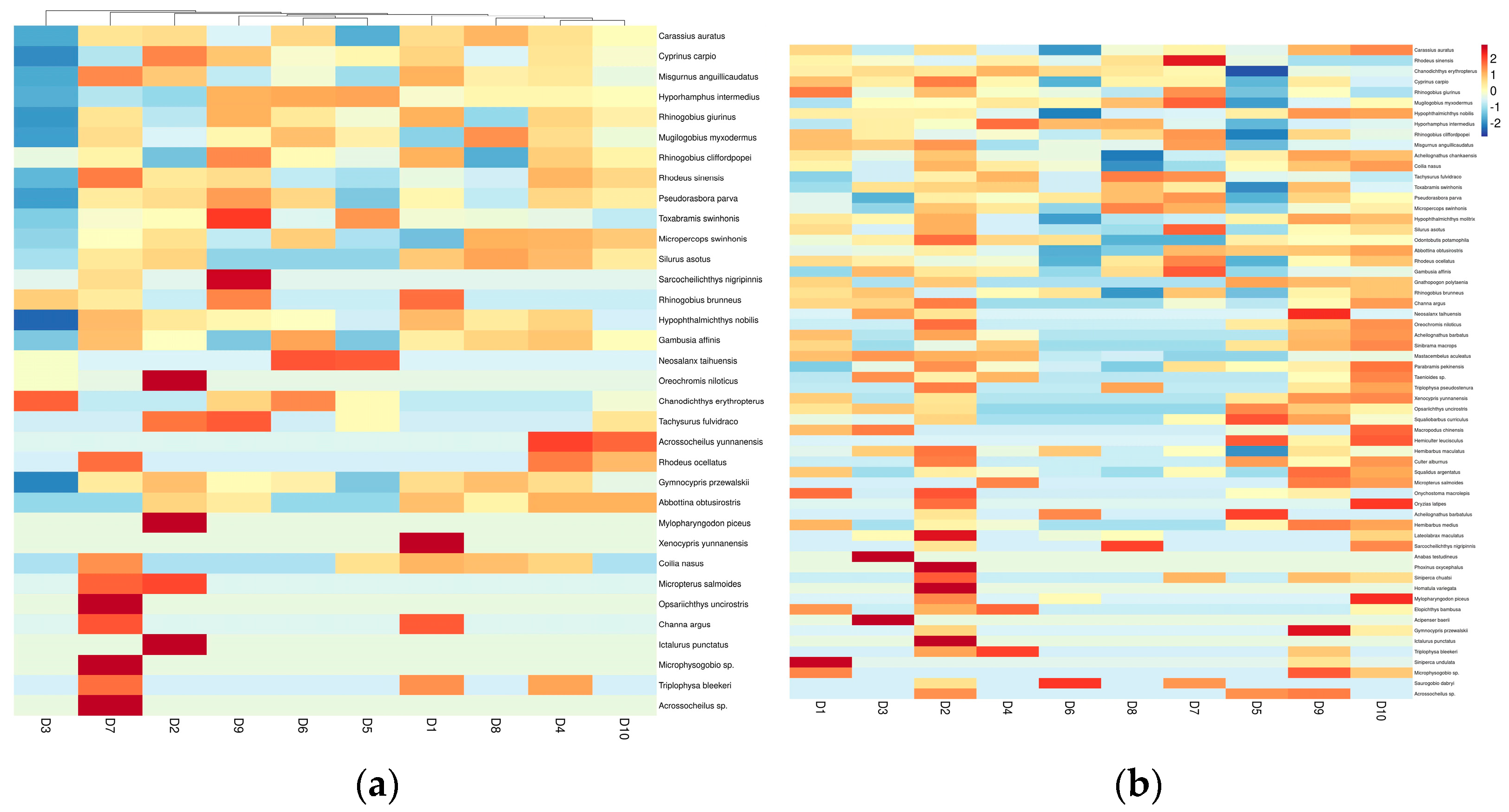
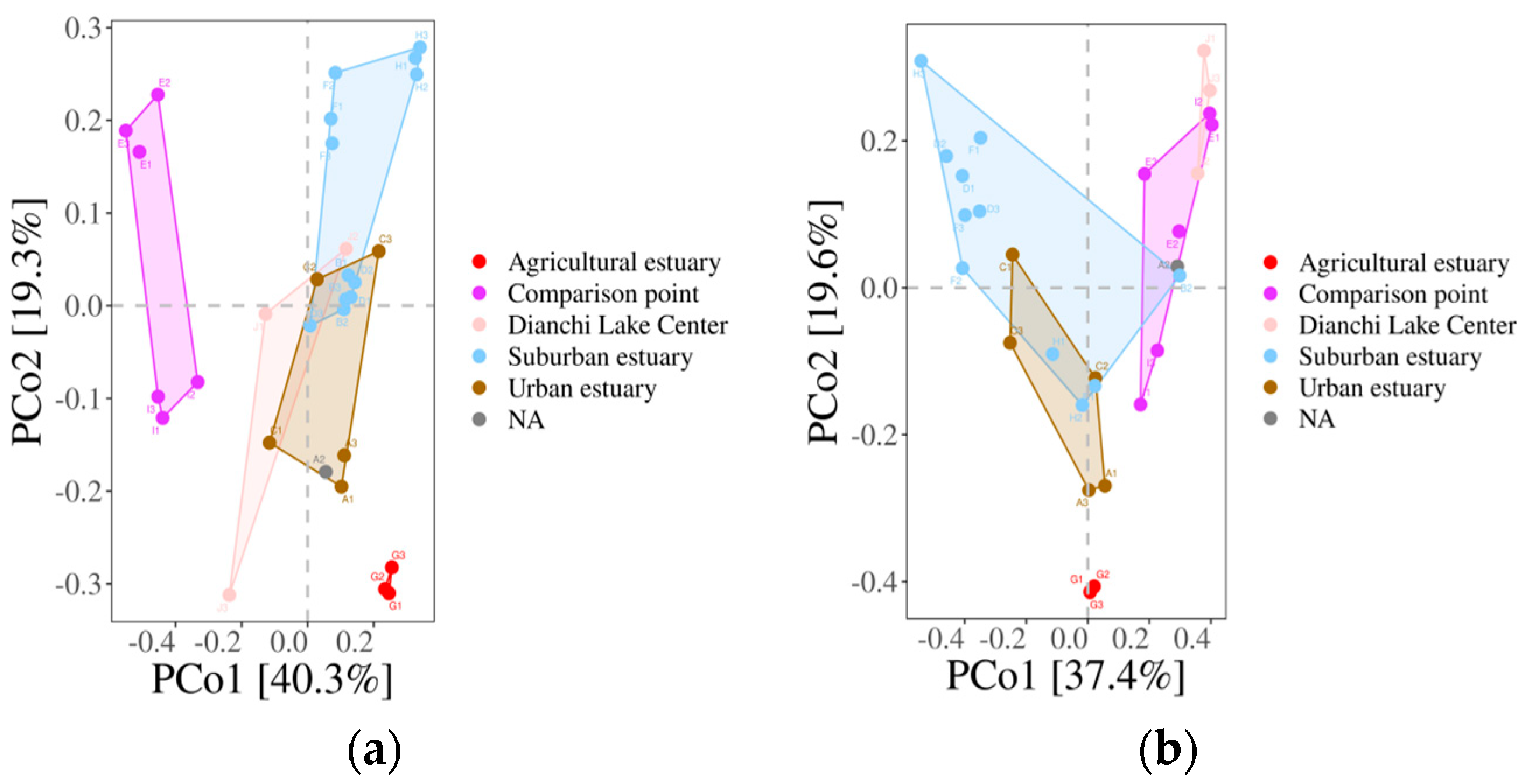
3.6.3. The β-Diversity of Fish Populations within the Dianchi Basin
4. Discussion
4.1. Fish Species Composition Monitored Using eDNA Techniques
4.2. Effects of Changes in Environmental Factors on Fish Diversity in the Dianchi Basin during Different Periods
4.3. Prospects and Limitations
4.4. Comparison of Previous Monitoring Data in the Dianchi Basin with the Present Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qin, W.; Jia, W.-F.; Hang, X.-H. Fish Diversity Protection and Sustainable Development of Fisheries. Freshw. Fish. 1999, 29, 8–11. [Google Scholar]
- Bonar, S.A.; Mercado-Silva, N.; Hubert, W.A.; Beard, T.D., Jr.; Dave, G.; Kubečka, J.; Graeb, B.D.; Lester, N.P.; Porath, M.; Winfield, I.J. Standard methods for sampling freshwater fish: Opportunities for international collaboration. Fisheries 2017, 42, 150–156. [Google Scholar] [CrossRef]
- Bayley, P.-B.; Peterson, J.-T. An approach to estimate the probability of the presence and richness of fish species. Trans. Am. Fish. Soc. 2001, 130, 620–633. [Google Scholar] [CrossRef]
- Kang, Z.; Zhang, Y.; Wu, Y.; Xie, D.; Xue, J.; Hua, J. Application of environmental DNA metabarcoding in biodiversity research and monitoring. Biotechnol. Bull. 2022, 38, 299–310. [Google Scholar]
- Xu, L.-F.; Yao, D.-D.; Yang, Y.-W.; Guo, X.-C.; Li, J.-Y.; Jiang, H.-B.; An, M.; Dong, X.-H.; Shao, L. Analysis of Fish Diversity in Artificial Lakes in Karst Plateau Based on eDNA Macro Barcode Technology. J. South. Agric. 2022, 53, 11. [Google Scholar]
- Wang, R.-X.; Yang, G.; Geng, Z.; Zhao, F.; Feng, X.-S.; Zhang, T. Application of Environmental DNA Technology in Fish Diversity Analysis in the Yangtze River Estuary. J. Aquat. Biol. 2023, 47, 365–375. [Google Scholar]
- Olds, B.P.; Jerde, C.L.; Renshaw, M.A.; Li, Y.; Evans, N.T.; Turner, C.R.; Deiner, K.; Mahon, A.R.; Brueseke, M.A.; Shirey, P.D.; et al. Estimating species richness using environmental DNA. Ecol. Evol. 2016, 6, 4214–4226. [Google Scholar] [CrossRef]
- Zhu, Y.; Han, L.-P.; Liu, L.-J.; Qiu, Z.-R.; Wang, Y.-X.; Liu, Y.-X.; He, Y.; Wang, H. Study on the Evaluation of Water Environment Quality in Dianchi Lake in 2018. Environ. Sci. J. 2020, 39, 77–85. [Google Scholar]
- Wen, H.; Cai, J.-L.; Su, Y. Characteristics of algal communities and their relationship with water environmental factors during the flood season of rivers entering the Dianchi Lake Basin. Lake Sci. 2011, 23, 40–48. [Google Scholar]
- Dong, J.; Li, G.-B.; Song, L.-R. Evolution characteristics of planktonic algae functional groups in Fuxian Lake, Erhai Lake, and Dianchi Lake since the 1960s. Lake Sci. 2014, 26, 735–742. [Google Scholar]
- Evans, N.T.; Li, Y.; Renshaw, M.A.; Olds, B.P.; Deiner, K.; Turner, C.R.; Jerde, C.L.; Lodge, D.M.; Lamberti, G.A.; Pfrender, M.E. Fish community assessment with Edna metabarcoding: Effects of sampling design and bioinformatic filtering. Can. J. Fish. Aquat. Sci. 2017, 74, 1362–1374. [Google Scholar] [CrossRef]
- Ruppert, K.-M.; Kline, R.-J.; Rahman, M.-S. Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 2019, 17, e00547. [Google Scholar] [CrossRef]
- Cheang, C.C.; Lee, B.Y.; Ip, B.H.Y.; Yiu, W.H.; Tsang, L.M.; Ang, P.O., Jr. Fish and crustacean biodiversity in an outer maritime estuary of the Pearl River Delta revealed by environmental DNA. Mar. Pollut. Bull. 2020, 161, 111707. [Google Scholar] [CrossRef] [PubMed]
- Balasingham, K.-D.; Walter, R.-P.; Mandrak, N.-E.; Heath, D.-D. Environmental DNA detection of rare and invasive fish species in two Great Lakes tributaries. Mol. Ecol. 2018, 27, 112–127. [Google Scholar] [CrossRef]
- Xu, C.-C.; Yen, I.-J.; Bowman, D.; Turner, C.R. Spider Web DNA: A New Spin on Noninvasive Genetics of Predator and Prey. PLoS ONE 2015, 10, e0142503. [Google Scholar] [CrossRef]
- Yuan, G.; Ru, H.-J.; Liu, X.-Q. 2007–2008 Fish Diversity and Resource Status in Yunnan Plateau Lakes. J. Lake Sci. 2010, 22, 837–841. [Google Scholar]
- Jerde, C.L.; Chadderton, W.L.; Mahon, A.R.; Renshaw, M.A.; Corush, J.; Budny, M.L.; Mysorekar, S.; Lodge, D.M. Detection of Asian carp DNA as part of a Grea Lakesbasin-wide surveillance program. Can. J. Fish. Aquat. Sci. 2013, 70, 522–526. [Google Scholar] [CrossRef]
- Zhang, L.-H. Establishment of a monitoring system for ecological compensation mechanism in the Dianchi Lake Basin. Water Resour. Hydropower Express 2019, 40, 53–56. [Google Scholar]
- Zhang, X.-Z. The Impact of Characteristic Pollution Sources in the Dianchi Lake Basin on the Water Quality and Plankton Community of Rivers Entering the Lake. Master’s Thesis, Shanghai Jiao TongUniversity, Shanghai, China, 2016. [Google Scholar]
- Wei, R.; Jin, Z.-J.; Zhang, X.-Z.; Huang, K.; Li, J.-H.; Kong, D.-p.; Yang, F.-L.; Zhou, B.-X. The impact of comprehensive treatment projects on ecological restoration and water quality of the Cailian River. Environ. Sci. Technol. 2016, 39, 174–179. [Google Scholar]
- Sun, Y.-X.; Wu, G.; Hu, H.; Guo, F.; Wu, Y.; Guo, Y. Evaluation of effluent quality of Kunming sewage treatment plant based on compliance assurance rate. China Environ. Sci. 2013, 33, 1113–1119. [Google Scholar]
- Zheng, L.-Q.; He, S.-Z. Soil phosphorus desorption pathways under different land use patterns in the Dianchi Lake Basin. Chin. J. Ecol. Agric. 2012, 20, 855–860. [Google Scholar] [CrossRef]
- Zheng, Y.-X.; Li, Z.-J.; Ni, J.-B. Research on Industrial Water Pollution Investigation Based on GIS: A Case Study of Dianchi Lake Basin. Environ. Prot. Sci. 2012, 38, 20–24. [Google Scholar]
- Ding, H.-X. Analysis and Research on the Emission Status of Industrial Pollutants in Kunming City from 1991 to 2007. Environ. Sci. J. 2009, 28, 53–56. [Google Scholar]
- Huang, W.-H.; Xiong, H.-J.; Deng, H.; Liu, J.; Li, J.-J.; Zhang, N.-M.; Bao, L. Distribution of inorganic phosphorus in soil under different land use patterns in typical areas on the south bank of Dianchi Lake. Environ. Pollut. Prev. 2018, 6, 689–692. [Google Scholar]
- Chen, J.-J. Analysis of Water Quality of Main Rivers Entering Dianchi Lake. J. Yunnan Agric. Univ. 2005, 4, 569–572. Available online: https://kns.cnki.net/kcms2/article/abstract?v=T-ziT3f7Rg9FLi2XjdfXnhzh6Xvv9RUOrKy7T0WbodPy0P3zcc7_ZbxxTYwwRv2BLQe-oMunHm10AMopp6HtDoBqrdE60cEhuAQFyoMzgWrEgkg1WiDbymhsBcnyYDNl&uniplatform=NZKPT&flag=copy (accessed on 2 January 2023).
- Chen, Y.-C. Application of Environmental DNA Technology in Fish Resources Investigation in the Upper Reach of Liuchong River. Master’s Thesis, Xinan University, Chongqing, China, 2020. [Google Scholar]
- Taberlet, P.; Bonin, A.; Zinger, L. Environmental DNA-for Biodiversity Research and Monitoring; Oxford University Press: New York, NY, USA, 2018. [Google Scholar]
- Miya, M.; Sato, Y.; Fukunaga, T.; Sado, T.; Poulsen, J.Y.; Sato, K.; Minamoto, T.; Yamamoto, S.; Yamanaka, H.; Araki, H.; et al. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: Detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2015, 2, 150088. [Google Scholar] [CrossRef] [PubMed]
- Evans, N.T.; Olds, B.P.; Renshaw, M.A.; Turner, C.R.; Li, Y.; Jerde, C.L.; Mahon, A.R.; Pfrender, M.E.; Lamberti, G.A.; Lodge, D.M. Quantification of mesocosm fish and amphibian species diversity via environmental DNA metabarcoding. Mol. Ecol. Resour. 2016, 16, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Qiwen, X.; Jianghua, Y.; Lijuan, Z.; Xiaowei, Z.; Chunsheng, L. Using environmental DNA metabarcoding to monitor community diversity of protists in sediments. Asian J. Ecotoxicol. 2022, 17, 175–186. [Google Scholar]
- Wanwan, Z.; Yuwei, X.; Jianghua, Y.; Yanan, Y.; Di, L.; Yong, Z.; Hongxia, Y.; Xiaowei, Z. Application and prospects of metabarcoding in environmental monitoring of phytoplankton community. Asian J. Ecotoxicol. 2017, 12, 15–24. [Google Scholar]
- Morard, R.; Darling, K.F.; Mahé, F.; Audic, S.; Ujiié, Y.; Weiner, A.K.; André, A.; Seears, H.A.; Wade, C.M.; Quillévéré, F.; et al. PFR2: A curated database of planktonic foraminifera 18S ribosomal DNA as a resource for studies of plankton ecology, biogeography and evolution. Mol. Ecol. 2015, 15, 1472–1485. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Ryberg, M.; Kristiansson, E.; Abarenkov, K.; Larsson, K.H.; Kõljalg, U. Taxonomic reliability of DNA sequences in public sequence databases: A fungal perspective. PLoS ONE 2006, 1, e59. [Google Scholar] [CrossRef]
- Chen, X.-Y. Yunnan Fish List. Zool. Res. 2013, 34, 281–337. [Google Scholar]
- Ji, F.; Han, D.; Yan, L.; Yan, S.; Zha, J.; Shen, J. Assessment of benthic invertebrate diversity and river ecological status along an urbanized gradient using environmental DNA metabarcoding and a traditional survey method. Sci. Total Environ. 2022, 806, 150587. [Google Scholar] [CrossRef] [PubMed]
- Lanz’en, A. Benthic eDNA metabarcoding provides accurate assessments of impact from oil extraction and ecological insights. Ecol. Indic. 2021, 130, 108064. [Google Scholar] [CrossRef]
- Chen, W.-J.; He, G.; Wu, B.; Fang, C.-L.; Lin, P.-C. Spatial distribution characteristics and resource assessment of fish in the Tongjiang waterway of Poyang Lake. J. Lake Sci. 2017, 29, 923–931. [Google Scholar]
- Rodríguez, M.-A.; Lewis, W.-M. Structure of fish assemblages along environmental gradients in floodplain lakes of the orinoco river. Ecol. Monogr. 1997, 67, 109–128. [Google Scholar] [CrossRef]
- Luo, J.-S. Assessing Fish Diversity in Plateau Lake in Central Yunnan Province Using Environmental DNA. Master’s Thesis, Yunnan University, Kunming, China, 2019. [Google Scholar]
- Liu, J.-K.; Cao, W.-X. Fish resources of the Yangtze River basin and the tactics for their conservation. Resour. Environ. Yangtze Basin 1992, 1, 17–23. [Google Scholar]


| Latitude and Longitude of the Sampling Points | |
|---|---|
| Panlong River | 102.696451′ E, 24.959603′ N |
| Baoxian River | 102.722900′ E, 24.924700′ N |
| Cailian River | 102.664900′ E, 24.962800′ N |
| Laoyu River | 102.763900′ E, 24.826300′ N |
| Nanchong River | 102.740700′ E, 24.779500′ N |
| Da River | 102.713800′ E, 24.773400′ N |
| Chai River | 102.688600′ E, 24.691600′ N |
| Dongda River | 102.648900′ E, 24.670000′ N |
| Dachun River | 102.635800′ E, 24.684700′ N |
| Dianchi Lake | 102.691236′ E, 24.802030′ N |
| Primer Name | Base Sequence |
|---|---|
| Teleo | F: ACACCGCCCGTCACTCT |
| R: CTTCCGGTACACTTACCAT | |
| MiFish | F: GTCGGTAAAACTCGTGCCAGC |
| R: CATAGTGGGGTATCTAATCCCAGTTTG | |
| Meta | F: TCGTGCCAGCCACCGCGGTTA |
| R: ATAGTGGGGTATCTAATCCCAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, K.; Li, X.; Meng, H.; Lin, Y.; Shen, L.; Ling, Z.; Zhang, X.; Xu, S. Spatial and Temporal Variation in the Fish Diversity in Dianchi Lake and the Influencing Factors. Water 2023, 15, 4244. https://doi.org/10.3390/w15244244
Zhao K, Li X, Meng H, Lin Y, Shen L, Ling Z, Zhang X, Xu S. Spatial and Temporal Variation in the Fish Diversity in Dianchi Lake and the Influencing Factors. Water. 2023; 15(24):4244. https://doi.org/10.3390/w15244244
Chicago/Turabian StyleZhao, Kaisong, Xiaoqin Li, Han Meng, Yuanyuan Lin, Liang Shen, Zhen Ling, Xiaowei Zhang, and Shan Xu. 2023. "Spatial and Temporal Variation in the Fish Diversity in Dianchi Lake and the Influencing Factors" Water 15, no. 24: 4244. https://doi.org/10.3390/w15244244
APA StyleZhao, K., Li, X., Meng, H., Lin, Y., Shen, L., Ling, Z., Zhang, X., & Xu, S. (2023). Spatial and Temporal Variation in the Fish Diversity in Dianchi Lake and the Influencing Factors. Water, 15(24), 4244. https://doi.org/10.3390/w15244244






