The Importance of Enterococci in the Monitoring of Fecal Pollution in River Water in Forests and Urban Areas
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and Sampling
2.2. Microbiological and Physicochemical Analyses
2.2.1. Physicochemical Parameters
2.2.2. Identification of Fecal Enterococci via the Culture-Based Method and Determination of Their Multidrug Resistance and Virulence Profiles
2.2.3. Identification of Enterococcus Strains via Fluorescence in Situ Hybridization (FISH)
2.2.4. Statistical Analysis
3. Results
3.1. Physicochemical Parameters of River Water and Wastewater Samples
3.2. Fecal Enterococci in River Water and Wastewater
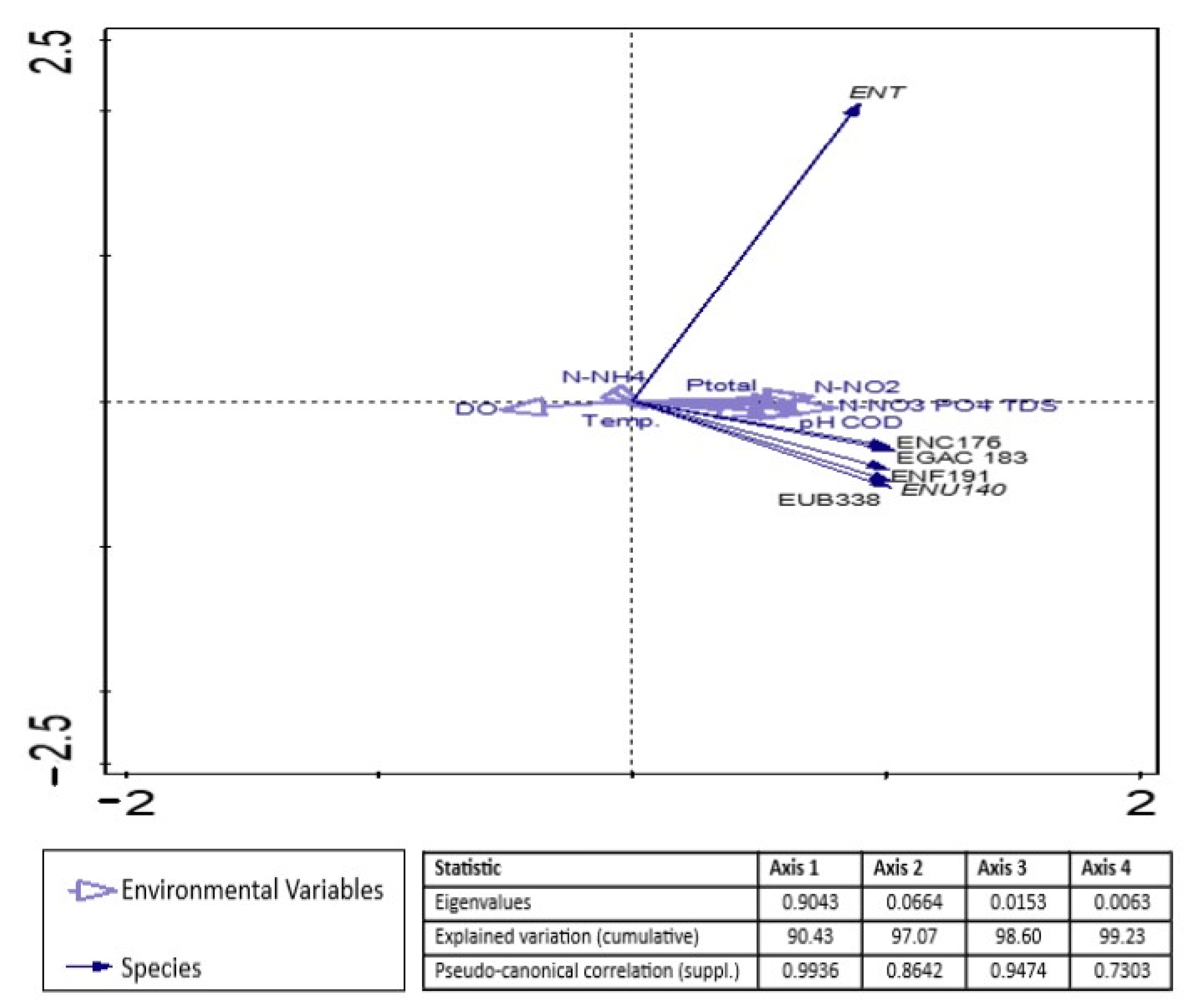
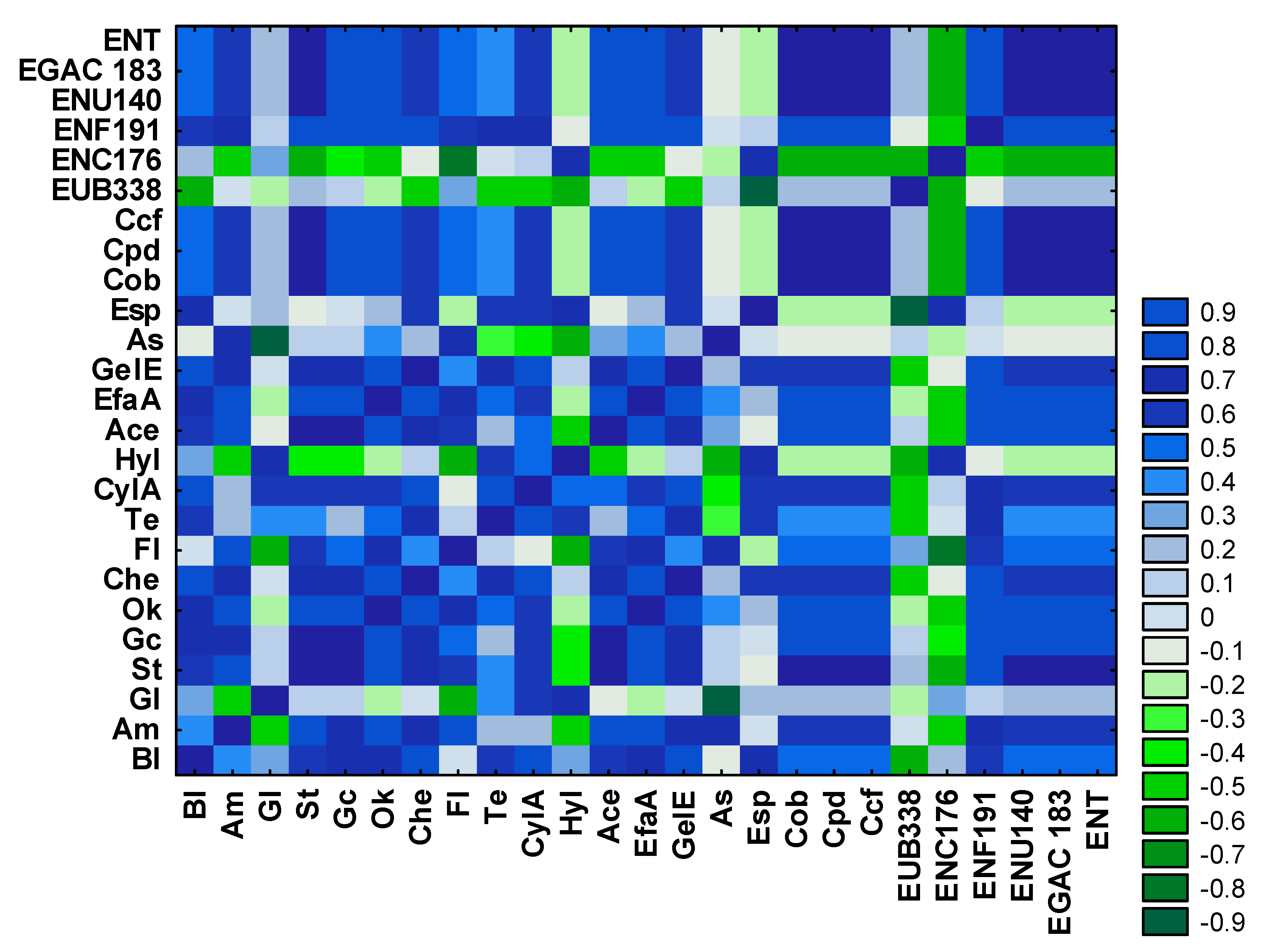
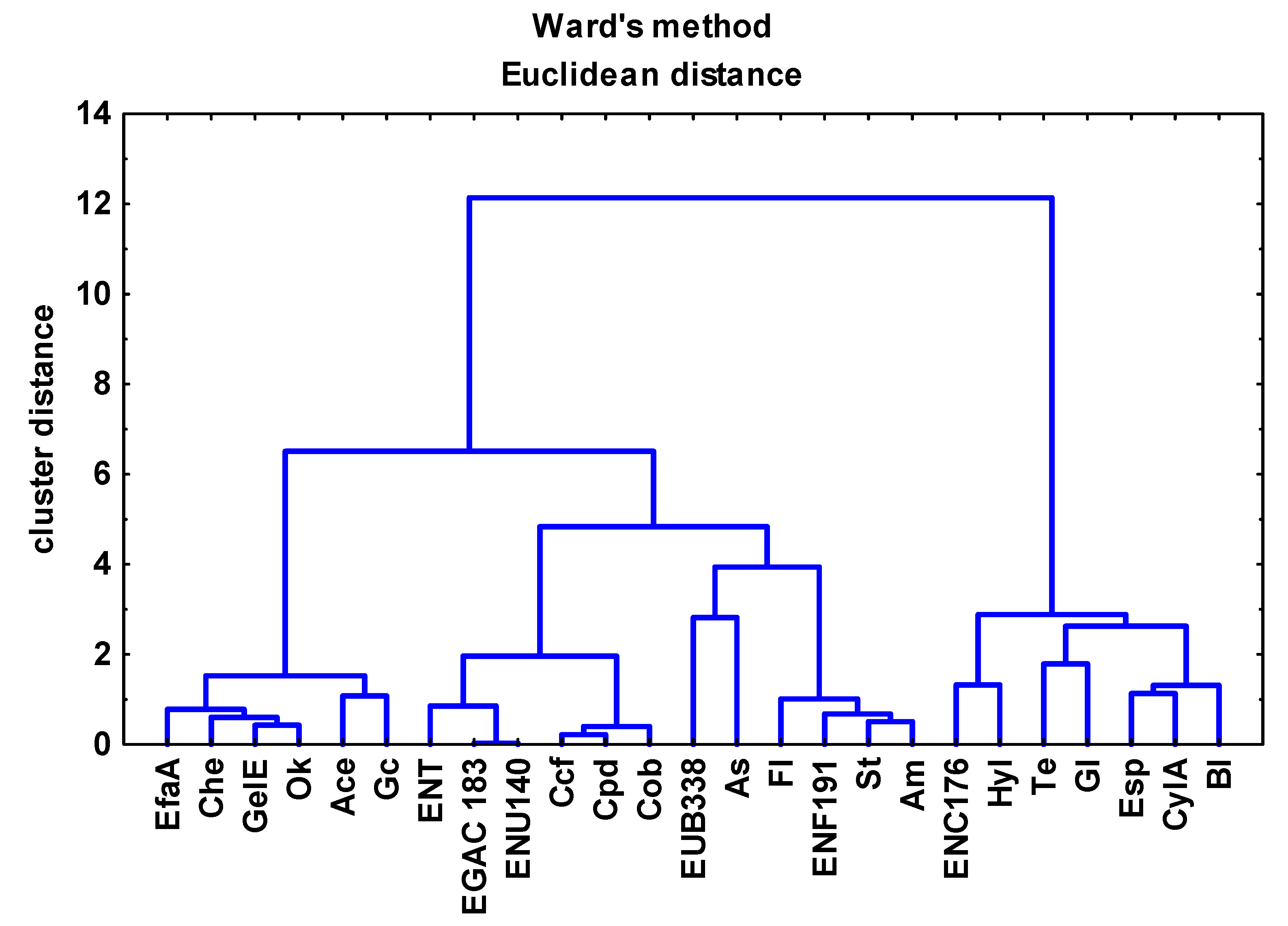
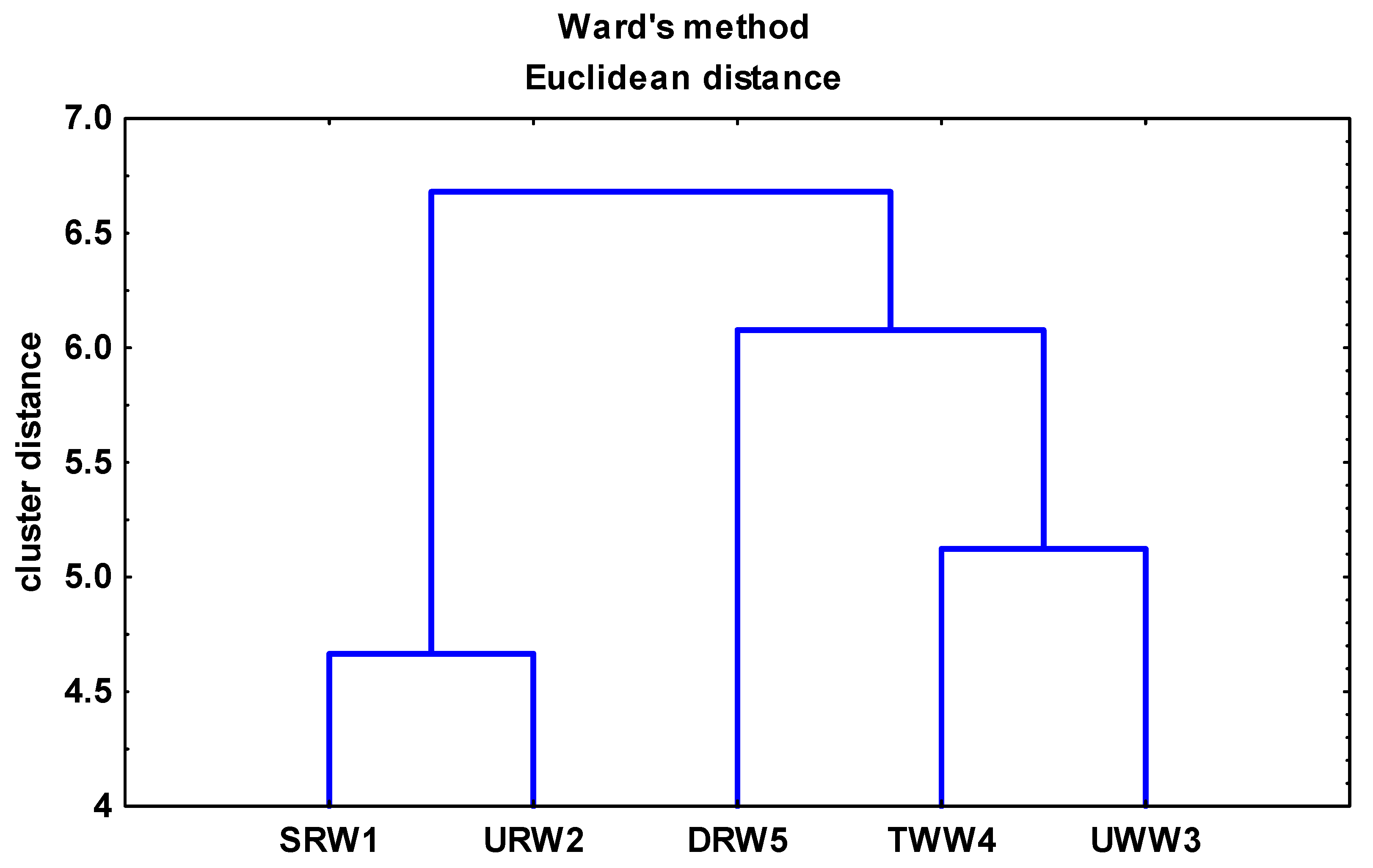
3.3. Analysis of the Relationships between Enterococcal Counts in Sampling Sites and the Physicochemical Parameters of River Water and Wastewater
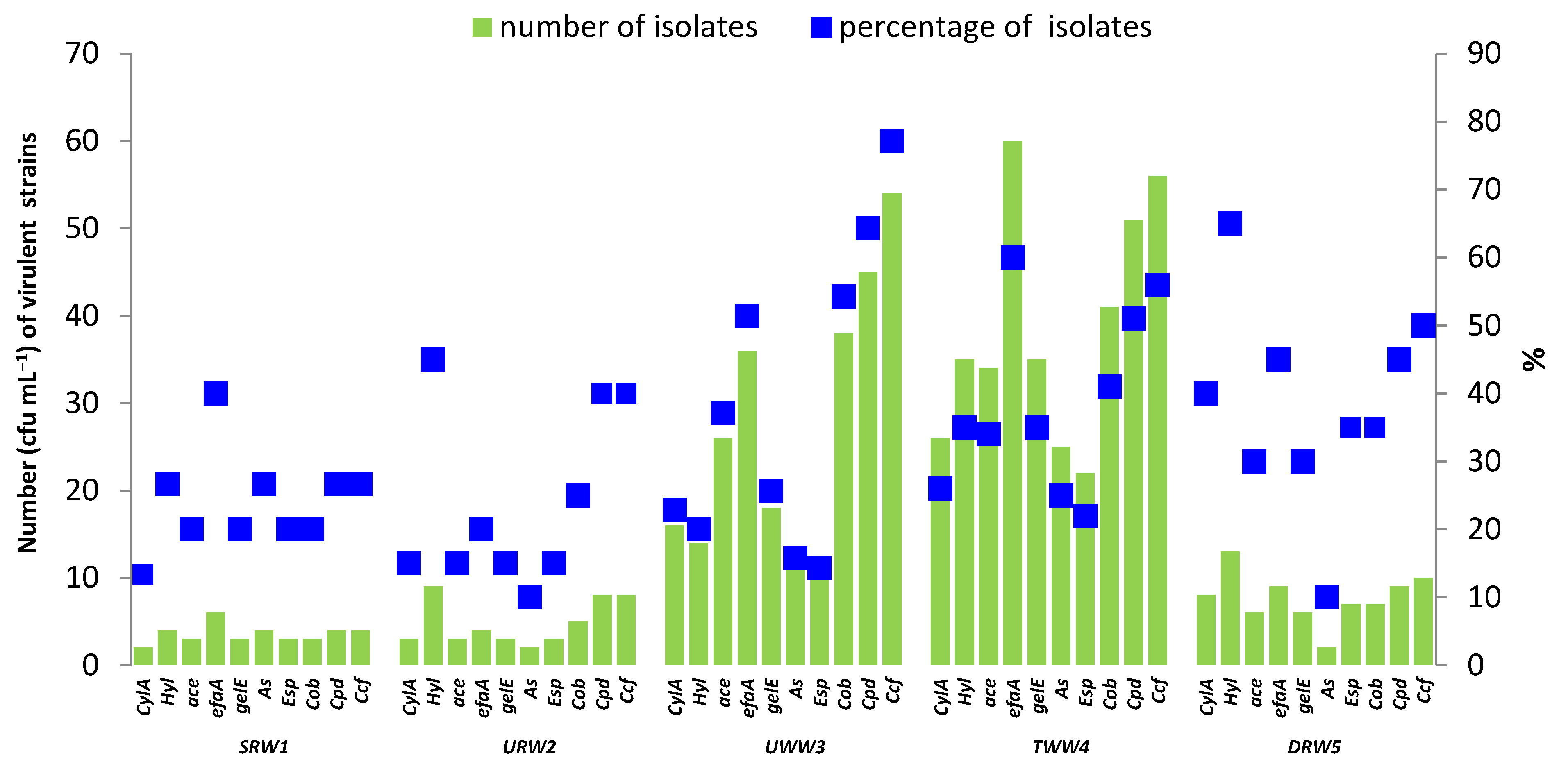
3.4. Antibiotic Resistance and Virulence Factors of Enterococci Isolated from River Water and Wastewater
3.5. Removal of Microorganisms during Wastewater Treatment
4. Discussion
4.1. Characterization of Bacteria Identified in River Water and Wastewater in the FISH Assay
4.2. Antibiotic Resistance and Virulence Factors of Enterococci Isolated from River Water and Wastewater
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mekonnen, M.M.; Hoekstra, A.Y. Four billion people facing severe water scarcity. Sci. Adv. 2016, 2, e1500323. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Liu, Z.; Wu, J.; Pan, X.; Fang, Z.; Li, J.; Bret, B.A. Future global urban water scarcity and potential solutions. Nat. Commun. 2021, 12, 4667. [Google Scholar] [CrossRef]
- Lenart-Boroń, A.; Boroń, P.; Kulik, K.; Prajsnar, J.; Żelazny, M.; Chmiel, M.J. Anthropogenic pollution gradient along a mountain river affects bacterial community composition and genera with potential pathogenic species. Sci. Rep. 2022, 12, 18140. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Mao, G.; Liu, J.; Gao, G.; Zou, C.; Bartlam, M.G.; Wang, Y. Spatial-temporal changes of bacterioplankton community along an exhorheic river. Front. Microbiol. 2016, 7, 250. [Google Scholar] [CrossRef]
- Gotkowska-Płachta, A. The Prevalence of virulent and multidrug-resistant enterococci in river water and in treated and untreated municipal and hospital wastewater. Int. J. Environ. Res. Public Health 2021, 18, 563. [Google Scholar] [CrossRef] [PubMed]
- Gotkowska-Płachta, A.; Gołaś, I.; Korzeniewska, E.; Koc, J.; Rochwerger, A.; Solarski, K. Evaluation of the distribution of fecal indicator bacteria in a river system depending on different types of land use in the southern watershed of the Baltic Sea. Environ. Sci. Pollut. Res. 2016, 23, 4073–4085. [Google Scholar] [CrossRef]
- Korzeniewska, E.; Harnisz, M. Culture-dependent and culture-independent methods in evaluation of emission of Enterobacteriaceae from sewage to the air and surface water. Water Air Soil. Poll. 2012, 223, 4039–4046. [Google Scholar] [CrossRef]
- Rolbiecki, D.; Korzeniewska, E.; Czatzkowska, M.; Harnisz, M. The Impact of Chlorine Disinfection of Hospital Wastewater on Clonal Similarity and ESBL-Production in Seected Bacteria of the Family Enterobacteriaceae. Int. J. Environ. Res. Public. Health 2022, 19, 13868. [Google Scholar] [CrossRef]
- Nhantumbo, C.; Cangi Vaz, N.; Rodrigues, M.; Manuel, C.; Rapulua, S.; Langa, J.; Nhantumbo, H.; Joaquim, D.; Dosse, M.; Sumbana, J.; et al. Assessment of Microbial Contamination in the Infulene River Basin, Mozambique. Water 2023, 15, 219. [Google Scholar] [CrossRef]
- Kebede, G.; Mushi, D.; Linke, R.B.; Dereje, O.; Lakew, A.; Hayes, D.S.; Farnleitner, A.H.; Graf, W. Macroinvertebrate indices versus microbial fecal pollution characteristics for water quality monitoring reveals contrasting results for an Ethiopian river. Ecol. Indic. 2020, 108, 105733. [Google Scholar] [CrossRef]
- Aarestrup, F.M.; Butaye, P.; Witte, W. Nonhuman reservoirs of enterococci. In The Enterococci: Pathogenesis. Molecular Biology and Antibiotic Resistance; Gilmore, M.S., Clewell, D.B., Courvalin, P., Dunny, G.M., Murray, B.E., Rice, L.B., Eds.; ASM Press: Washington, DC, USA, 2002; pp. 55–99. [Google Scholar] [CrossRef]
- Gołaś, I.; Gotkowska-Płachta, A.; Potorski, J.A. Microorganisms as Sanitary State Bioindicators of Flowing Waters in Poland. In Polish River Basins and Lakes—Part II. The Handbook of Environmental Chemistry; Korzeniewska, E., Harnisz, M., Eds.; Springer: Cham, Switzerland, 2020; Volume 87. [Google Scholar] [CrossRef]
- Fatoba, D.O.; Abia, A.L.K.; Amoako, D.G.; Essack, S.Y. Rethinking Manure Application: Increase in Multidrug-Resistant Enterococcus spp. in Agricultural Soil Following Chicken Litter Application. Microorganisms 2021, 9, 885. [Google Scholar] [CrossRef] [PubMed]
- Iweriebor, B.C.; Gaqavu, S.; Obi, L.C.; Nwodo, U.U.; Okoh, A.I. Antibiotic susceptibilities of Enterococcus species isolated from hospital and domestic wastewater effluents in Alice, Eastern Cape Province of South Africa. Int. J. Environ. Res. Public Health 2015, 12, 4231–4246. [Google Scholar] [CrossRef] [PubMed]
- Łuczkiewicz, A.; Jankowska, K.; Bray, R.; Kulbat, E.; Quant, B.; Sokolowska, A.; Olanczuk-Neyman, K. Antimicrobial resistance of fecal indicators in disinfected wastewater. Water Sci. Technol. 2011, 64, 2352–2361. [Google Scholar] [CrossRef] [PubMed]
- Varela, A.R.; Ferro, G.; Vredenburg, J.; Yanık, M.; Vieira, L.; Rizzo, L.; Lameiras, C.; Manaia, C.M. Vancomycin resistant enterococci: From the hospital effluent to the urban wastewater treatment plant. Sci. Total Environ. 2013, 450–451, 155–161. [Google Scholar] [CrossRef]
- Cagnoli, G.; Bertelloni, F.; Interrante, P.; Ceccherelli, R.; Marzoni, M.; Ebani, V.V. Antimicrobial-Resistant Enterococcus spp. in Wild Avifauna from Central Italy. Antibiotics 2022, 11, 852. [Google Scholar] [CrossRef]
- Caneschi, A.; Bardhi, A.; Barbarossa, A.; Zaghini, A. The Use of Antibiotics and Antimicrobial Resistance in Veterinary Medicine, a Complex Phenomenon: A Narrative Review. Antibiotics 2023, 12, 487. [Google Scholar] [CrossRef]
- Nowakiewicz, A.; Zięba, P.; Gnat, S.; Trościańczyk, A.; Osińska, M.; Łagowski, D.; Kosior-Korzecka, U.; Puzio, I. A significant number of multi-drug resistant Enterococcus faecalis in wildlife animals; long-term consequences and new or known reservoirs of resistance? Sci. Total Environ. 2020, 705, 135830. [Google Scholar] [CrossRef]
- World Health Organization. List of Bacteria for Which New Antibiotics Are Urgently Needed. 2017. Available online: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf (accessed on 10 December 2019).
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef]
- Byappanahalli, M.N.; Nevers, M.B.; Korajkic, A.; Staley, Z.R.; Harwood, V.J. Enterococci in the environment. Microbiol. Mol. Biol. Rev. 2012, 76, 685–706. [Google Scholar] [CrossRef]
- Rogers, S.W.; Moorman, T.B.; Ong, S.K. Fluorescent in situ hybridization and micro-autoradiography applied to ecophysiology in soil. Soil. Sci. Soc. Am. J. 2007, 71, 620–631. [Google Scholar] [CrossRef]
- Lew, S.; Lew, M.; Mieszczynski, T.; Szarek, J. Selected fluorescent techniques for identification of the physiological state of individual water and soil bacterial cells-review. Folia Microbiol. 2010, 55, 107–118. [Google Scholar] [CrossRef]
- Lew, S.; Glińska-Lewczuk, K.; Burandt, P.; Grzybowski, M.; Obolewski, K. Fecal bacteria in coastal lakes: An anthropogenic contamination or natural element of microbial diversity? Ecol. Indic. 2023, 152, 110370. [Google Scholar] [CrossRef]
- Davis, B.C.; Keenum, I.; Calarco, J.; Liguori, K.; Milligan, E.; Pruden, A.; Harwood, V.J. Towards the standardization of Enterococcus culture methods for waterborne antibiotic resistance monitoring: A critical review of trends across studies. Water Res. X 2022, 17, 100161. [Google Scholar] [CrossRef]
- Ming, H.; Ma, Y.; Gu, Y.; Su, J.; Guo, J.; Li, J.; Li, X.; Jin, Y.; Fan, J. Enterococci may not present the pollution of most enteric pathogenic bacteria in recreational seawaters of Xinghai bathing Beach, China. Ecol. Indic. 2020, 110, 105938. [Google Scholar] [CrossRef]
- Huachang, H.; Jianwen, Q.; Liang, Y. Environmental factors influencing the distribution of total and fecal coliform bacteria in six water storage reservoirs in the Pearl River Delta Region, China. J. Environ. Sci. Technol. 2010, 22, 663–668. [Google Scholar] [CrossRef]
- Wilkes, G.; Edge, T.A.; Gannon, V.P.J.; Jokinen, C.; Lyautey, E.; Neumann, N.F.; Ruecker, N.; Scott, A.; Sunohara, M.; Topp, E.; et al. Associations among pathogenic bacteria, parasites, and environmental and land use factors in multiple mixed-use watersheds. Water Res. 2011, 45, 5807–5825. [Google Scholar] [CrossRef]
- İzgördü, Ö.K.; Darcan, C.; Kariptaş, E. Overview of VBNC, a survival strategy for microorganisms. 3 Biotech 2022, 12, 307. [Google Scholar] [CrossRef]
- Heim, S.; Lleo, M.D.M.; Bonato, B.; Guzman, C.A.; Canepari, P. The viable but nonculturable state and starvation are different stress responses of Enterococcus faecalis, as determined by proteome analysis. J. Bacteriol. 2002, 184, 6739–6745. [Google Scholar] [CrossRef]
- Oliver, J.D. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microb. Rev. 2010, 34, 415–425. [Google Scholar] [CrossRef]
- Napier, M.D.; Haugland, R.; Poole, C.; Dufour, A.P.; Stewart, J.R.; Weber, D.J.; Warma, M.; Lavendre, S.; Wade, T.J. Exposure to human-associated fecal indicators and self-reported illness among swimmers at recreational beaches: A cohort study. Environ. Health 2017, 16, 103. [Google Scholar] [CrossRef]
- Amann, R.; Snaidr, J.; Wagner, M.; Ludwig, W.; Schleifer, K.H. In situ visualization of high genetic diversity in a natural microbial community. J. Bacteriol. 1996, 178, 3496–3500. [Google Scholar] [CrossRef] [PubMed]
- Huber, D.; Voith von Voithenberg, L.; Kaigala, G.V. Fluorescence in situ hybridization (FISH): History, limitations and what to expect from micro-scale FISH? Micro Nano Eng. 2018, 1, 15–24. [Google Scholar] [CrossRef]
- Wellinghausen, N.; Bartel, M.; Essig, A.; Poppert, S. Rapid identification of clinically relevant Enterococcus species by fluorescence in situ hybridization. J. Clin. Microbiol. 2007, 45, 3424–3426. [Google Scholar] [CrossRef] [PubMed]
- Bertaux, J.; Gloger, U.; Schmid, M.; Hartmann, A.; Scheu, S. Routine fluorescence in situ hybridization in soil. J. Microbiol. Methods. 2007, 69, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Pernthaler, J.; Glöckner, F.O.; Schönhuber, W.; Amann, R. Fluorescence in situ hybridization with rRNA-targeted oligonucleotide probes. Methods Microbiol. 2001, 30, 207–210. [Google Scholar] [CrossRef]
- Bouvier, T.; Del Giorgio, P.A. Factors influencing the detection of bacterial cells using fluorescence in situ hybridization (FISH): A quantitative review of published reports. FEMS Microbiol. 2003, 44, 3–15. [Google Scholar] [CrossRef]
- PN-EN ISO 19458:2007; Water Quality: Sampling for Microbiological Analysis. Polish Committee for Standardization: Warsaw, Poland, 2007. (In Polish)
- Hermanowicz, W.; Dożańska, W.; Dojlido, J.; Koziorowski, B. Physicochemical Methods of Water and Wastewater Examination; Arkady: Warsaw, Poland, 1999. (In Polish) [Google Scholar]
- PN-EN ISO 7899-2:2004; Water Quality—Detection and Enumeration of Intestinal Enterococci—Part 2: Membrane Filtration Method. PKN Warsaw: Płock, Poland, 2004.
- Facklam, R.R.; Collins, M.D. Identification of enterococcus species isolated from human infections by a conventional test scheme. J. Clin. Microbiol. 1989, 27, 731–734. [Google Scholar] [CrossRef] [PubMed]
- Ke, D.; Picard, F.J.; Martineau, F.; Menard, C.; Roy, P.H.; Ouellette, M.; Bergeron, M.G. Development of a PCR assay for rapid detection of enterococci. J. Clin. Microbiol. 1999, 37, 3497–3503. [Google Scholar] [CrossRef]
- Deasy, B.M.; Rea, M.C.; Fitzgerald, G.F.; Cogan, T.M.; Beresford, T.P. A rapid PCR based method to distinguish between Lactococcus and Enterococcus. Syst. Appl. Microbiol. 2000, 23, 510–522. [Google Scholar] [CrossRef]
- Vankerckhoven, V.; Van Autgaerden, T.; Vael, C.; Lammens, C.; Chapelle, S.; Rossi, R.; Jabes, D.; Goossens, H. Development of a Multiplex PCR for the Detection of asa1, gelE, cylA, esp, and hyl Genes in Enterococci and Survey for Virulence Determinants among European Hospital Isolates of Enterococcus faecium. J. Clin. Microbiol. 2004, 42, 4473–4479. [Google Scholar] [CrossRef]
- Mannu, L.; Paba, A.; Daga, E.; Comunian, R.; Zanetti, S.; Duprè, I.; Sechi, L.A. Comparison of the incidence of virulence determinants and antibiotic resistance between Enterococcus faecium strains of dairy, animal and clinical origin. Int. J. Food Microbiol. 2003, 88, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Shankar, V.; Baghdayan, A.S.; Huycke, M.M.; Lindahl, G.; Gilmore, M.S. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect. Immun. 1999, 67, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Eaton, T.J.; Gasson, M.J. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 2001, 67, 1628–1635. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters. 2012. Available online: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/EUCAST_breakpoints_v_2.0_120101.pdf (accessed on 20 January 2020).
- CLSI. Clinical and Laboratory Standards institute. In Performance Standards for Antimicrobial Disk Susceptibility Tests: Approved Standard—Eleventh Edition; CLSI Document 2012, M02-A11; CLSI: Wayne, PA, USA, 2012. [Google Scholar]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef]
- Monteiro, S.; Santos, R. Incidence of enterococci resistant to clinically relevant antibiotics in environmental waters and in reclaimed waters used for irrigation. J. Water Health 2020, 18, 911–924. [Google Scholar] [CrossRef]
- Loy, A.; Maixner, F.; Wagner, M.; Horn, M. ProbeBase—An online resource for rRNA-targeted oligonucleotide probes: New features. Nucleic Acids Res. 2007, 35, D800–D804. [Google Scholar] [CrossRef]
- Gescher, D.M.; Kovacevic, D.; Schmiedel, D.; Siemoneit, S.; Mallmann, C.; Halle, E.; Göbel, U.B.; Moter, U.B. Fluorescence in situ hybridisation (FISH) accelerates identification of Gram-positive cocci in positive blood cultures. Int. J. Antimicrob. Agents 2008, 32, S51–S59. [Google Scholar] [CrossRef]
- Daims, H.; Brühl, A.; Amann, R.; Schleifer, K.H.; Wagner, M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: Development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 1999, 22, 434–444. [Google Scholar] [CrossRef]
- Stoecker, K.; Dorninger, C.; Daims, H.; Wagner, M. Double labeling of oligonucleotide probes for fluorescence in situ hybridization (DOPE-FISH) improves signal intensity and increases rRNA accessibility. Appl. Environ. Microbiol. 2010, 76, 922–926. [Google Scholar] [CrossRef]
- Amann, R.I.; Binder, B.J.; Olson, R.J.; Chisholm, S.W.; Devereux, R.; Stahl, D.A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 1990, 56, 1919–1925. [Google Scholar] [CrossRef]
- Wallner, G.; Amann, R.; Beisker, W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 1993, 14, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Ter Braak, C.J.F.; Šmilauer, P. Reference Manual and Cano Draw for Windows User’s Guide: Software for Canonical Community Ordination (Version 4.5); Microcomputer Power: Ithaca, NY, USA, 2002. [Google Scholar]
- Stępień-Pyśniak, D.; Hauschild, T.; Nowaczek, A.; Marek, A.; Dec, M. Wild birds as a potential source of known and novel multilocus sequence types of antibiotic-resistant enterococcus faecalis. J. Wildl. Dis. 2018, 54, 219–228. [Google Scholar] [CrossRef]
- León-Sampedro, R.; del Campo, R.; Rodriguez-Baños, M.; Lanza, V.F.; Pozuelo, M.J.; Francés-Cuesta, C.; Tedim, A.P.; Freitas, A.R.; Novais, C.; Peixe, L.; et al. Phylogenomics of Enterococcus faecalis from wild birds: New insights into host-associated differences in core and accessory genomes of the species. Environ. Microbiol. 2019, 21, 3046–3062. [Google Scholar] [CrossRef]
- Council of the European Communities. Council Directive 91/271/ECC of Concerning Urban Waste Water Treatment. Off. J. Eur. Communities 1991, 135, 40–52. [Google Scholar]
- Czatzkowska, M.; Wolak, I.; Harnisz, M.; Korzeniewska, E. Impact of Anthropogenic Activities on the Dissemination of ARGs in the Environment-A Review. Int. J. Environ. Res. Public. Health 2022, 19, 12853. [Google Scholar] [CrossRef] [PubMed]
- European Center for Disease Prevention and Control. Surveillance Atlas of Infectious Diseases. Available online: https://www.ecdc.europa.eu/en/surveillance-atlas-infectious-diseases (accessed on 28 April 2023).
- Hricová, K.; Röderová, M.; Fryčák, P.; Pauk, V.; Kurka, O.; Mezerová, K.; Štosová, T.; Bardoň, J.; Milde, D.; Kučová, P.; et al. Prevalence of Vancomycin-Resistant Enterococci and Antimicrobial Residues in Wastewater and Surface Water. Life 2021, 11, 1403. [Google Scholar] [CrossRef]
- Lata, P.; Ram, S.; Agrawal, M.; Shanker, R. Enterococci in river Ganga surface waters: Propensity of species distribution, dissemination of antimicrobial-resistance and virulence-markers among species along landscape. BMC Microbiol. 2009, 9, 140. [Google Scholar] [CrossRef]
- Zieliński, W.; Hubeny, J.; Buta-Hubeny, M.; Rolbiecki, D.; Harnisz, M.; Paukszto, Ł.; Korzeniewska, E. Metagenomics analysis of probable transmission of determinants of antibiotic resistance from wastewater to the environment—A case study. Sci. Total Environ. 2022, 827, 154354. [Google Scholar] [CrossRef]
- Strateva, T.; Atanasova, D.; Savov, E.; Petrova, G.; Mitov, I. Incidence of virulence determinants in clinical Enterococcus faecalis and Enterococcus faecium isolates collected in Bulgaria. Braz. J. Infect. Dis. 2016, 20, 127–133. [Google Scholar] [CrossRef]
- Creti, R.; Imperi, M.; Bertuccini, L.; Fabretti, F.; Orefici, G.; Di Rosa, R.; Baldassarri, L. Survey for virulence determinants among Enterococcus faecalis isolated from different sources. J. Med. Microbiol. 2004, 53, 1320. [Google Scholar] [CrossRef]
- Ferguson, D.M.; Talavera, G.N.; Hernández, L.A.; Weisberg, S.B.; Ambrose, R.F.; Jay, J.A. Virulence genes among Enterococcus faecalis and Enterococcus faecium isolated from coastal beaches and human and nonhuman sources in Southern California and Puerto Rico. J. Pathog. 2016, 2016, 3437214. [Google Scholar] [CrossRef] [PubMed]
- Georges, M.; Odoyo, E.; Matano, D.; Tiria, F.; Kyany’a, C.; Mbwika, D.; Mutai, W.C.; Musila, L. Determination of Enterococcus faecalis and Enterococcus faecium Antimicrobial Resistance and Virulence Factors and Their Association with Clinical and Demographic Factors in Kenya. J. Pathog. 2022, 2022, 3129439. [Google Scholar] [CrossRef] [PubMed]




| Probe | Sequence (5’–3’) | Bacteria | Time (h)/% FA a | Position | References |
|---|---|---|---|---|---|
| ENC176 | CA GTT CTC TGC GTC TAC CTC | Enterococcus spp. b | 1.5/30 | 23S rRNA | [36] |
| ENF191 | GAA AGC GCC TTT CAC TCT TAT GC | Enterococcus faecalis | 1.5/30 | 16S rRNA | [36] |
| ENU140 | TTC ACA CAA TCG TAA CAT CCT | Enterococcus faecium | 1.5/30 | 23S rRNA | [36] |
| EGAC183 | CAA CTT TCT TCC ATG CGG AAA AT | Enterococcus gallinarumc | 3/30 | 16S rRNA | [36] |
| EUB338 | GCT GCC TCC CGT AGG AGT | Domain Eubacteria | 1.5/35 | 16S rRNA | [58] |
| NON338 | ACT CCT ACG GGA GGC AGC | Negative control | 3/35 | 16S rRNA | [59] |
| Parameter | Sampling Sites Mean ± SD | Differences (p) between | ||||
|---|---|---|---|---|---|---|
| SRW1 | URW2 | UWW3 | TWW4 | DRW5 | Site/Seasons | |
| Temp | 12.3 ± 2.8 | 14.15 ± 4.50 | 13.60 ± 5.36 | 14.94 ± 6.88 | 14.88 ± 7.06 | 0.96/0.001 * |
| pH | 7.28 ± 0.43 | 7.40 ± 0.37 | 8.22 ± 0.22 | 8.36 ± 0.30 | 7.23 ± 0.33 | 0.007 */0.324 |
| DO | 10.30 ± 2.8 | 8.57 ± 0.98 | 0.50 ± 0.15 | 8.96 ± 0.96 | 8.18 ± 0.59 | 0.023 */0.178 |
| NH4-N | 0.06 ± 0.05 | 0.07 ± 0.02 | 78.08 ± 12.56 | 5.94 ± 2.44 | 0.19 ± 0.04 | 0.002 */0.951 |
| NO2-N | 0.01 ± 0.004 | 0.01 ± 0.004 | 1.01 ± 0.002 | 1.98 ± 0.50 | 0.05 ± 0.03 | 0.002 */0.854 |
| NO3-N | 2.41 ± 0.27 | 0.38 ± 0.55 | 1.14 ± 0.05 | 2.03 ± 0.71 | 0.18 ± 0.03 | 0.005 */0.807 |
| PO4-P | 0.26 ± 0.07 | 0.23 ± 0.07 | 33.79 ± 9.33 | 1.96 ± 1.03 | 0.29 ± 0.19 | 0.005 */0.550 |
| PT | 0.48 ± 0.25 | 0.44 ± 0.31 | 40.14 ± 7.22 | 2.81 ± 1.69 | 0.79 ± 0.17 | 0.003 */0.730 |
| COD | 9.0 ± 8.11 | 17.50 ± 5.19 | 389.94 ± 216.31 | 35.41 ± 2.96 | 22.10 ± 3.88 | 0.002 */0.748 |
| TDS | 0.29 ± 0.06 | 0.22 ± 0.02 | 1.77 ± 0.20 | 1.66 ± 0.030 | 0.21 ± 0.05 | 0.004 */0.767 |
| Sampling Sites | ||||||
|---|---|---|---|---|---|---|
| Method | SRW1 | URW2 | UWW3 | TWW4 | DRW5 | |
| FISH (×106 cell 1 mL−1) | EUB338 | 2.0 ± 1.5 a (0.9–4.0) b | 5.1 ± 2.3 (2.7–7.7) | 216.1 ± 95.5 (124.26–304.9) | 45.2 ± 6.9 (37.2–52.6) | 5.5 ± 2.7 (2.1–7.9) |
| ENC176 | 0.05 ± 0.01 (0.04–0.07) 2.4% c | 0.11 ± 0.05 (0.07–0.17) 2.2% | 3.20 ± 1.74 (1.75–5.54) 1.5% | 0.88 ± 0.12 (0.70–0.97) 1.9% | 0.15 ± 0.06 (0.058–0.196) 2.7% | |
| ENF191 | 0.006 ± 0.002 (0.003–0.008) 0.3% | 0.017 ± 0.006 (0.01–0.02) 0.3% | 1.36 ± 0.67 (0.67–2.26) 0.6% | 0.38 ± 0.20 (0.27–0.69) 0.8% | 0.020 ± 0.006 (0.012–0.027) 0.4% | |
| ENU140 | 0.002 ± 0.001 (0.001–0.002) 0.08% | 0.004 ± 0.002 (0.002–0.006) 0.09% | 0.74 ± 0.68 (0.15–1.42) 0.3% | 0.091 ± 0.035 (0.064–0.14) 0.2% | 0.006 ± 0.003 (0.002–0.007) 0.11% | |
| EGAC183 | 0.0003 ± 0.0001 (0.0002–0.0005) 0.02% | 0.0009 ± 0.0004 (0.0003–0.0012) 0.02% | 0.17 ± 0.16 (0.03–0.37) 0.1% | 0.021 ± 0.0085 (0.012–0.032) 0.05% | 0.0012 ± 0.0003 (0.001–0.002) 0.02% | |
| Culture method (×103 CFU 1 mL−1) b | ENT | 0.0003 ± 0.0003 (0.0–0.0007) | 0.0019 ± 0.0013 (0.0008–0.0034) | 22.60 ± 15.86 (10.12–45.00) | 0.61 ± 0.67 (0.19–1.60) | 0.0039 ± 00.42 (0.0006–0.0101) |
| No. of Antibiotics to Which an Isolate Was Resistant (a) | No. of Tested Antibiotics (b) | MAR Index (a/b) | No. of MAR Isolates (%) | ||||
|---|---|---|---|---|---|---|---|
| SRW1 (n = 15) | URW2 (n = 20) | UWW3 (n = 70) | TWW4 (n = 100) | DRW5 (n = 20) | |||
| 9 | 9 | 1.0 | 0 | 0 | 1 (1.4) | 6 (6.0) | 0 |
| 8 | 9 | 0.8 | 0 | 0 | 2 (2.9) | 4 (4.0) | 0 |
| 7 | 9 | 0.7 | 0 | 1 (5.0) | 5 (7.1) | 14 (14.0) | 2 (10.0) |
| 6 | 9 | 0.6 | 1 (6.7) | 0 | 6 (8.6) | 18 (15.0) | 1 (5.0) |
| 5 | 9 | 0.5 | 1 (6.7) | 1 (5.0) | 17 (24.3) | 8 (8.0) | 2 (10.0) |
| 4 | 9 | 0.4 | 2 (13.3) | 2 (10.0) | 11 (15.7) | 4 (4.0) | 2 (10.0) |
| 3 | 9 | 0.3 | 4 (26.6) | 1 (5.0) | 4 (5.7) | 7 (7.0) | 6 (30.0) |
| 2 | 9 | 0.2 | 1 (6.7) | 4 (20) | 9 (12.6) | 18 (18.0) | 3 (15.0) |
| 0.56 | 9 (60) | 10 (50) | 55 (78.6) | 79 (79.0) | 16 (80.0) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gotkowska-Płachta, A.; Gołaś, I. The Importance of Enterococci in the Monitoring of Fecal Pollution in River Water in Forests and Urban Areas. Water 2023, 15, 3708. https://doi.org/10.3390/w15213708
Gotkowska-Płachta A, Gołaś I. The Importance of Enterococci in the Monitoring of Fecal Pollution in River Water in Forests and Urban Areas. Water. 2023; 15(21):3708. https://doi.org/10.3390/w15213708
Chicago/Turabian StyleGotkowska-Płachta, Anna, and Iwona Gołaś. 2023. "The Importance of Enterococci in the Monitoring of Fecal Pollution in River Water in Forests and Urban Areas" Water 15, no. 21: 3708. https://doi.org/10.3390/w15213708





