Oxygen Uptake Rate as an Indicator of the Substrates Utilized by Candidatus Accumulibacter
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Bioreactor for Obtaining a Ca. Accumulibacter-Enriched Culture
2.2. Determination of the Oxygen Uptake Rates
- OURsum is the total specific oxygen uptake rate
- OURend is the endogenous specific oxygen uptake rate.
2.3. Determination of Dynamics of Substrate and Phosphate in Batch Experiments
2.4. Molecular Techniques
2.5. Analytical Techniques
2.6. Statistical Data Processing
3. Results
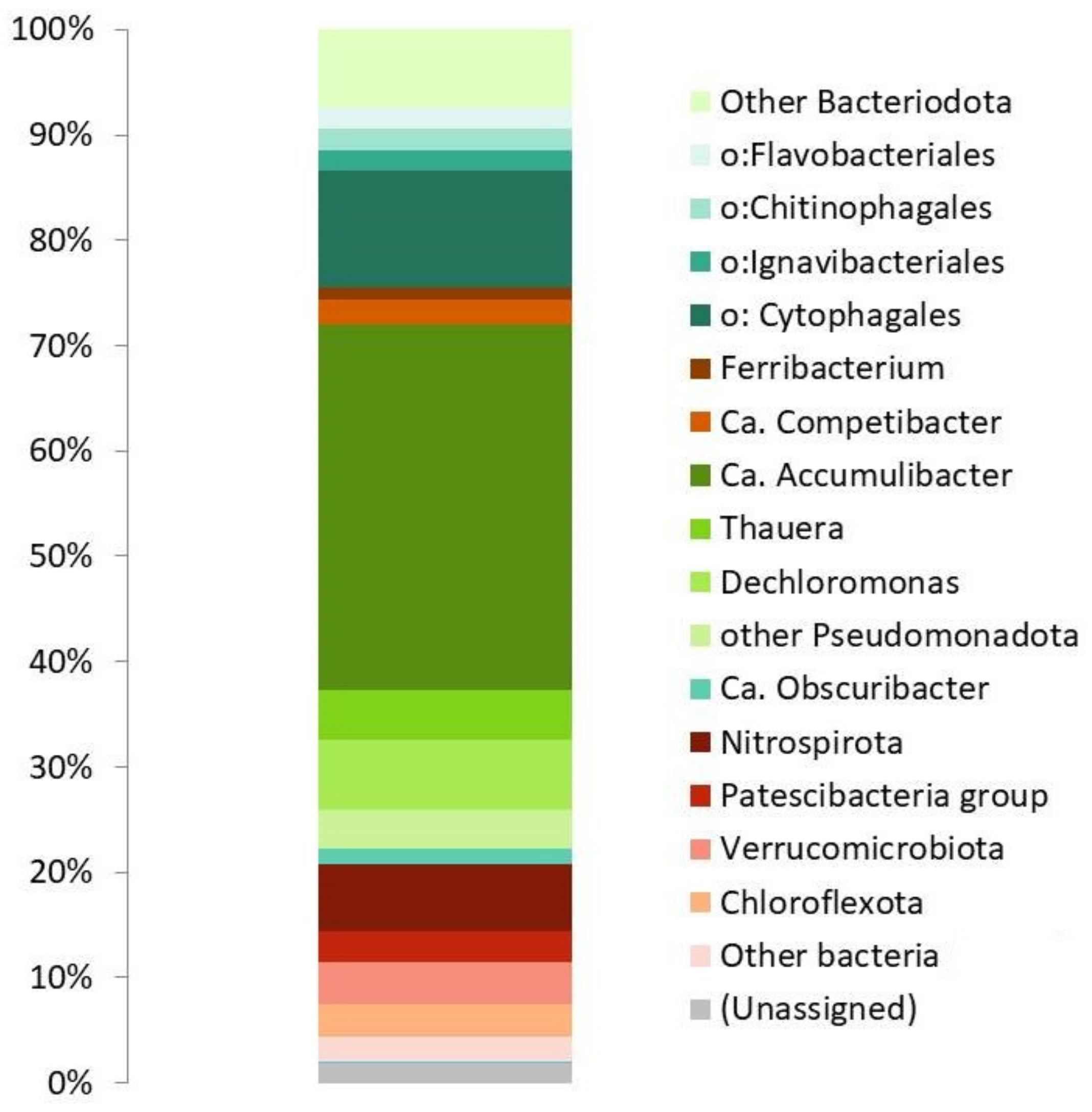
3.1. Bioreactor Operation and Microbial Community Composition
3.2. Oxygen Consumption
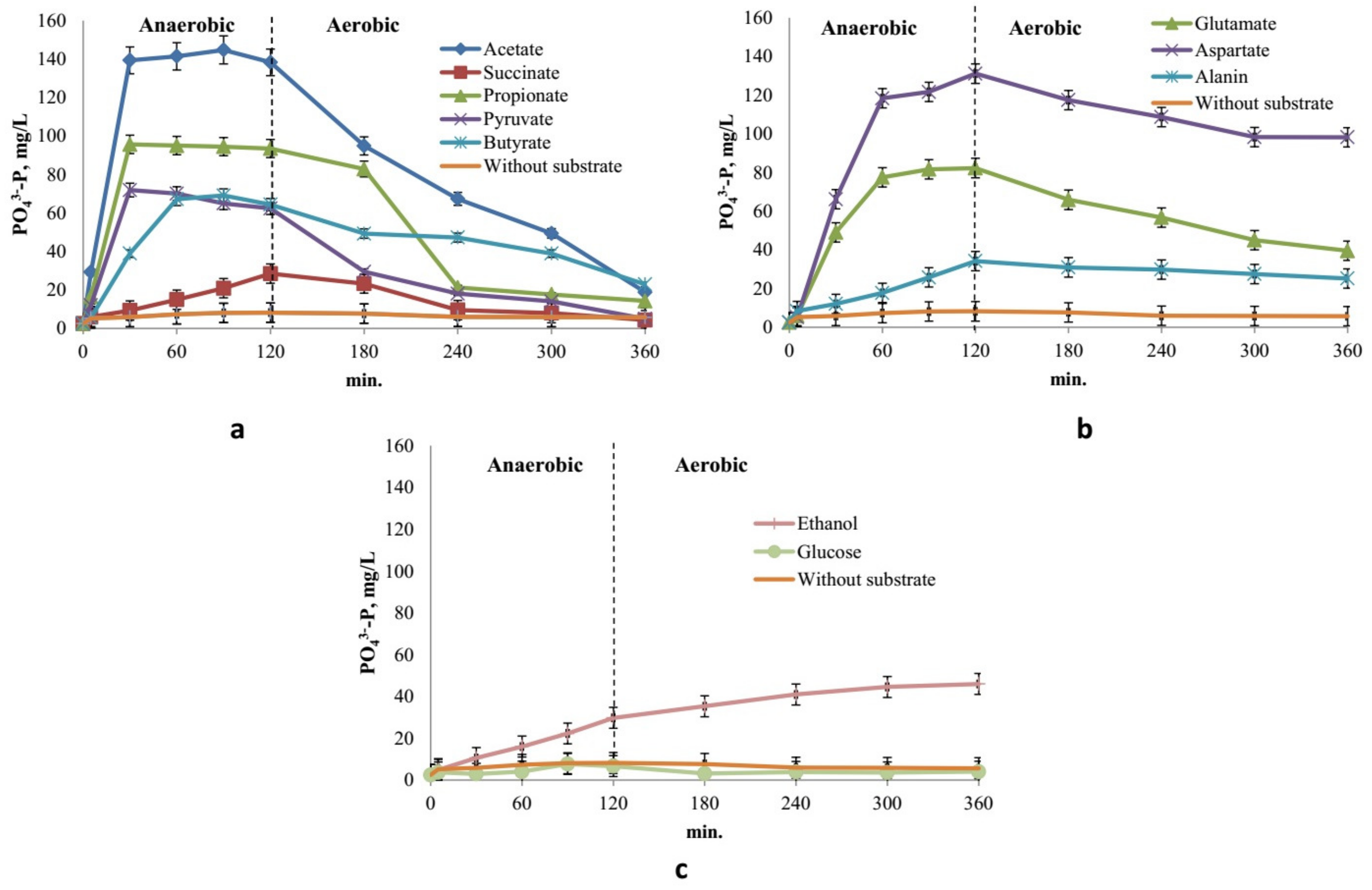
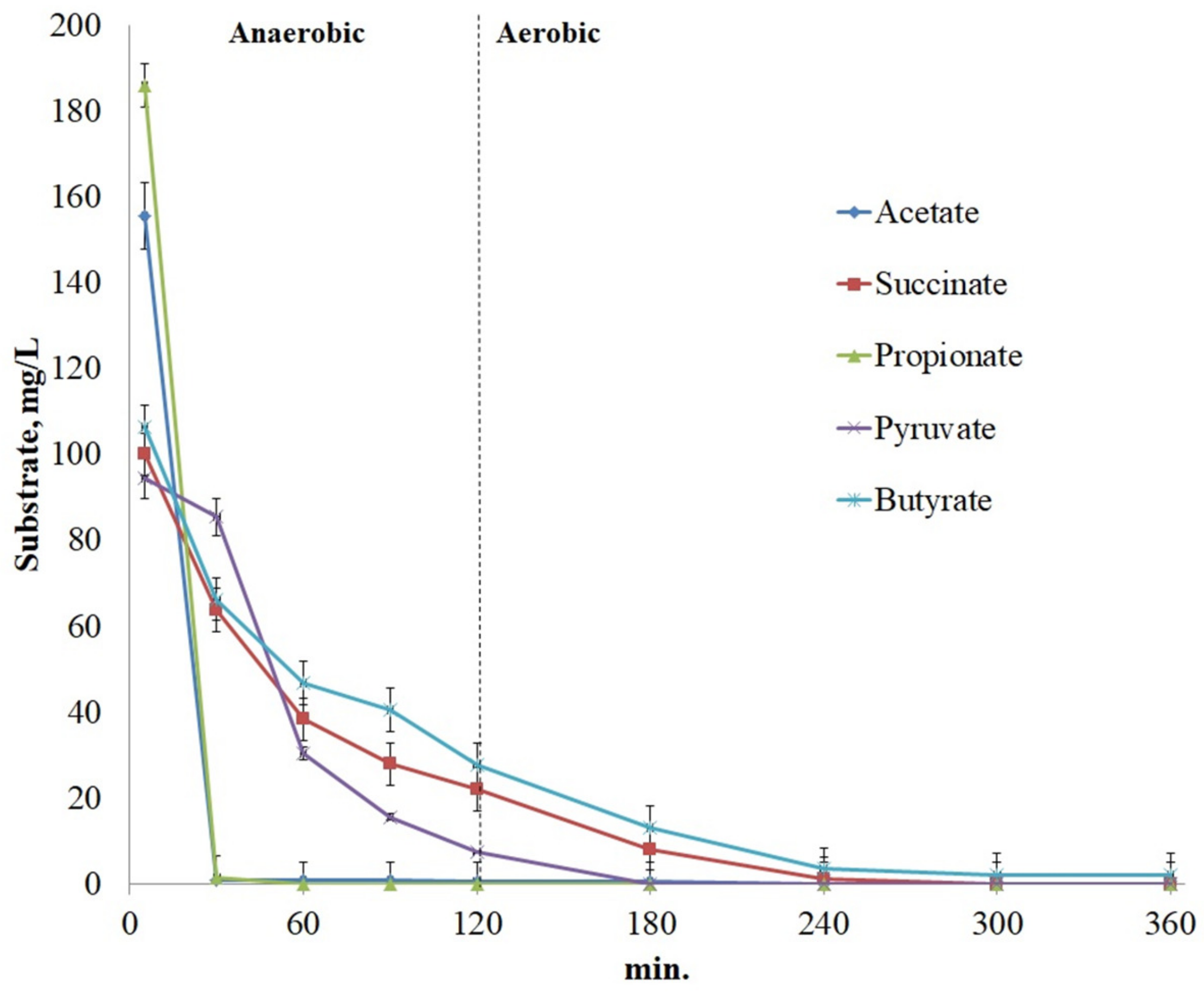
3.3. Cycle of Phosphate Release/Uptake under Anaerobic/Aerobic Conditions
3.4. Abilities of Ca. Accumulibacter Transport Systems
4. Discussion
4.1. Response of Ca. Accumulibacter to the Substrates
4.1.1. Volatile Fatty Acids
4.1.2. Pyruvate
4.1.3. Succinate
4.1.4. Amino Acids
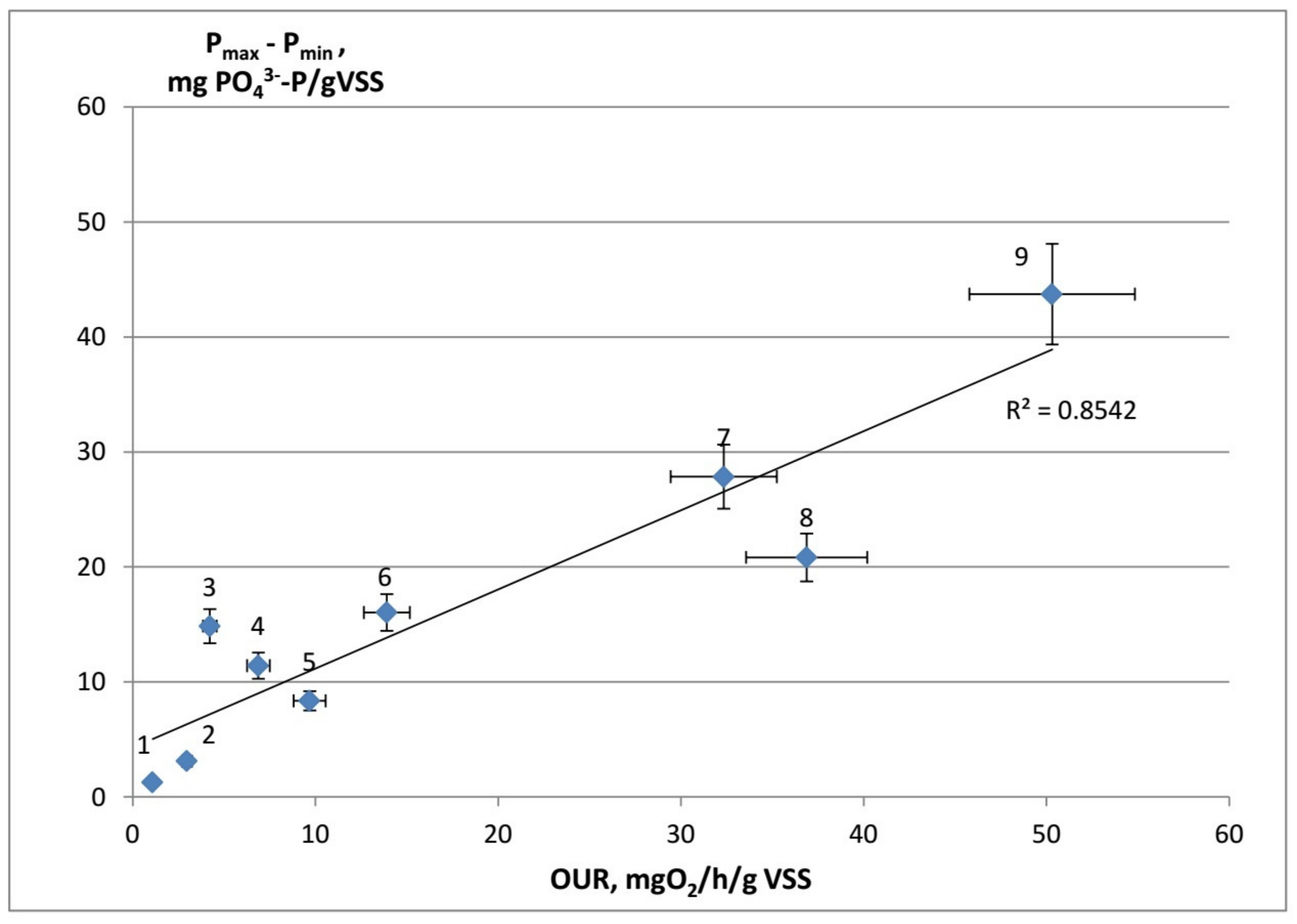
4.2. Correlation between OUR and Phosphate Uptake during the Anaerobic Stage
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wentzel, M.C.; Loewenthal, R.E.; Ekama, G.A.; Marais, G.V.R. Enhanced polyphosphate organism cultures in activated sludge systems—Part 1: Enhanced culture development. Water SA 1988, 14, 81–92. [Google Scholar]
- Schuler, A.J.; Jenkins, D. Enhanced biological phosphorus removal from wastewater by biomass with different phosphorus contents, part I: Experimental Results and Comparison with Metabolic Models. Water Environ. Res. 2003, 75, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Welles, L.; Abbas, B.; Sorokin, D.Y.; Lopez-Vazquez, C.M.; Hooijmans, C.M.; van Loosdrecht, M.C.M.; Brdjanovic, D. Metabolic response of ‘Candidatus Accumulibacter phosphatis’ clade II to changes in the influent P/C ratio. Front. Microbiol. 2017, 7, 2121. [Google Scholar] [CrossRef] [PubMed]
- Oehmen, A.; Lemos, P.C.; Carvalho, G.; Yuan, Z.; Keller, J.; Blackall, L.L.; Reis, M.A.M. Advances in enhanced biological phosphorus removal: From micro to macro scale. Water Res. 2007, 41, 2271–2300. [Google Scholar] [CrossRef] [PubMed]
- Dorofeev, A.G.; Nikolaev, Y.A.; Mardanov, A.V.; Pimenov, N.V. Role of phosphate-accumulating bacteria in biological phosphorus removal from wastewater. Appl. Biochem. Microbiol. 2020, 56, 3–18. [Google Scholar] [CrossRef]
- Zhao, W.; Bi, X.; Peng, Y.; Bai, M. Research advances of the phosphorus-accumulating organisms of Candidatus Accumulibacter, Dechloromonas and Tetrasphaera: Metabolic mechanisms, applications and influencing factors. Chemosphere 2022, 307 Pt 1, 135675. [Google Scholar] [CrossRef] [PubMed]
- Paez-Watson, T.; van Loosdrecht, M.C.M.; Wahl, S.A. Predicting the impact of temperature on metabolic fluxes using resource allocation modelling: Application to polyphosphate accumulating organisms. Water Res. 2023, 228, 119365. [Google Scholar] [CrossRef] [PubMed]
- Dorofeev, A.G.; Nikolaev, Y.A.; Mardanov, A.V.; Pimenov, N.V. Cyclic metabolism as a mode of microbial existence. Microbiology 2019, 88, 402–415. [Google Scholar] [CrossRef]
- Marques, R.; Santos, J.; Nguyen, H.; Carvalho, G.; Noronha, J.P.; Nielsen, P.H.; Reis, M.A.M.; Oehmen, A. Metabolism and ecological niche of Tetrasphaera and Ca. Accumulibacter in enhanced biological phosphorus removal. Water Res. 2017, 122, 159–171. [Google Scholar] [CrossRef]
- Brand, V.R.; Crosby, L.D.; Criddle, C.S. Niche differentiation among three closely related Competibacteraceae clades at a fullscale activated sludge wastewater treatment plant and putative linkages to process performance. Appl. Environ. Microbiol. 2019, 85, e02301–e02318. [Google Scholar] [CrossRef]
- Kong, Y.; Nielsen, J.L.; Nielsen, P.H. Microautoradiographic study of Rhodocyclus-related polyphosphate-accumulating bacteria in full-scale enhanced biological phosphorus removal plants. Appl. Environ. Microbiol. 2004, 70, 5383–5390. [Google Scholar] [CrossRef] [PubMed]
- Levantesi, C.; Serafim, L.S.; Crocetti, G.R.; Lemos, P.C.; Rossetti, S.; Blackall, L.L.; Reis, M.A.M.; Tandoi, V. Analysis of the microbial community structure and function of a laboratory scale enhanced biological phosphorus removal reactor. Environ. Microbiol. 2002, 4, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Pijuan, M.; Baeza, J.A.; Casas, C.; Lafuente, J. Response of an EBPR population developed in an SBR with propionate to different carbon sources. Water Sci. Technol. 2004, 50, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Begum, S.A.; Batista, J.R. Impact of butyrate on microbial selection in en- hanced biological phosphorus removal systems. Environ. Technol. 2014, 35, 2961–2972. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, J.; Oehmen, A.; Le, C.; Geng, Y.; Zhou, Y. Butyrate can support PAOs but not GAOs in tropical climates. Water Res. 2021, 193, 116884. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Chen, H.; Chen, L.; Deng, X.; Hu, Z.; Wang, C.; Wei, C.; Qiu, G.; Wuertz, S. Glycine adversely affects enhanced biological phosphorus removal. Water Res. 2022, 209, 117894. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Kristiansen, R.; Vestergaard, M.; Wimmer, R.; Nielsen, P.H. Intracellular accumulation of glycine in polyphosphate-accumulating organisms in activated sludge, a novel storage mechanism under dynamic anaerobic-aerobic conditions. Appl. Environ. Microbiol. 2015, 81, 4809–4818. [Google Scholar] [CrossRef] [PubMed]
- Oyserman, B.O.; Noguera, D.R.; del Rio, T.G.; Tringe, S.G.; McMahon, K.D. Metatranscriptomic insights on gene expression and regulatory controls in Candidatus Accumulibacter phosphatis. ISME J. 2016, 10, 810–822. [Google Scholar] [CrossRef]
- Qiu, G.; Liu, X.; Saw, N.; Law, Y.; Zuniga-Montanez, R.; Thi, S.S.; Nguyen, T.Q.N.; Nielsen, P.H.; Williams, R.B.H.; Wuertz, S. Metabolic traits of Candidatus Accumulibacter clade IIF strain SCELSE-1 using amino acids as carbon sources for enhanced biological phosphorus removal. Environ. Sci. Technol. 2020, 54, 2448–2458. [Google Scholar] [CrossRef]
- Ziliani, A.; Bovio-Winkler, P.; Cabezas, F.; Etchebehere, C.; Garcia, H.A.; López-Vázquez, C.M.; Brdjanovic, D.; van Loosdrecht, M.C.M.; Rubio-Rincón, F.J. Putative metabolism of Ca. Accumulibacter via the utilization of glucose. Water Res. 2023, 229, 119446. [Google Scholar] [CrossRef]
- McMahon, K.D.; He, S.; Oehmen, A. The microbiology of phosphorus removal. In Microbial Ecology of Activated Sludge; Seviour, R., Nielsen, P.H., Eds.; IWA: London, UK, 2010; pp. 281–319. [Google Scholar]
- Smolders, G.J.; van der Meij, J.; van Loosdrecht, M.C.M.; Heijnen, J.J. Stoichiometric model of the aerobic metabolism of the biological phosphorus removal process. Biotechnol. Bioeng. 1994, 44, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Welles, L.; Lopez-Vazquez, C.M.; Hooijmans, C.M.; van Loosdrecht, M.C.M.; Brdjanovic, D. Impact of salinity on the aerobic metabolism of phosphate-accumulating organisms. Appl. Microbiol. Biotechnol. 2015, 99, 3659–3672. [Google Scholar] [CrossRef] [PubMed]
- Pelevina, A.V.; Berestovskaya, Y.Y.; Grachev, V.A.; Dorofeeva, I.K.; Sorokin, V.V.; Dorofeev, A.G.; Kallistova, A.Y.; Nikolaev, Y.A.; Kotlyarov, R.Y.; Beletskii, A.V.; et al. A Microbial Consortium Removing Phosphates under Conditions of Cyclic Aerobic-Anaerobic Cultivation. Microbiology 2021, 90, 66–77. [Google Scholar] [CrossRef]
- Pelevina, A.V.; Grouzdev, E.V.; Berestovskaya, Y.Y.; Dorofeev, A.G.; Nikolaev, Y.A.; Kallistova, A.Y.; Beletsky, A.V.; Ravin, N.V.; Pimenov, N.V.; Mardanov, A.V. New insight into the granule formation in the reactor for enhanced biological phosphorus removal. Front. Microbiol. 2023, in press. [Google Scholar]
- Brdjanovic, D.; van Loosdrecht, M.C.M.; Hooijmans, C.M.; Alaerts, G.J.; Heijnen, J.J. Temperature effects on physiology of biological phosphorus removal. J. Environ. Eng. 1997, 123, 144–153. [Google Scholar] [CrossRef]
- Schuler, A.J.; Jenkins, D. Enhanced biological phosphorus removal from wastewater by biomass with different phosphorus contents, part II: Anaerobic adenosine triphosphate utilization and acetate uptake rates. Water Environ. Res. 2003, 75, 512–522. [Google Scholar] [CrossRef] [PubMed]
- Bassin, J.P.; Winkler, M.-K.H.; Kleerebezem, R.; Dezotti, M.; van Loosdrecht, M.C.M. Improved phosphate removal by selective sludge discharge in aerobic granular sludge reactors. Biotechnol. Bioeng. 2012, 109, 1919–1928. [Google Scholar] [CrossRef] [PubMed]
- Acevedo, B.; Murgui, M.; Borr’as, L.; Barat, R. New insights in the metabolic behaviour of PAO under negligible poly-P reserves. Chem. Eng. J. 2017, 311, 82–90. [Google Scholar] [CrossRef]
- Smolders, G.J.F.; van der Meij, J.; van Loosdrecht, M.C.M.; Heijnen, J.J. Model of the Anaerobic Metabolism of the Biological Phosphorus Removal Process: Stoichiometry and pH Influence. Biotechnol. Bioeng. 1994, 43, 461–470. [Google Scholar] [CrossRef]
- Liu, W.-T.; Mino, T.; Matsuo, T.; Nakamura, K. Biological phosphorus removal processes—Effect of pH on anaerobic substrate metabolism. Water Sci. Technol. 1996, 34, 25–32. [Google Scholar]
- Ren, T.; Chi, Y.; Wang, Y.; Shi, X.; Jin, X.; Jin, P. Diversified metabolism makes novel Thauera strain highly competitive in low carbon wastewater treatment. Water Res. 2021, 206, 117742. [Google Scholar] [CrossRef] [PubMed]
- Oehmen, A.; Zeng, R.J.; Yuan, Z.; Keller, J. Anaerobic metabolism of propionate by polyphosphate-accumulating organisms in enhanced biological phosphorus removal systems. Biotechnol. Bioeng. 2005, 1, 43–53. [Google Scholar] [CrossRef] [PubMed]
- L’opez-V’azquez, C.M.; Hooijmans, C.M.; Brdjanovic, D.; Gijzen, H.J.; van Loosdrecht, M.C.M. A practical method for quantification of phosphorus- and glycogen accumulating organism populations in activated sludge systems. Water Environ. Res. 2007, 79, 2487–2498. [Google Scholar] [CrossRef] [PubMed]
- Diaz, R.; Mackey, B.; Chadalavada, S.; Kainthola, J.; Heck, P.; Goel, R. Enhanced Bio-P removal: Past, present, and future—A comprehensive review. Chemosphere 2022, 309, 136518. [Google Scholar] [CrossRef] [PubMed]
- Ribas, D.; Soares-Silva, I.; Vieira, D.; Sousa-Silva, M.; Sá-Pessoa, J.; Azevedo-Silva, J.; Viegas, S.C.; Arraiano, C.M.; Diallinas, G.; Paiva, S.; et al. The acetate uptake transporter family motif “NPAPLGL (M/S)” is essential for substrate uptake. Fungal Genet. Biol. 2019, 122, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gimenez, R.; Nuñez, M.F.; Badia, J.; Aguilar, J.; Baldoma, L. The gene yjcG, cotranscribed with the gene acs, encodes an acetate permease in Escherichia coli. J. Bacteriol. 2003, 185, 6448–6455. [Google Scholar] [CrossRef] [PubMed]
- Jolkver, E.; Emer, D.; Ballan, S.; Krämer, R.; Eikmanns, B.J.; Marin, K. Identification and characterization of a bacterial transport system for the uptake of pyruvate, propionate, and acetate in Corynebacterium glutamicum. J. Bacteriol. 2009, 191, 940–948. [Google Scholar] [CrossRef]
- Barr, J.J.; Dutilh, B.E.; Skennerton, C.T.; Fukushima, T.; Hastie, M.L.; Gorman, J.J.; Tyson, G.W.; Bond, P.L. Metagenomic and metaproteomic analyses of Accumulibacter phosphatis-enriched floccular and granular biofilm. Environ. Microbiol. 2016, 18, 273–287. [Google Scholar] [CrossRef]
- Liu, X.-W.; Wang, H.-H.; Chen, J.-Y.; Li, X.-T.; Chen, G.-Q. Biosynthesis of poly (3-hydroxybutyrate-co-3-hydroxyvalerate) by recombinant Escherichia coli harboring propionyl-CoA synthase gene (prpE) or propionate permease gene (prpP). Biochem. Eng. J. 2009, 43, 72–77. [Google Scholar] [CrossRef]
- Arun, V.; Mino, T.; Matsuo, T. Metabolism of carboxylic acids located in and around the glycolytic pathway and the TCA cycle in the biological phosphorus removal process. In Proceedings of the Fourteenth Biennial Conference of the International Association on Water Pollution Research and Control, Brighton, UK, 18–21 July 1988. [Google Scholar] [CrossRef]
- Satoh, H.; Ramey, W.D.; Koch, F.A.; Oldham, W.K.; Mino, T.; Matsuo, T. Anaerobic substrate uptake by the enhanced biological phosphorus removal activated sludge treating real sewage. Water Sci. Technol. 1996, 34, 9–16. [Google Scholar] [CrossRef]
- Skory, C.D.; Hector, R.E.; Gorsich, S.W.; Rich, J.O. Analysis of a functional lactate permease in the fungus Rhizopus. Enzyme Microb. Technol. 2010, 46, 43–50. [Google Scholar] [CrossRef]
- Song, H.; Lee, S.Y. Production of succinic acid by bacterial fermentation. Enzyme Microbial Technol. 2006, 39, 352–361. [Google Scholar] [CrossRef]
- Wainaina, S.; Lukitawesa; Awasthi, K.M.; Taherzadeh, M.J. Bioengineering of anaerobic digestion for volatile fatty acids, hydrogen or methane production: A critical review. Bioengineered 2019, 10, 437–458. [Google Scholar] [CrossRef] [PubMed]
- Putri, D.N.; Sahlan, M.; Montastruc, L.; Meyer, M.; Negny, S.; Hermansyah, H. Progress of fermentation methods for bio-succinic acid production using agro-industrial waste by Actinobacillus succinogenes. Energy Rep. 2020, 6, 234–239. [Google Scholar] [CrossRef]
- Hosie, A.H.F.; Poole, P.S. Bacterial ABC transporters of amino acids. Res. Microbiol. 2001, 152, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yan, Z.; Wang, P.; Zheng, X.; Fan, J. Comparative metagenomics reveals the microbial diversity and metabolic potentials in the sediments and surrounding seawaters of Qinhuangdao mariculture area. PLoS ONE 2020, 15, e0234128. [Google Scholar] [CrossRef] [PubMed]
- Oehmen, A.; Yuan, Z.; Blackall, L.L.; Keller, J. Comparison of acetate and propionate uptake by polyphosphate accumulating organisms and glycogen accumulating organisms. Biotechnol. Bioeng. 2005, 91, 162–168. [Google Scholar] [CrossRef]

| Substrate | OUR, mg O2/(g TSS h) | Anaerobic Phase | Aerobic Phase | ||
|---|---|---|---|---|---|
| Pmax, mg PO43−-P/L | P-Release/Substrate-Uptake, P-mol/S-mol | Average P Release Rate in the First 30 min, mg PO43−-P/(L h) | Pmax − Pmin, mg PO43−-P/L | ||
| Acetate | 32.2 | 144.7 | 1.4 | 273.6 | 125.9 |
| Pyruvate | 23.6 | 65.0 | 1.7 | 138.8 | 60 |
| Propionate | 20.7 | 94.5 | 1.6 | 186.2 | 80.2 |
| Butyrate | 8.9 | 69.1 | 2.1 | 72.8 | 46.2 |
| Succinate 1 | 6.2 | 28.5 | 1.3 | 13.7 | 24.1 |
| d,l-Aspartate | 4.4 | 131.1 | 2.0 2 | 127.3 | 32.9 |
| l-Glutamate | 2.7 | 82.3 | 2.1 2 | 93.1 | 42.8 |
| d,l-Alanine | 1.9 | 34.2 | 0.3 2 | 19.2 | 9 |
| d-Glucose | 1.1 | 7.7 | - | - | 3.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dorofeev, A.; Pelevina, A.; Nikolaev, Y.; Berestovskaya, Y.; Gruzdev, E.; Mardanov, A.; Pimenov, N. Oxygen Uptake Rate as an Indicator of the Substrates Utilized by Candidatus Accumulibacter. Water 2023, 15, 3657. https://doi.org/10.3390/w15203657
Dorofeev A, Pelevina A, Nikolaev Y, Berestovskaya Y, Gruzdev E, Mardanov A, Pimenov N. Oxygen Uptake Rate as an Indicator of the Substrates Utilized by Candidatus Accumulibacter. Water. 2023; 15(20):3657. https://doi.org/10.3390/w15203657
Chicago/Turabian StyleDorofeev, Alexander, Anna Pelevina, Yuri Nikolaev, Yulia Berestovskaya, Evgeny Gruzdev, Andrey Mardanov, and Nikolai Pimenov. 2023. "Oxygen Uptake Rate as an Indicator of the Substrates Utilized by Candidatus Accumulibacter" Water 15, no. 20: 3657. https://doi.org/10.3390/w15203657





