Characterization of a Thermophilic and Inhibitor-Tolerant GH1 β-Glucosidase Present in a Hot Spring
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Metagenomic DNA Extraction
2.2. Metagenomic Sequencing and Synthesis of β-Glucosidase Gene
2.3. Sequence Analysis of β-Glucosidase Gene
2.4. Culture Medium
2.5. Gene Amplification and Construction of Recombinant Vector
2.6. Heterologous expression of ThBg2
2.7. Purification of Recombinant Proteins
2.8. Enzyme Assay of β-Glucosidase
2.9. Determination of Optimum Temperature and Thermostability of ThBg2
2.10. Determination of Optimum pH and pH Stability of ThBg2
2.11. Effects of Metal Ions and Other Chemicals on the Activity of ThBg2
2.12. Effect of Glucose Concentration on Enzymatic Activity
2.13. Kinetic Parameters of ThBg2
2.14. Hydrolysis of Corn Stalks by ThBg2
2.15. Statistical Analysis
3. Results
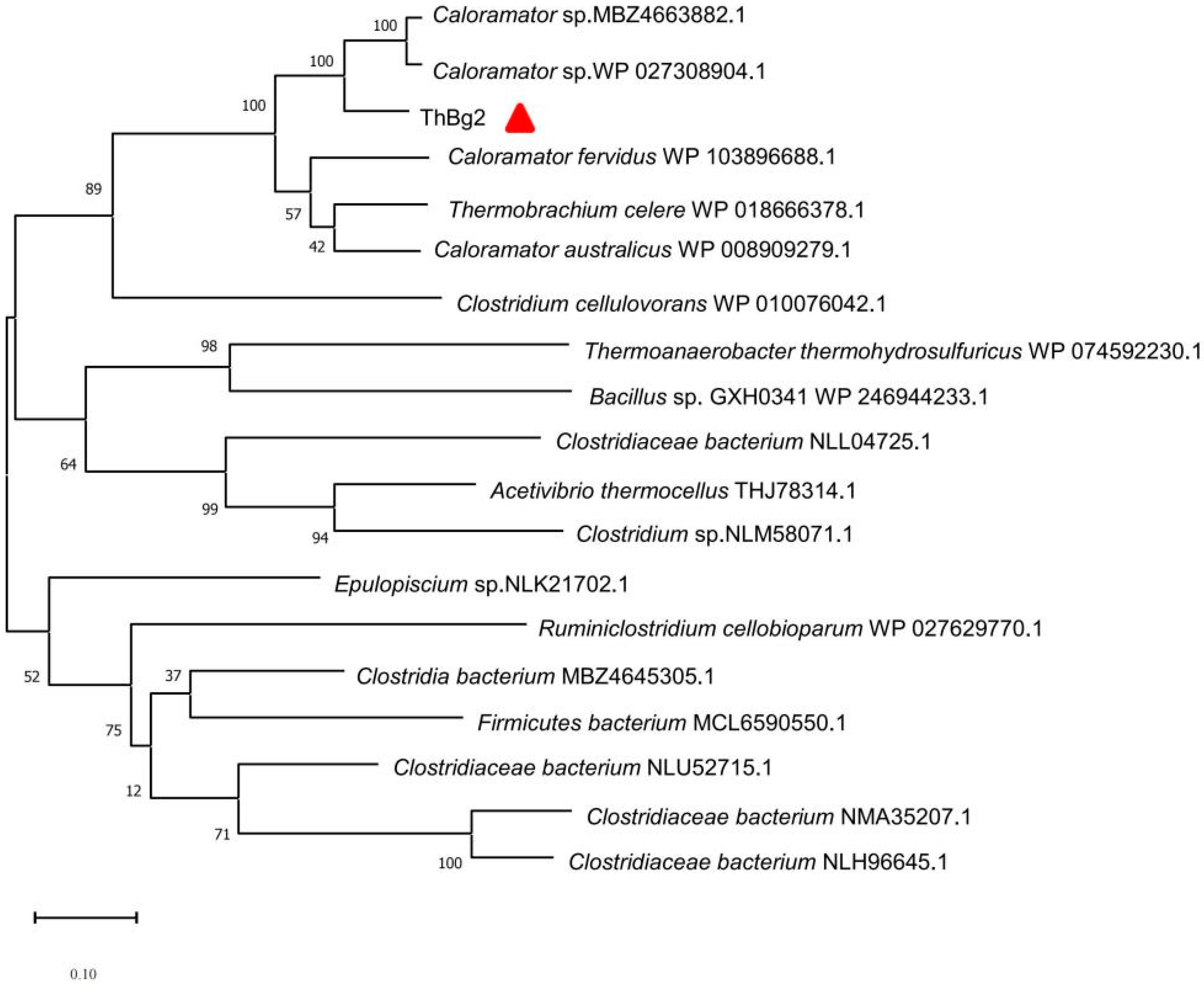
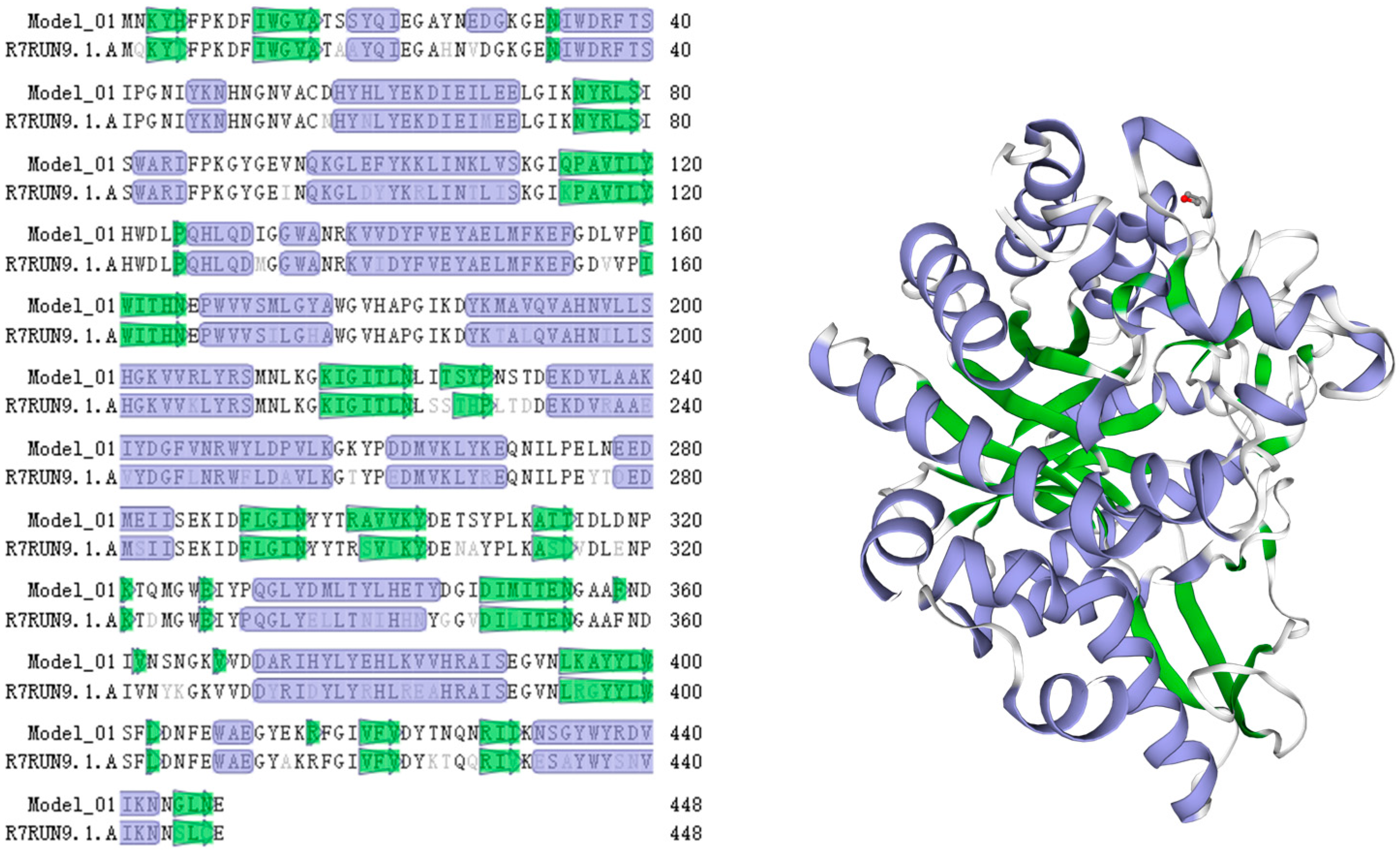
3.1. Sequence Analysis
3.2. Expression and Purification of ThBg2
3.3. Optimal Temperature and Thermal Stability of ThBg2
3.4. Optimal pH and pH Stability of ThBg2
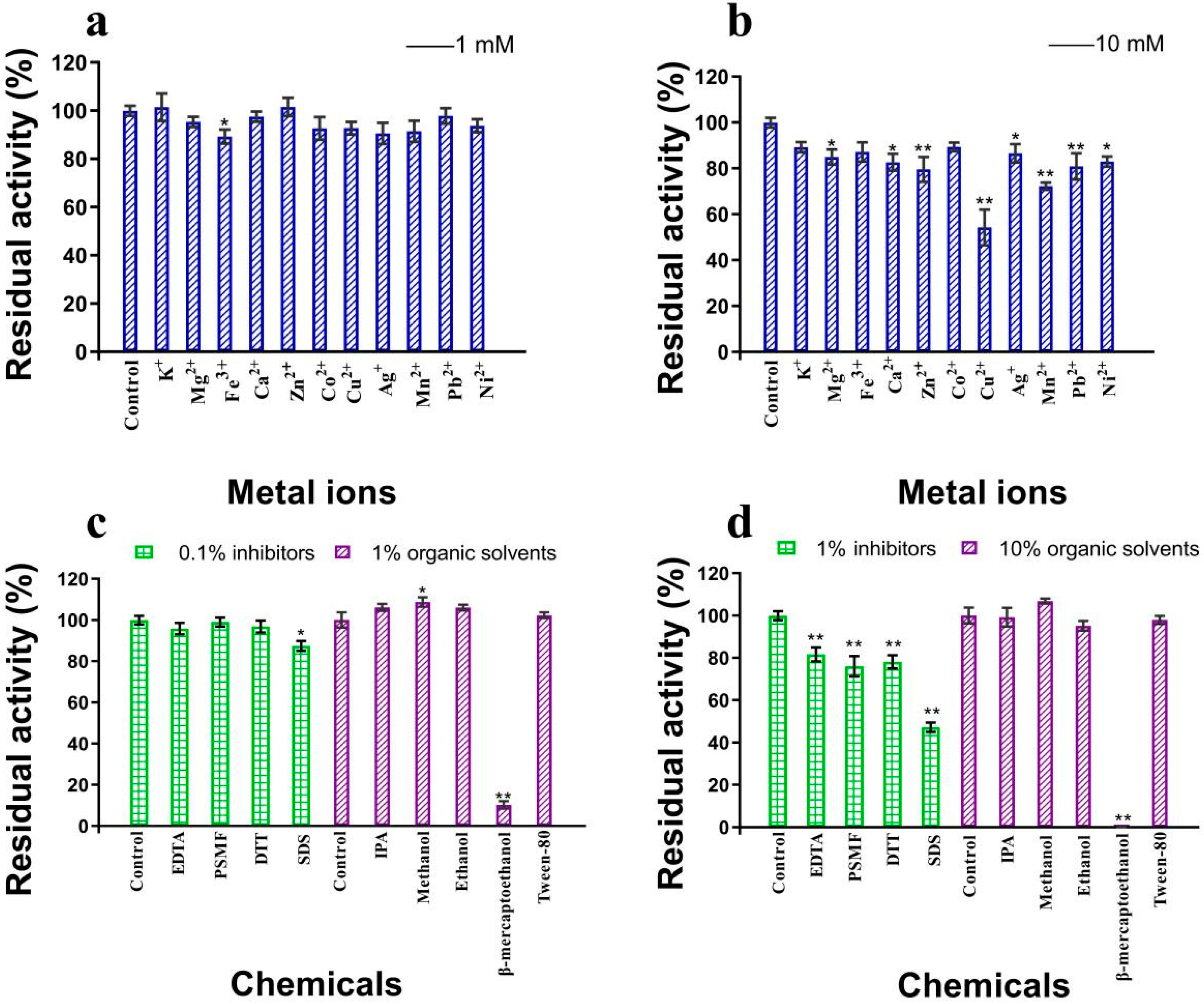
3.5. Effects of Metal Ions and Other Chemicals on the Activity of β-Glucosidase
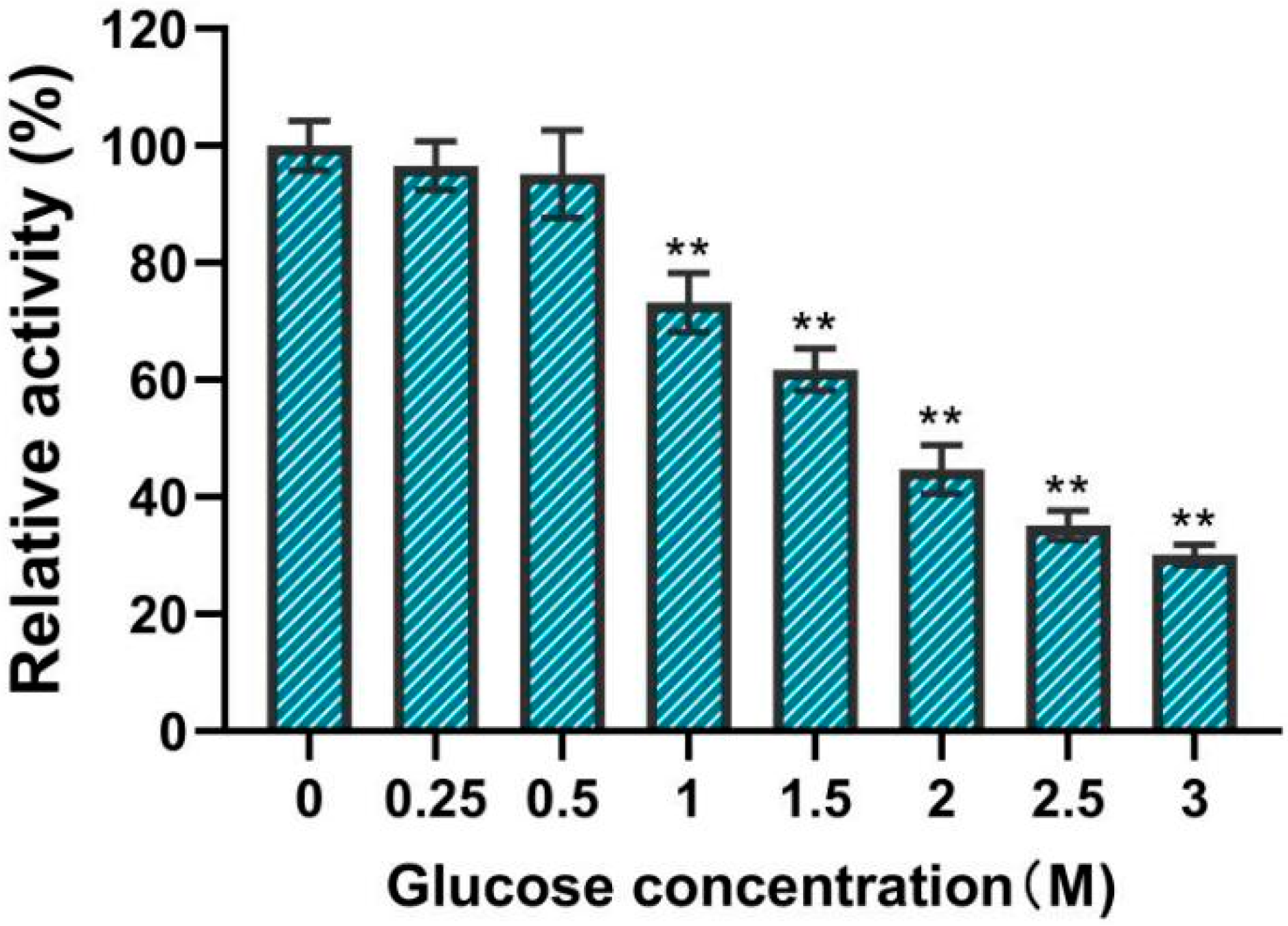
3.6. Effect of Glucose Concentration on Enzymatic Activity
3.7. Kinetic Properties of ThBg2
3.8. Analysis of Hydrolysis of Corn Stalks by ThBg2
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Ginni, G.; Kavitha, S.; Yukesh, K.R. Valorization of agricultural residues: Different biorefinery routes. J. Environ. Chem. Eng. 2021, 9, 105435. [Google Scholar] [CrossRef]
- Rakhee, K.; Mondher, T.N. Bifunctional xylanases and their potential use in biotechnology. J. Ind. Microbiol. Biotechnol. 2008, 3, 635–644. [Google Scholar] [CrossRef]
- McKendry, P. Energy production from biomass (part 1): Overview of biomass. Bioresour. Technol. 2002, 83, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Lynd, L.R.; Weimer, P.J.; van Zyl, W.H.; Pretorius, I.S. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef]
- Gao, D.; Chundawat, S.P.S.; Liu, T. Strategy for Identification of Novel Fungal and Bacterial Glycosyl Hydrolase Hybrid Mixtures that can Efficiently Saccharify Pretreated Lignocellulosic Biomass. BioEnergy Res. 2010, 3, 67–81. [Google Scholar] [CrossRef][Green Version]
- Bokkenheuser, V.D.; Shackleton, C.H.; Winter, J. Hydrolysis of dietary flavonoid glycosides by strains of intestinal Bacteroides from humans. Biochem. J. 1987, 248, 953–956. [Google Scholar] [CrossRef]
- Freer, S.N. Kinetic characterization of a beta-glucosidase from a yeast, Candida wickerhamii. J. Biol. Chem. 1993, 268, 9337–9342. [Google Scholar] [CrossRef]
- Shallom, D.; Shoham, Y. Microbial hemicellulases. Curr. Opin. Microbiol. 2003, 6, 219–228. [Google Scholar] [CrossRef]
- Zhang, P.Z.; Zhang, R.; Sirisena, S.; Gan, R.; Fang, Z.X. Beta-glucosidase activity of wine yeasts and its impacts on wine volatiles and phenolics: A mini-review. Food Microbiol. 2021, 100, 103859. [Google Scholar] [CrossRef]
- Abdella, A.; El-Baz, A.F.; Ibrahim, I.A.; Mahrous, E.E.; Yang, S.T. Biotransformation of soy flour isoflavones by Aspergillus niger NRRL 3122 β-glucosidaseenzyme. Nat. Prod. Res. 2018, 32, 2382–2391. [Google Scholar] [CrossRef]
- Ahmed, S.S.; Akhter, M.; Sajjad, M.; Gul, R.; Khurshid, S. Soluble production, characterization, and structural aesthetics of an industrially important thermostable β-glucosidase from Clostridium thermocellum in Escherichia coli. BioMed Res. Int. 2019, 2019, 9308593. [Google Scholar] [CrossRef]
- Yin, Y.R.; Sang, P.; Yang, F.L.; Li, T.; Yang, R.F.; Liu, H.Y.; Luo, Z.L.; Li, W.J.; Yang, L.Q. Characterization of a Cu2+, SDS, alcohol and glucose tolerant GH1 beta-glucosidase from Bacillus sp. CGMCC 1.16541. Antonie Van Leeuwenhoek 2020, 113, 1467–1477. [Google Scholar] [CrossRef]
- Xie, Y.F.; Han, X.M.; Lu, F.P. Expression, purification and enzymatic properties of β-glucosidase from Lactobacillus paracasei. China Biotechnol. 2019, 39, 72–79. [Google Scholar] [CrossRef]
- Ravin, N.V.; Mardanov, A.V.; Skryabin, K.G. Metagenomics as a tool for the investigation of uncultured microorganisms. Russ. J. Genet. 2015, 51, 431–439. [Google Scholar] [CrossRef]
- Banik, J.J.; Brady, S.F. Recent application of metagenomic approaches toward the discovery of antimicrobials and other bioactive small molecules. Curr. Opin. Microbiol. 2010, 13, 603–609. [Google Scholar] [CrossRef]
- Tuffin, M.; Anderson, D.; Heath, C.; Cowan, D.A. Metagenomic gene discovery: How far have we moved into novel sequence space? Biotechnol. J. 2009, 4, 1671–1683. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; Huang, Q.; Chang, L.; Sun, Q.; Zhou, J.; Lu, H. Cloning and characterization of two beta-glucosidase/xylosidase enzymes from yak rumen metagenome. Appl. Biochem. Biotechnol. 2012, 166, 72–86. [Google Scholar] [CrossRef]
- Jiang, C.; Li, S.-X.; Luo, F.-F.; Jin, K.; Wang, Q.; Hao, Z.-Y.; Wu, L.-L.; Zhao, G.-C.; Ma, G.-F.; Shen, P.-H.; et al. Biochemical characterization of two novel β-glucosidase genes by metagenome expression cloning. Bioresour. Technol. 2011, 102, 3272–3278. [Google Scholar] [CrossRef]
- Wang, Q.; Qian, C.; Zhang, X.Z.; Liu, N.; Yan, X.; Zhou, Z. Characterization of a novel thermostable β-glucosidase from a metagenomic library of termite gut. Enzym. Microb. Technol. 2012, 51, 319–324. [Google Scholar] [CrossRef]
- Lima, R.A.T.; De Oliveira, G.; Souza, A.A.; Lopes, F.A.C.; Santana, R.H.; Istvan, P.; Quirino, B.F.; Barbosa, J.; De Freitas, S.; Garay, A.V.; et al. Functional and structural characterization of a novel GH3 β-glucosidase from the gut metagenome of the Brazilian Cerrado termite Syntermes wheeleri. Int. J. Biol. Macromol. 2020, 165 Pt A, 822–834. [Google Scholar] [CrossRef]
- Mai, Z.; Wang, L.; Zeng, Q. Characterization of a novel isoflavone glycoside-hydrolyzing β-glucosidase from mangrove soil metagenomic library. Biochem. Biophys. Res. Commun. 2021, 569, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, N.; Yang, H.; Zhao, F.; Yu, Y.; Tian, Y.; Lu, X. Cloning and characterization of a new β-glucosidase from a metagenomic library of rumen of cattle feeding with Miscanthus sinensis. BMC Biotechnol. 2014, 14, 85. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Fu, Q.; Li, W.; Wang, B.; Yin, X.; Liu, S.; Xu, Z.; Niu, Q. Identification and characterization of a novel β-glucosidase via metagenomic analysis of Bursaphelenchus xylophilus and its microbial flora. Sci. Rep. 2017, 7, 14850. [Google Scholar] [CrossRef] [PubMed]
- Averhoff, B.; Müller, V. Exploring research frontiers in microbiology: Recent advances in halophilic and thermophilic extremophiles. Res. Microbiol. 2010, 161, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Wang, S.; Dong, H.; Jiang, H.; Briggs, B.R.; Peacock, J.P.; Huang, Q.; Huang, L.; Wu, G.; Zhi, X.; et al. A Comprehensive Census of Microbial Diversity in Hot Springs of Tengchong, Yunnan Province China Using 16S rRNA Gene Pyrosequencing. PLoS ONE 2013, 8, e53350. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Sumitani, J.-I.; Nam, Y.-W.; Nishimaki, T.; Tani, S.; Wakagi, T.; Kawaguchi, T.; Fushinobu, S. Crystal structures of glycoside hydrolase family 3 β-glucosidase 1 from Aspergillus aculeatus. Biochem. J. 2013, 452, 211–221. [Google Scholar] [CrossRef]
- Nakaya, A.; Katayama, T.; Itoh, M.; Hiranuka, K.; Kawashima, S.; Moriya, Y.; Goto, S. KEGG OC: A large-scale automatic construction of taxonomy-based ortholog clusters. Nucleic Acids Res. 2012, 41, 353–357. [Google Scholar] [CrossRef]
- Tatusov, R.L. The COG database: New developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001, 29, 22–28. [Google Scholar] [CrossRef]
- Finn, R.D.; Tate, J.; Mistry, J.; Coggill, P.C.; Sammut, S.J.; Hotz, H.R.; Bateman, A. The Pfam protein families database. Nucleic Acids Res. 2007, 36, 281–288. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Badieyan, S.; Bevan, D.R.; Zhang, C. Study and design of stability in GH5 cellulases. Biotechnol. Bioeng. 2012, 109, 31–44. [Google Scholar] [CrossRef]
- Vieille, C.; Zeikus, G.J. Hyperthermophilic enzymes: Sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 2001, 65, 1–43. [Google Scholar] [CrossRef]
- Kufner, K.; Lipps, G. Construction of a chimeric thermoacidophilic beta-endoglucanase. BMC Biochem. 2013, 14, 11. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jiang, C.-J.; Chen, G.; Huang, J.; Huang, Q.; Jin, K.; Shen, P.-H.; Li, J.-F.; Wu, B. A novel β-glucosidase with lipolytic activity from a soil metagenome. Folia Microbiol. 2011, 56, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Xu, X.; Qian, L.; Shi, P.; Bai, Y.; Luo, H.; Ma, R.; Yao, B. Engineering a highly active thermophilic β-glucosidase to enhance its pH stability and saccharifification performance. Biotechnol. Biofuels 2016, 9, 147. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Weng, H.; Xi, Y. Purification and characterization of a novel endo-β-1,4-glucanase from the thermoacidophilic Aspergillus terreus. Biotechnol. Lett. 2008, 30, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Pang, Q.; Zhao, L.; Fan, S.; Shi, H. Thermoanaerobacterium thermosaccharolyticum β-glucosidase: A glucose-tolerant enzyme with high specific activity for cellobiose. Biotechnol. Biofuels 2012, 5, 31. [Google Scholar] [CrossRef]
- Ariaeenejad, S.; Nooshi-Nedamani, S.; Rahban, M.; Kavousi, K.; Pirbalooti, A.G.; Mirghaderi, S.; Mohammadi, M.; Mirzaei, M.; Salekdeh, G.H. A Novel High Glucose-Tolerant β-Glucosidase: Targeted Computational Approach for Metagenomic Screening. Front. Bioeng. Biotechnol. 2020, 8, 813. [Google Scholar] [CrossRef]
- Navarro, C.A.; Bernath, D.V.; Jerez, C.A. Heavy metal resistance strategies of acidophilic bacteria and their acquisition: Importance for biomining and bioremediation. Biol. Res. 2013, 46, 363–371. [Google Scholar] [CrossRef]
- Kaushal, G.; Rai, A.K.; Singh, S.P. A novel β-glucosidase from a hot-spring metagenome shows elevated thermal stability and tolerance to glucose and ethanol. Enzym. Microb. Technol. 2021, 145, 109764. [Google Scholar] [CrossRef]
- Garcia, N.F.L.; da Silva Santos, F.R.; Gonçalves, F.A.; da Paz, M.F.; Fonseca, G.G.; Leite, R.S.R. Production of β-glucosidase on solid-state fermentation by Lichtheimia ramosa in agroindustrial residues: Characterization and catalytic properties of the enzymatic extract. Electron. J. Biotechnol. 2015, 18, 314–319. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, H.; Lin, C.C.; Xu, B. Optimization of rice wine fermentation process based on the simultaneous saccharifification and fermentation kinetic model. Chin. J. Chem. Eng. 2016, 24, 1406–1412. [Google Scholar] [CrossRef]
- Sun, S.; Sun, S.; Cao, X.; Sun, R. The role of pretreatment in improving the enzymatic hydrolysis of lignocellulosic materials. Bioresour. Technol. 2016, 199, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, X.; Wei, W.; Xu, J.; Wang, W.; Xie, Z.; Zhang, Z.; Jiang, H.; Wang, Q.; Wei, C. A novel efficient β-glucanase from a paddy soil microbial metagenome with versatile activities. Biotechnol. Biofuels 2016, 9, 36. [Google Scholar] [CrossRef]
- Alves, L.F.; Meleiro, L.P.; Silva, R.N.; Westmann, C.A.; Guazzaroni, M.E. Novel Ethanol- and 5-Hydroxymethyl Furfural-Stimulated β-Glucosidase Retrieved From a Brazilian Secondary Atlantic Forest Soil Metagenome. Front. Microbiol. 2018, 9, 2556. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, T.; Miyazaki, K.; Yaoi, K. Characterization of a Novel β-Glucosidase from a Compost Microbial Metagenome with Strong Transglycosylation Activity. J. Biol. Chem. 2013, 288, 18325–18334. [Google Scholar] [CrossRef]
- Koppram, R.; Tomás-Pejó, E.; Xiros, C.; Olsson, L. Lignocellulosic ethanol production at high-gravity: Challenges and perspectives. Trends Biotechnol. 2014, 32, 46–53. [Google Scholar] [CrossRef]
- Lu, J.; Du, L.; Wei, Y.; Hu, Y.; Huang, R. Expression and characterization of a novel highly glucose-tolerant β-glucosidase from a soil metagenome. Acta Biochim. Biophys. Sin. 2013, 45, 664–673. [Google Scholar] [CrossRef]
- Zhao, L.; Xie, J.; Zhang, X.; Cao, F.; Pei, J. Overexpression and characterization of a glucose-tolerant β-glucosidase from Thermotoga thermarum DSM 5069T with high catalytic efficiency of ginsenoside Rb1 to Rd. J. Mol. Catal. B Enzym. 2013, 95, 62–69. [Google Scholar] [CrossRef]
- Limauro, D.; Bartolucci, S.; Pedone, E.; Fusco, F.A.; Contursi, P.; Fiorentino, G. Biochemical characterization of a novel thermostable β-glucosidase from Dictyoglomus turgidum. Int. J. Biol. Macromol. 2018, 113, 783–791. [Google Scholar] [CrossRef]
- de Giuseppe, P.O.; Souza, T.A.C.B.; Souza, F.H.M.; Zanphorlin, L.M.; Machado, C.B.; Ward, R.J.; Jorge, J.A.; Furriel, R.P.M.; Murakami, M.T. Structural basis for glucose tolerance in GH1 β-glucosidases. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 1631–1639. [Google Scholar] [CrossRef] [PubMed]
- de Lima, L.H.F.; Fernandez-Quintéro, M.L.; Rocha, R.E.O.; Mariano, D.C.B.; de Melo-Minardi, R.C.; Liedl, K.R. Conformational flexibility correlates with glucose tolerance for point mutations in β-glucosidases—A computational study. J. Biomol. Struct. Dyn. 2021, 39, 1621–1634. [Google Scholar] [CrossRef] [PubMed]
- Tomme, P.; Tilbeurgh, H.; Pettersson, G.; Damme, J.; Vandekerckhove, J.; Knowles, J.; Teeri, T.; Claeyssens, M. Studies of the cellulolytic system of Trichoderma reesei qm 9414: Analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur. J. Biochem. 1988, 170, 575–581. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.-Y.; Wu, P.; Wu, X.-C.; Zhu, Q.-R.; Zhu, Q.; Zheng, H.-Z.; Zhu, D.; Lv, Z.-H.; Yin, Y.-R. Characterization of a Thermophilic and Inhibitor-Tolerant GH1 β-Glucosidase Present in a Hot Spring. Water 2023, 15, 3389. https://doi.org/10.3390/w15193389
Huang Y-Y, Wu P, Wu X-C, Zhu Q-R, Zhu Q, Zheng H-Z, Zhu D, Lv Z-H, Yin Y-R. Characterization of a Thermophilic and Inhibitor-Tolerant GH1 β-Glucosidase Present in a Hot Spring. Water. 2023; 15(19):3389. https://doi.org/10.3390/w15193389
Chicago/Turabian StyleHuang, Yu-Ying, Pei Wu, Xing-Ci Wu, Qian-Ru Zhu, Qian Zhu, Hong-Zhao Zheng, Dan Zhu, Zhi-Hua Lv, and Yi-Rui Yin. 2023. "Characterization of a Thermophilic and Inhibitor-Tolerant GH1 β-Glucosidase Present in a Hot Spring" Water 15, no. 19: 3389. https://doi.org/10.3390/w15193389
APA StyleHuang, Y.-Y., Wu, P., Wu, X.-C., Zhu, Q.-R., Zhu, Q., Zheng, H.-Z., Zhu, D., Lv, Z.-H., & Yin, Y.-R. (2023). Characterization of a Thermophilic and Inhibitor-Tolerant GH1 β-Glucosidase Present in a Hot Spring. Water, 15(19), 3389. https://doi.org/10.3390/w15193389




