Molecular Investigation of the Achnanthidium minutissimum Complex (Bacillariophyceae) from the Transbaikal Region with the Description of Three New Species
Abstract
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kulikovskiy, M.; Lange-Bertalot, H.; Witkowski, A.; Khursevich, G. Achnanthidium sibiricum (Bacillariophyceae), a new species from bottom sediments in Lake Baikal. Algol. Stud. 2011, 136/137, 77–87. [Google Scholar] [CrossRef]
- Kulikovskiy, M.; Lange-Bertalot, H.; Metzeltin, D.; Witkowski, A. Lake Baikal: Hotspot of Endemic Diatoms I. Iconogr. Diatomol. 2012, 23, 7–608. [Google Scholar]
- Kulikovskiy, M.; Lange-Bertalot, H.; Witkowski, A. Gliwiczia gen. nov. a new monoraphid diatom genus from Lake Baikal with a description of four species new for science. Phytotaxa 2013, 109, 1–16. [Google Scholar] [CrossRef]
- Kulikovskiy, M.; Lange-Bertalot, H.; Kuznetsova, I. Lake Baikal: Hotspot of Endemic Diatoms II. Iconogr. Diatomol. 2015, 26, 1–656. [Google Scholar]
- Kulikovskiy, M.; Glushchenko, A.; Genkal, S.; Kuznetsova, I.; Kociolek, J.P. Platebaikalia—A new monoraphid diatom genus from ancient Lake Baikal with comments on the genus Platessa. Fottea. Olomouc. 2020, 20, 58–67. [Google Scholar] [CrossRef]
- Kulikovskiy, M.; Glushchenko, A.; Kuznetsova, I.; Genkal, S.; Kociolek, J.P. Gololobovia gen. nov.—A new genus from Lake Baikal with comments on pore occlusion in monoraphid diatoms. Phycologia 2020, 59, 616–633. [Google Scholar] [CrossRef]
- Kulikovskiy, M.; Glushchenko, A.; Genkal, S.; Kuznetsova, I.; Kociolek, J.P. Revision of Monoraphid Diatom Genus Platessa with Description of Platesiberia gen. nov. from Ancient Lake Baikal. Water 2022, 14, 2957. [Google Scholar] [CrossRef]
- Novais, M.H.; Jüttner, I.; Van de Vijver, B.; Morais, M.M.; Hoffmann, L.; Ector, L. Morphological variability within the Achnanthidium minutissimum species complex (Bacillariophyta): Comparison between the type material of Achnanthes minutissima and related taxa, and new freshwater Achnanthidium species from Portugal. Phytotaxa 2015, 224, 101–139. [Google Scholar] [CrossRef]
- Foged, N.; Håkansson, H.; Flower, R.J. Some diatoms from Siberia especially from Lake Baikal. Diatom Res. 1993, 8, 231–279. [Google Scholar] [CrossRef]
- Pomazkina, G.; Votyakova, N. Microphytobenthos of South Baikal. In Diatoms—Indicators of Environment and Climate Changing; Proceedings of the V Diatoms Conference, Irkutsk, Russia, 16–20 March 1993; Likhoshway, E., Ed.; LIN SB RAS: Irkutsk, Russia, 1993; pp. 48–50. [Google Scholar]
- Skvortzow, B.W. Bottom diatoms from Olchon Gate of Baikal Lake, Siberia. Philipp. J. Sci. 1937, 62, 293–377. [Google Scholar]
- Round, F.E.; Bukhtiyarova, L. Four new genera based on Achnanthes (Achnanthidium) together with a re-definition of Achnanthidium. Diatom Res. 1996, 11, 345–361. [Google Scholar] [CrossRef]
- Pinseel, E.; Vanormelingen, P.; Hamilton, P.B.; Vyverman, W.; Van de Vijver, B.; Kopalová, K. Molecular and morphological characterization of the Achnanthidium minutissimum complex (Bacillariophyta) in Petuniabukta (Spitsbergen, High Arctic) including the description of A. digitatum sp. nov. Eur. J. Phycol. 2017, 52, 264–280. [Google Scholar] [CrossRef]
- Windler, M.; Leinweber, K.; Bartulos, C.R.; Philipp, B.; Kroth, P.G. Biofilm and capsule formation of the diatom Achnanthidium minutissimum are affected by a bacterium. J. Phycol. 2015, 51, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Monnier, O.; Lange-Bertalot, H.; Rimet, F.; Hoffmann, L.; Ector, L. Achnanthidium atomoides sp. nov., a new diatom from the Grand-Duchy of Luxembourg. Vie Milieu/Life Environ. 2004, 54, 127–136. [Google Scholar]
- Morales, E.A.; Ector, L.; Fernández, E.; Novais, M.H.; Hlúbiková, D.; Hamilton, P.B.; Blanco, S.; Vis, M.L.; Kociolek, J.P. The genus Achnanthidium Kütz. (Achnanthales, Bacillariophyceae) in Bolivian streams: A report of taxa found in recent investigations. Algol. Stud. 2011, 136/137, 89–130. [Google Scholar] [CrossRef]
- Novais, M.H.; Hlúbiková, D.; Morais, M.; Hoffmann, L.; Ector, L. Morphology and ecology of Achnanthidium caravelense (Bacillariophyceae), a new species from Portuguese rivers. Algol. Stud. 2011, 136/137, 131–150. [Google Scholar] [CrossRef]
- Van de Vijver, B.; Jarlman, A.; Lange-Bertalot, H.; Mertens, A.; de Haan, M.; Ector, L. Four new European Achnanthidium species (Bacillariophyceae). Algol. Stud. 2011, 136/137, 193–210. [Google Scholar] [CrossRef]
- Wojtal, A.Z.; Ector, L.; Van de Vijver, B.; Morales, E.A.; Blanco, S.; Piatek, J.; Smieja, A. The Achnanthidium minutissimum complex (Bacillariophyceae) in southern Poland. Algol. Stud. 2011, 136/137, 211–238. [Google Scholar] [CrossRef]
- Van de Vijver, B.; Kopalová, K. Four Achnanthidium species (Bacillariophyta) formerly identified as Achnanthidium minutissimum from the Antarctic Region. Eur. J. Taxon. 2014, 79, 1–19. [Google Scholar] [CrossRef]
- Krahn, K.J.; Wetzel, C.E.; Ector, L.; Schwalb, A. Achnanthidium neotropicum sp. nov., a new freshwater diatom from Lake Apastepeque in El Salvador (Central America). Phytotaxa 2018, 382, 89–101. [Google Scholar] [CrossRef]
- Miao, M.; Li, Z.; Hwang, E.A.; Kim, H.K.; Lee, H.; Kim, B.H. Two new benthic diatoms of the genus Achnanthidium (Bacillariophyceae) from the Hangang River, Korea. Diversity 2020, 12, 285. [Google Scholar] [CrossRef]
- Potapova, M.; Hamilton, P.B. Morphological and ecological variation within the Achnanthidium minutissimum (Bacillariophyceae) species complex. J. Phycol. 2007, 43, 561–575. [Google Scholar] [CrossRef]
- Hlúbiková, D.; Ector, L.; Hoffmann, L. Examination of the type material of some diatom species related to Achnanthidium minutissimum (Kütz.) Czarn. (Bacillariophyceae). Algol. Stud. 2011, 136, 19–43. [Google Scholar] [CrossRef]
- Van de Vijver, B.; Ector, L.; Beltrami, M.E.; de Haan, M.; Falasco, E.; Hlúbiková, D.; Jarlman, A.; Kelly, M.; Novais, M.H.; Wojtal, A.Z. A critical analysis of the type material of Achnanthidium lineare W. Sm. (Bacillariophyceae). Algol. Stud. 2011, 136/137, 167–191. [Google Scholar] [CrossRef]
- Marquardt, G.C.; Costa, L.F.; Bicudo, D.C.; de M Bicudo, C.E.; Blanco, S.; Wetzel, C.E.; Ector, L. Type analysis of Achnanthidium minutissimum and A. catenatum and description of A. tropicocatenatum sp. nov. (Bacillariophyta), a common species in Brazilian reservoirs. Plant Ecol. Evol. 2017, 150, 313–330. [Google Scholar] [CrossRef]
- Guillard, R.R.L.; Lorenzen, C.J. Yellow-green algae with chlorophyllide c1,2. J. Phycol. 1972, 8, 10–14. [Google Scholar] [CrossRef]
- Zimmermann, J.; Jahn, R.; Gemeinholzer, B. Barcoding diatoms: Evaluation of the V4 subregion on the 18S rRNA gene, including new primers and protocols. Org. Divers. Evol. 2011, 11, 173–192. [Google Scholar] [CrossRef]
- Alverson, A.J.; Jansen, R.K.; Theriot, E.C. Bridging the Rubicon: Phylogenetic analysis reveals repeated colonizations of marine and fresh waters by thalassiosiroid diatoms. Mol. Phylogenet. Evol. 2007, 45, 193–210. [Google Scholar] [CrossRef]
- Daugbjerg, N.; Andersen, R.A. A molecular phylogeny of the heterokont algae based on analyses of chloroplast-encoded rbcL sequence data. J. Phycol. 1997, 33, 1031–1041. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Katoh, K.; Toh, H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 2010, 26, 1899–1900. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A Rapid Bootstrap Algorithm for the RAxML Web Servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef]
- Van de Vijver, B.; Wetzel, C.E.; Ector, L. Analysis of the type material of Achnanthidium jackii Rabenhorst (Bacillariophyta, Achnanthidiaceae). Not. Algarum 2018, 55, 1–4. [Google Scholar]
- Ponader, K.C.; Potapova, M.G. Diatoms from the genus Achnanthidium in flowing waters of the Appalachian Mountains (North America): Ecology, distribution and taxonomic notes. Limnologica 2007, 37, 227–241. [Google Scholar] [CrossRef]
- Maltsev, Y.; Maltseva, S.; Kociolek, J.P.; Jahn, R.; Kulikovskiy, M. Biogeography of the cosmopolitan terrestrial diatom Hantzschia amphioxys sensu lato based on molecular and morphological data. Sci. Rep. 2021, 11, 4266. [Google Scholar] [CrossRef]

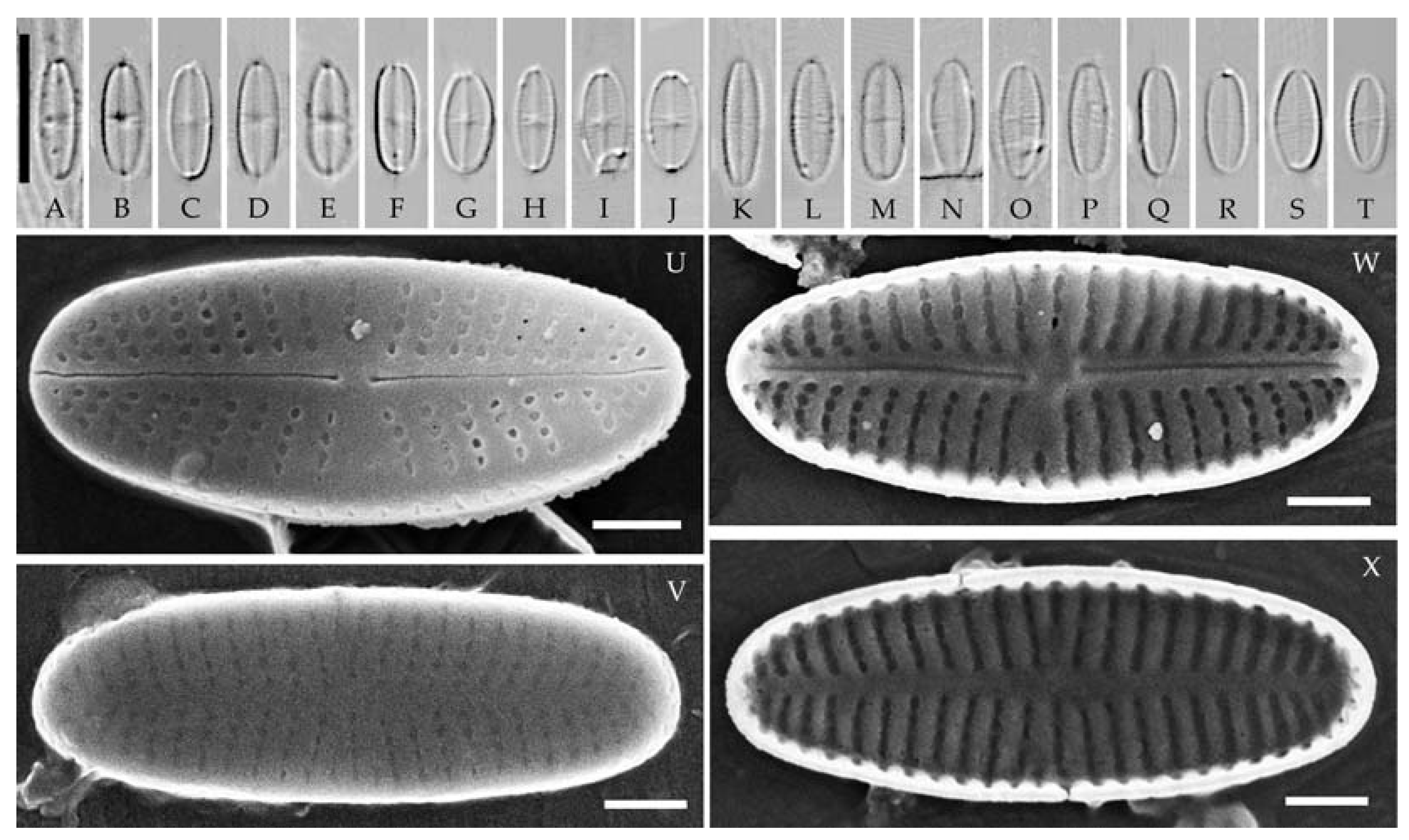


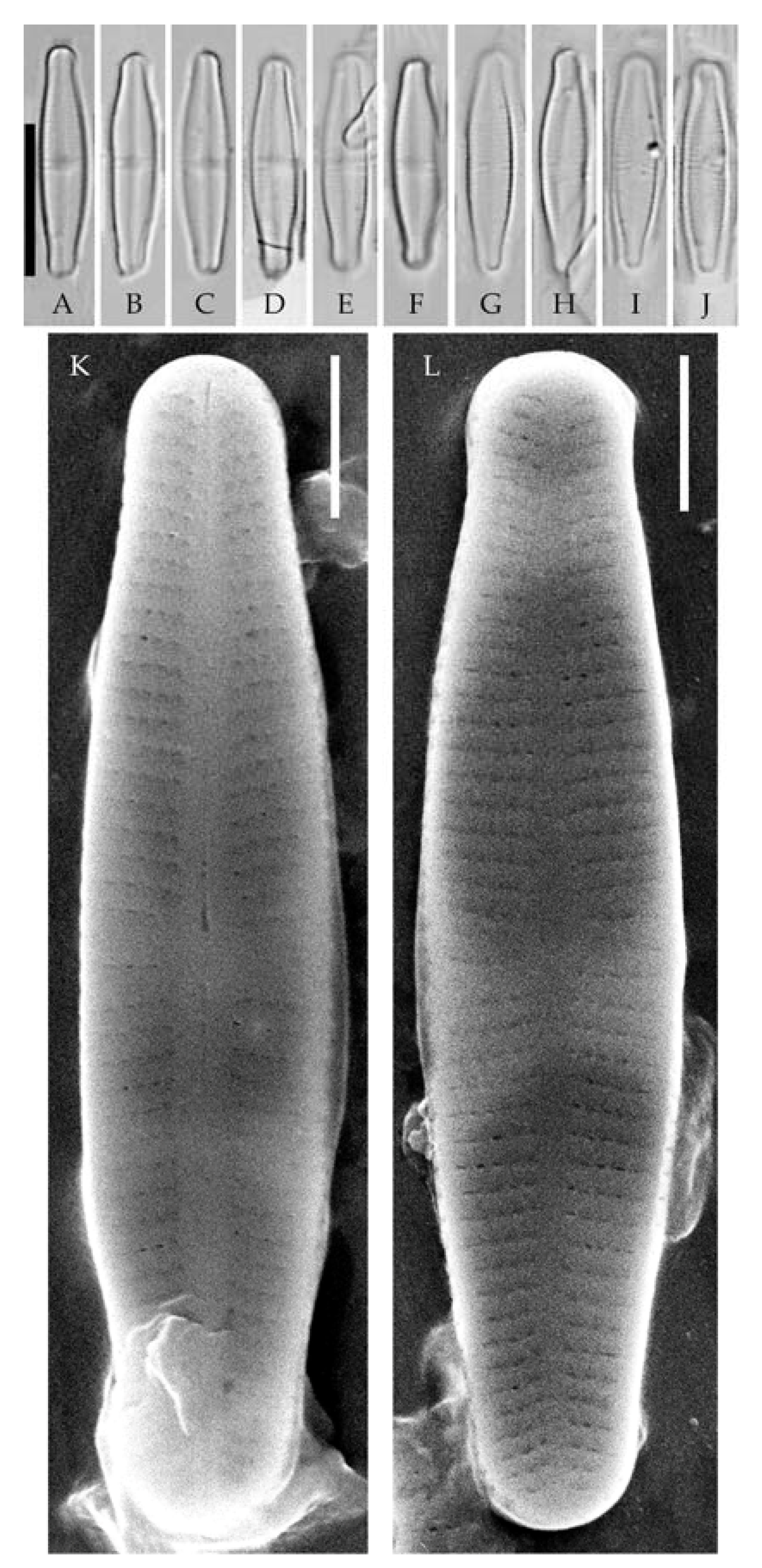
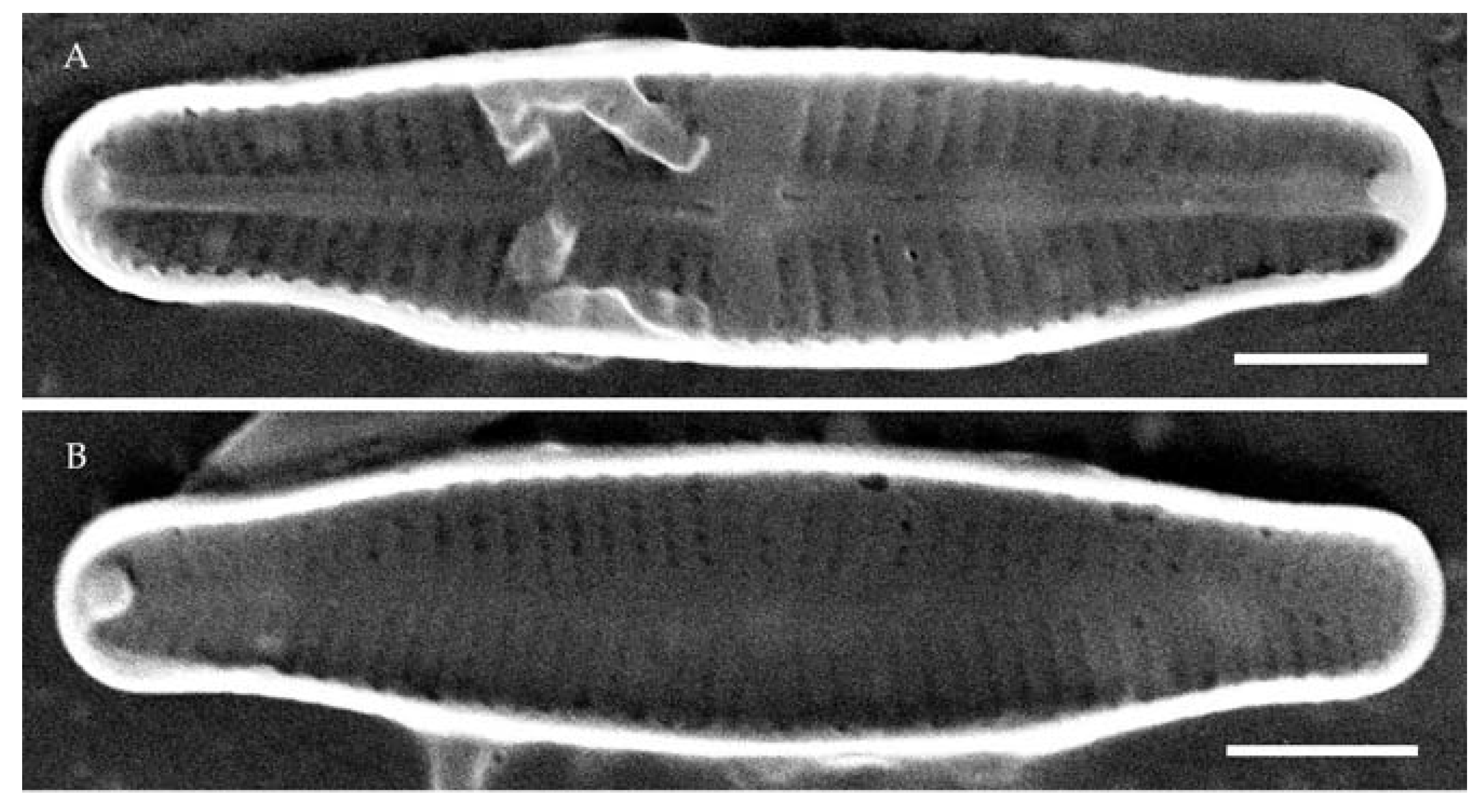
| Strain | Sampling Locality | Collection Date | Latitude | Longitude | pH | Water Temperature, °C | Sample Type | Slide No. |
|---|---|---|---|---|---|---|---|---|
| Achnanthidium baicalonanum sp. nov. B387 | Lake Frolikha | 26 July 2012 | 55°26′15.2″ N | 110°01′16.1″ E | N/A | 18.2 | benthos | B387 |
| Achnanthidium baicalonanum sp. nov. B397 | puddle by the road on the way from Lake Ilchir | 7 July 2014 | 51°56′49.3″ N | 100°49′34.8″ E | 7.66 | 11.6 | benthos | B397 |
| Achnanthidium baicalonanum sp. nov. B398 | puddle by the road on the way from Lake Ilchir | 7 July 2014 | 51°56′49.3″ N | 100°49′34.8″ E | 7.66 | 11.6 | benthos | B398 |
| Achnanthidium cf. minutissimum B443 | Lake Nizhnee Vesy | 19 July 2014 | 55°27′29.9″ N | 110°16′08.2″ E | 5.02 | 13.9 | plankton | B443 |
| Achnanthidium angustum sp. nov. B522 | Ehe-Ugui river | 5 June 2014 | 51°41′66″ N | 101°40′547″ E | 7.90 | 9.5 | epilithic periphyton | B522 (01313) |
| Achnanthidium angustum sp. nov. B543 | Ehe-Ugui river | 5 June 2014 | 51°41′66″ N | 101°40′547″ E | 7.90 | 9.5 | epilithic periphyton | B543 (01334) |
| Achnanthidium obscurum sp. nov. B722 | Levaya Frolikha river | 21 July 2014 | 55°22′57.2″ N | 110°07′43.7″ E | N/A | 12.0 | sand near the river bank with detritus | B722 (01512) |
| Achnanthidium obscurum sp. nov. B723 | Levaya Frolikha river | 21 July 2014 | 55°22′57.2″ N | 110°07′43.7″ E | N/A | 12.0 | sand near the river bank with detritus | B723 (01513) |
| Achnanthidium baicalonanum sp. nov. | A. straubianum | A. nanum | A. atomoides | A. duriense | |
|---|---|---|---|---|---|
| Valve shape | linear-elliptic to elliptic | elliptic | linear to linear-elliptic | linear-elliptic | elliptic to linear-elliptic |
| Valve apices | broadly rounded | broadly rounded | broadly rounded to slightly subrostrate | broadly rounded | broadly rounded to slightly protracted |
| Valve length, μm | 5.8–8.7 | 6.5–8.5 | 6.4–10.9 | 4.3–11.6 | 5.0–9.7 |
| Valve width, μm | 2.1–3.4 | 2.6–3.7 | 1.9–2.8 | 2.2–3.2 | 2.0–2.7 |
| Raphe valve | |||||
| Distal raphe ends | straight or slightly bent to one side | straight | straight | more or less straight | straight or slightly bent to one side |
| Axial area | narrow linear | narrow lanceolate | narrow linear | narrow linear | narrow linear |
| Central area | indistinct and asymmetrical | almost absent | indistinct | bowtie-shaped | indistinct |
| Striae pattern | radiate | weakly radiate to parallel | radiate | radiate and curved | almost parallel to weakly radiate |
| Striae density in 10 μm | 27–39 | 27–30 | 28–36 | 25–32 | 35 |
| Areolae | rounded to transapically elongated | rounded to elongated to rectangular | rounded to elongated | rounded | rounded or square |
| Rapheless valve | |||||
| Axial area | narrow linear | narrow lanceolate | narrow linear | narrow linear | narrow linear |
| Central area | absent | almost absent | indistinct | absent | indistinct |
| Striae pattern | weakly radiate | weakly radiate to parallel | parallel to weakly radiate | parallel | almost parallel to weakly radiate |
| Striae density in 10 μm | 31–35 | 27–30 | 28–36 | 23–28 | 35 |
| Areolae | rounded to transapically elongated | rounded to elongated to rectangular | rounded to elongated | rounded | rounded or square, sometimes slit-like |
| Source | this study | [24] | [8] | [15] | [8] |
| Achnanthidium obscurum sp. nov. | A. lineare | A. minutissimum | A. maritimo-antarcticum | A. indistinctum | |
|---|---|---|---|---|---|
| Valve shape | linear to linear-lanceolate | linear to narrow lanceolate | linear-lanceolate | linear-lanceolate | narrow lanceolate |
| Valve apices | slightly protracted and rounded | broadly rounded | subrostrate to subcapitate | rostrate to subcapitate | rostrate |
| Valve length, μm | 10.1–12.0 | 9.0–13.5 | 8.9–18.5 | 12–15 | 8.5–13.0 |
| Valve width, μm | 2.5–3.3 | 2.2–2.8 | 2.5–3.0 | 2.3–2.7 | 1.8–2.2 |
| Raphe valve | |||||
| Distal raphe ends | straight, teardrop-shaped | long, almost straight | straight or slightly bent | straight, short | straight, short |
| Axial area | narrow linear | narrow linear | narrow linear | linear | narrow linear |
| Central area | asymmetrical, more or less rectangular | broad rectangular fascia | variable | irregular | small, indistinct |
| Striae pattern | radiate | radiate | radiate | weakly radiate | radiate |
| Striae density in 10 μm | 31–33 | 28–32 | 28–32 | 30–33 | 36 |
| Areolae | rounded to transapically elongated | rounded to slit-like | rounded to transapically elongated | rounded to square to rectangular | rounded to elongated to rectangular |
| Rapheless valve | |||||
| Axial area | narrow linear | narrow lanceolate | narrow linear | narrow linear | narrow linear |
| Central area | absent | almost absent to small elliptical | absent | weakly elliptical | almost absent |
| Striae pattern | radiate | radiate | radiate | weakly radiate | radiate |
| Striae density in 10 μm | 31–36 | 28–32 | 28–32 | 30–32 | 30–35 |
| Areolae | rounded to transapically elongated | rounded to slit-like | rounded to transapically elongated | rounded to square to rectangular | rounded to elongated to rectangular |
| Source | this study | [25] | [23] | [20] | [20] |
| Achnanthidium angustum sp. nov. | A. lineare | A. minutissimum | A. jackii | A. indistinctum | |
|---|---|---|---|---|---|
| Valve shape | narrow linear-lanceolate | linear to narrow lanceolate | linear-lanceolate | linear-lanceolate to weakly lanceolate | narrow lanceolate |
| Valve apices | protracted and rounded | broadly rounded | subrostrate to subcapitate | rostrate | rostrate |
| Valve length, μm | 12.3–14.8 | 9.0–13.5 | 8.9–18.5 | 8.0–17.0 | 8.5–13.0 |
| Valve width, μm | 2.4–3.1 | 2.2–2.8 | 2.5–3.0 | 3.0–3.9 | 1.8–2.2 |
| Raphe valve | |||||
| Distal raphe ends | straight, long | long, almost straight | straight or slightly bent | straight, short | straight, short |
| Axial area | narrow linear | narrow linear | narrow linear | narrow linear | narrow linear |
| Central area | indistinct | broad rectangular fascia | variable | asymmetrical fascia | small, indistinct |
| Striae pattern | radiate | radiate | radiate | weakly radiate to almost parallel | radiate |
| Striae density in 10 μm | 31–37 | 28–32 | 28–32 | 25–30 | 36 |
| Areolae | rounded to transapically elongated | rounded to slit-like | rounded to transapically elongated | rounded to slit-like | rounded to elongated to rectangular |
| Rapheless valve | |||||
| Axial area | narrow lanceolate | narrow lanceolate | narrow linear | narrow linear-lanceolate | narrow linear |
| Central area | absent | almost absent to small elliptical | absent | absent to small fascia to asymmetrical | almost absent |
| Striae pattern | radiate | radiate | radiate | weakly radiate to almost parallel | radiate |
| Striae density in 10 μm | 31–35 | 28–32 | 28–32 | 28–30 | 30–35 |
| Areolae | rounded to transapically elongated | rounded to slit-like | rounded to transapically elongated | rounded to slit-like | rounded to elongated to rectangular |
| Source | this study | [25] | [23] | [36] | [20] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tseplik, N.; Genkal, S.; Maltsev, Y.; Kuznetsova, I.; Kulikovskiy, M. Molecular Investigation of the Achnanthidium minutissimum Complex (Bacillariophyceae) from the Transbaikal Region with the Description of Three New Species. Water 2023, 15, 3379. https://doi.org/10.3390/w15193379
Tseplik N, Genkal S, Maltsev Y, Kuznetsova I, Kulikovskiy M. Molecular Investigation of the Achnanthidium minutissimum Complex (Bacillariophyceae) from the Transbaikal Region with the Description of Three New Species. Water. 2023; 15(19):3379. https://doi.org/10.3390/w15193379
Chicago/Turabian StyleTseplik, Natalia, Sergey Genkal, Yevhen Maltsev, Irina Kuznetsova, and Maxim Kulikovskiy. 2023. "Molecular Investigation of the Achnanthidium minutissimum Complex (Bacillariophyceae) from the Transbaikal Region with the Description of Three New Species" Water 15, no. 19: 3379. https://doi.org/10.3390/w15193379
APA StyleTseplik, N., Genkal, S., Maltsev, Y., Kuznetsova, I., & Kulikovskiy, M. (2023). Molecular Investigation of the Achnanthidium minutissimum Complex (Bacillariophyceae) from the Transbaikal Region with the Description of Three New Species. Water, 15(19), 3379. https://doi.org/10.3390/w15193379







