A Mini-Review of Antibiotic Resistance Drivers in Urban Wastewater Treatment Plants: Environmental Concentrations, Mechanism and Perspectives
Abstract
1. Introduction
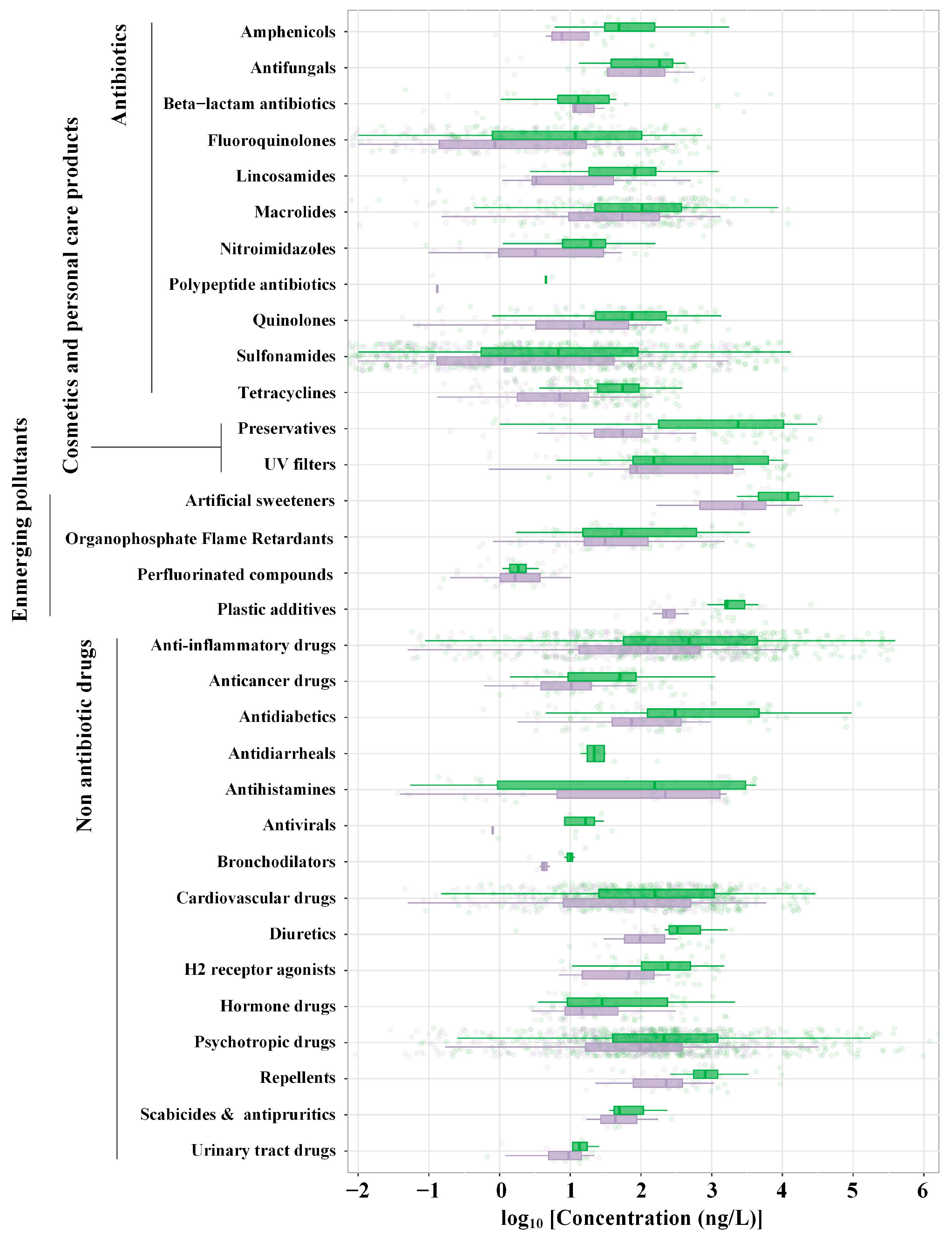
2. Antibiotics
3. Heavy Metals
4. Disinfectants, Cosmetics and Personal Care Products
5. Non-Antibiotic Drugs
6. Microplastics
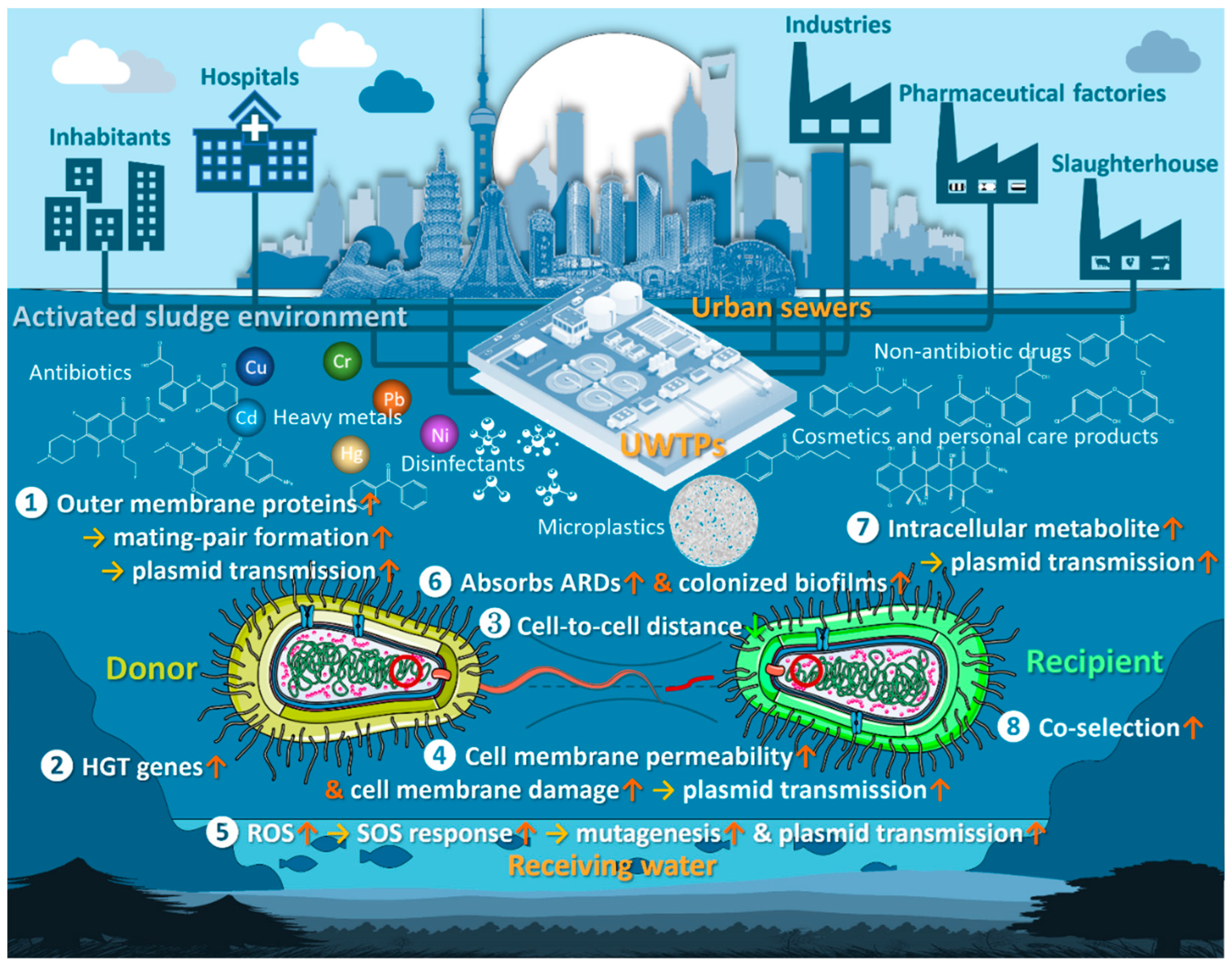
7. Mechanisms for Promoting Antibiotic Resistance in Bacteria
8. Current Bottleneck and Perspectives
- To conduct ARD exposure experiments at actual concentration levels consistent with the activated sludge environment.
- To evaluate the effects of long-term exposure to ARDs on strains.
- To reveal the mechanisms of joint antibiotic resistance promotion and the joint effects of ARDs in combination (synergistic, additive, antagonistic).
- To investigate the mechanisms of promoting antibiotic resistance to activated sludge microbiomes.
- To develop a high-throughput screening technique and assessment system for finding the mechanism promoting antibiotic resistance in numerous unassessed pollutants.
- To establish a quantitative risk assessment system for promoting antibiotic resistance to contaminants.
- To develop ARG-targeted control technology for the activated sludge microbiome in UWTPs.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Zieliński, W.; Korzeniewska, E.; Harnisz, M.; Drzymała, J.; Felis, E.; Bajkacz, S. Wastewater Treatment Plants as a Reservoir of Integrase and Antibiotic Resistance Genes – An Epidemiological Threat to Workers and Environment. Environ. Int. 2021, 156, 106641. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Wang, B.; Huang, K.; Yin, J.; Ren, X.; Wang, Z.; Zhang, X.-X. Correlations among Antibiotic Resistance Genes, Mobile Genetic Elements and Microbial Communities in Municipal Sewage Treatment Plants Revealed by High-Throughput Sequencing. Int. J. Environ. Res. Public Health 2023, 20, 3593. [Google Scholar] [CrossRef]
- Berglund, F.; Ebmeyer, S.; Kristiansson, E.; Larsson, D.G.J. Evidence for Wastewaters as Environments Where Mobile Antibiotic Resistance Genes Emerge. Commun. Biol. 2023, 6, 321. [Google Scholar] [CrossRef] [PubMed]
- Schages, L.; Lucassen, R.; Wichern, F.; Kalscheuer, R.; Bockmühl, D. The Household Resistome: Frequency of β-Lactamases, Class 1 Integrons, and Antibiotic-Resistant Bacteria in the Domestic Environment and Their Reduction during Automated Dishwashing and Laundering. Appl. Environ. Microbiol. 2020, 86, e02062-20. [Google Scholar] [CrossRef]
- Raymond, F.; Ouameur, A.A.; Déraspe, M.; Iqbal, N.; Gingras, H.; Dridi, B.; Leprohon, P.; Plante, P.-L.; Giroux, R.; Bérubé, È.; et al. The Initial State of the Human Gut Microbiome Determines Its Reshaping by Antibiotics. ISME J. 2016, 10, 707–720. [Google Scholar] [CrossRef]
- Hu, Y.; Yang, X.; Qin, J.; Lu, N.; Cheng, G.; Wu, N.; Pan, Y.; Li, J.; Zhu, L.; Wang, X.; et al. Metagenome-Wide Analysis of Antibiotic Resistance Genes in a Large Cohort of Human Gut Microbiota. Nat. Commun. 2013, 4, 2151. [Google Scholar] [CrossRef]
- Sommer, M.O.A.; Dantas, G.; Church, G.M. Functional Characterization of the Antibiotic Resistance Reservoir in the Human Microflora. Science 2009, 325, 1128–1131. [Google Scholar] [CrossRef]
- Khan, F.A.; Söderquist, B.; Jass, J. Prevalence and Diversity of Antibiotic Resistance Genes in Swedish Aquatic Environments Impacted by Household and Hospital Wastewater. Front. Microbiol. 2019, 10, 688. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, K.; Chen, W.; Chen, J.; Zheng, J.; Liu, C.; Cheng, L.; Zhou, W.; Shen, H.; Cao, X. Epidemiological Characteristics of Carbapenem-Resistant Enterobacteriaceae Collected from 17 Hospitals in Nanjing District of China. Antimicrob. Resist. Infect. Control 2020, 9, 15. [Google Scholar] [CrossRef]
- Chng, K.R.; Li, C.; Bertrand, D.; Ng, A.H.Q.; Kwah, J.S.; Low, H.M.; Tong, C.; Natrajan, M.; Zhang, M.H.; Xu, L.; et al. Cartography of Opportunistic Pathogens and Antibiotic Resistance Genes in a Tertiary Hospital Environment. Nat. Med. 2020, 26, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson-Palme, J.; Milakovic, M.; Švecová, H.; Ganjto, M.; Jonsson, V.; Grabic, R.; Udikovic-Kolic, N. Industrial Wastewater Treatment Plant Enriches Antibiotic Resistance Genes and Alters the Structure of Microbial Communities. Water Res. 2019, 162, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Calvo-Bado, L.; Murray, A.K.; Amos, G.C.A.; Hawkey, P.M.; Wellington, E.M.; Gaze, W.H. Novel Clinically Relevant Antibiotic Resistance Genes Associated with Sewage Sludge and Industrial Waste Streams Revealed by Functional Metagenomic Screening. Environ. Int. 2019, 132, 105120. [Google Scholar] [CrossRef] [PubMed]
- Guo, N.; Wang, Y.; Tong, T.; Wang, S. The Fate of Antibiotic Resistance Genes and Their Potential Hosts during Bio-Electrochemical Treatment of High-Salinity Pharmaceutical Wastewater. Water Res. 2018, 133, 79–86. [Google Scholar] [CrossRef]
- Aydin, S.; Ince, B.; Ince, O. Development of Antibiotic Resistance Genes in Microbial Communities during Long-Term Operation of Anaerobic Reactors in the Treatment of Pharmaceutical Wastewater. Water Res. 2015, 83, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Wang, J.; Li, X.; Cui, F. Insight into Effect of High-Level Cephalexin on Fate and Driver Mechanism of Antibiotics Resistance Genes in Antibiotic Wastewater Treatment System. Ecotoxicol. Environ. Saf. 2020, 201, 110739. [Google Scholar] [CrossRef] [PubMed]
- Vidovic, N.; Vidovic, S. Antimicrobial Resistance and Food Animals: Influence of Livestock Environment on the Emergence and Dissemination of Antimicrobial Resistance. Antibiotics 2020, 9, 52. [Google Scholar] [CrossRef]
- Li, N.; Liu, C.; Zhang, Z.; Li, H.; Song, T.; Liang, T.; Li, B.; Li, L.; Feng, S.; Su, Q.; et al. Research and Technological Advances Regarding the Study of the Spread of Antimicrobial Resistance Genes and Antimicrobial-Resistant Bacteria Related to Animal Husbandry. Int. J. Environ. Res. Public. Health 2019, 16, 4896. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, Y.; Cao, J.; Bi, Y.; Lv, N.; Liu, F.; Liang, S.; Shi, Y.; Jiao, X.; Gao, G.F.; et al. Antibiotic Resistance Gene Reservoir in Live Poultry Markets. J. Infect. 2019, 78, 445–453. [Google Scholar] [CrossRef]
- Wu, Z.; Che, Y.; Dang, C.; Zhang, M.; Zhang, X.; Sun, Y.; Li, X.; Zhang, T.; Xia, Y. Nanopore-Based Long-Read Metagenomics Uncover the Resistome Intrusion by Antibiotic Resistant Bacteria from Treated Wastewater in Receiving Water Body. Water Res. 2022, 226, 119282. [Google Scholar] [CrossRef]
- Xie, J.; Jin, L.; Wu, D.; Pruden, A.; Li, X. Inhalable Antibiotic Resistome from Wastewater Treatment Plants to Urban Areas: Bacterial Hosts, Dissemination Risks, and Source Contributions. Environ. Sci. Technol. 2022, 56, 7040–7051. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Cui, E.; Zuo, Y.; Cheng, W.; Chen, H. Fate of Antibiotic and Metal Resistance Genes during Two-Phase Anaerobic Digestion of Residue Sludge Revealed by Metagenomic Approach. Environ. Sci. Pollut. Res. 2018, 25, 13956–13963. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Deng, Y.; Ma, L.; Wang, Y.; Chan, L.Y.L.; Zhang, T. Exploration of the Antibiotic Resistome in a Wastewater Treatment Plant by a Nine-Year Longitudinal Metagenomic Study. Environ. Int. 2019, 133, 105270. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Yang, Y.; Deng, Y.; Huang, Y.; Li, L.; Chan, L.Y.L.; Zhang, T. An Assessment of Resistome and Mobilome in Wastewater Treatment Plants through Temporal and Spatial Metagenomic Analysis. Water Res. 2022, 209, 117885. [Google Scholar] [CrossRef]
- Brandis, G.; Larsson, J.; Elf, J. Antibiotic Perseverance Increases the Risk of Resistance Development. Proc. Natl. Acad. Sci. USA 2023, 120, e2216216120. [Google Scholar] [CrossRef]
- Ju, F.; Beck, K.; Yin, X.; Maccagnan, A.; McArdell, C.S.; Singer, H.P.; Johnson, D.R.; Zhang, T.; Bürgmann, H. Wastewater Treatment Plant Resistomes Are Shaped by Bacterial Composition, Genetic Exchange, and Upregulated Expression in the Effluent Microbiomes. ISME J. 2019, 13, 346–360. [Google Scholar] [CrossRef]
- Che, Y.; Xu, X.; Yang, Y.; Břinda, K.; Hanage, W.; Yang, C.; Zhang, T. High-Resolution Genomic Surveillance Elucidates a Multilayered Hierarchical Transfer of Resistance between WWTP- and Human/Animal-Associated Bacteria. Microbiome 2022, 10, 16. [Google Scholar] [CrossRef]
- Dai, D.; Brown, C.; Bürgmann, H.; Larsson, D.G.J.; Nambi, I.; Zhang, T.; Flach, C.-F.; Pruden, A.; Vikesland, P.J. Long-Read Metagenomic Sequencing Reveals Shifts in Associations of Antibiotic Resistance Genes with Mobile Genetic Elements from Sewage to Activated Sludge. Microbiome 2022, 10, 20. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Larsson, D.G.J. Concentrations of Antibiotics Predicted to Select for Resistant Bacteria: Proposed Limits for Environmental Regulation. Environ. Int. 2016, 86, 140–149. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Wright, M.S.; Stepanauskas, R.; McArthur, J.V. Co-Selection of Antibiotic and Metal Resistance. Trends Microbiol. 2006, 14, 176–182. [Google Scholar] [CrossRef]
- Xu, Y.; Xu, J.; Mao, D.; Luo, Y. Effect of the Selective Pressure of Sub-Lethal Level of Heavy Metals on the Fate and Distribution of ARGs in the Catchment Scale. Environ. Pollut. 2017, 220, 900–908. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Wu, S.; Men, Y. Exposure to Environmental Levels of Pesticides Stimulates and Diversifies Evolution in Escherichia coli toward Higher Antibiotic Resistance. Environ. Sci. Technol. 2020, 54, 8770–8778. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; He, Y.; Sun, J.; Li, J.; Bai, J.; Zhang, C. Chronic Exposure to an Environmentally Relevant Triclosan Concentration Induces Persistent Triclosan Resistance but Reversible Antibiotic Tolerance in Escherichia coli. Environ. Sci. Technol. 2019, 53, 3277–3286. [Google Scholar] [CrossRef]
- Maier, L.; Pruteanu, M.; Kuhn, M.; Zeller, G.; Telzerow, A.; Anderson, E.E.; Brochado, A.R.; Fernandez, K.C.; Dose, H.; Mori, H.; et al. Extensive Impact of Non-Antibiotic Drugs on Human Gut Bacteria. Nature 2018, 555, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Stedtfeld, R.D.; Guo, X.; Bhalsod, G.D.; Jeon, S.; Tiedje, J.M.; Li, H.; Zhang, W. Pharmaceutical Exposure Changed Antibiotic Resistance Genes and Bacterial Communities in Soil-Surface- and Overhead-Irrigated Greenhouse Lettuce. Environ. Int. 2019, 131, 105031. [Google Scholar] [CrossRef]
- Xia, J.; Sun, H.; Zhang, X.; Zhang, T.; Ren, H.; Ye, L. Aromatic Compounds Lead to Increased Abundance of Antibiotic Resistance Genes in Wastewater Treatment Bioreactors. Water Res. 2019, 166, 115073. [Google Scholar] [CrossRef]
- Imran, M.; Das, K.R.; Naik, M.M. Co-Selection of Multi-Antibiotic Resistance in Bacterial Pathogens in Metal and Microplastic Contaminated Environments: An Emerging Health Threat. Chemosphere 2019, 215, 846–857. [Google Scholar] [CrossRef]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular Mechanisms of Antibiotic Resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Durão, P.; Balbontín, R.; Gordo, I. Evolutionary Mechanisms Shaping the Maintenance of Antibiotic Resistance. Trends Microbiol. 2018, 26, 677–691. [Google Scholar] [CrossRef]
- Zhang, Q.-Q.; Ying, G.-G.; Pan, C.-G.; Liu, Y.-S.; Zhao, J.-L. Comprehensive Evaluation of Antibiotics Emission and Fate in the River Basins of China: Source Analysis, Multimedia Modeling, and Linkage to Bacterial Resistance. Environ. Sci. Technol. 2015, 49, 6772–6782. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, P.; Yang, Q. Occurrence and Diversity of Antibiotic Resistance in Untreated Hospital Wastewater. Sci. Total Environ. 2018, 621, 990–999. [Google Scholar] [CrossRef] [PubMed]
- Petrin, S.; Patuzzi, I.; Di Cesare, A.; Tiengo, A.; Sette, G.; Biancotto, G.; Corno, G.; Drigo, M.; Losasso, C.; Cibin, V. Evaluation and Quantification of Antimicrobial Residues and Antimicrobial Resistance Genes in Two Italian Swine Farms. Environ. Pollut. 2019, 255, 113183. [Google Scholar] [CrossRef] [PubMed]
- Sundararaman, S.; Saravanane, R. Effect of Loading Rate and HRT on the Removal of Cephalosporin and Their Intermediates during the Operation of a Membrane Bioreactor Treating Pharmaceutical Wastewater. Water Sci. Technol. 2010, 61, 1907–1914. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ternes, T.A.; Bonerz, M.; Herrmann, N.; Teiser, B.; Andersen, H.R. Irrigation of Treated Wastewater in Braunschweig, Germany: An Option to Remove Pharmaceuticals and Musk Fragrances. Chemosphere 2007, 66, 894–904. [Google Scholar] [CrossRef]
- Csanády, M.; Deák, Z. Trickling Filter Experiment for Purification of Antibiotic-Containing Hospital Sewage. Water Res. 1972, 6, 1541–1547. [Google Scholar] [CrossRef]
- Peng, X.; Tan, J.; Tang, C.; Yu, Y.; Wang, Z. Multiresidue Determination of Fluoroquinolone, Sulfonamide, Trimethoprim, and Chloramphenicol Antibiotics in Urban Waters in China. Environ. Toxicol. Chem. 2008, 27, 73. [Google Scholar] [CrossRef]
- Watkinson, A.J.; Murby, E.J.; Costanzo, S.D. Removal of Antibiotics in Conventional and Advanced Wastewater Treatment: Implications for Environmental Discharge and Wastewater Recycling. Water Res. 2007, 41, 4164–4176. [Google Scholar] [CrossRef]
- Lindberg, R.H.; Wennberg, P.; Johansson, M.I.; Tysklind, M.; Andersson, B.A.V. Screening of Human Antibiotic Substances and Determination of Weekly Mass Flows in Five Sewage Treatment Plants in Sweden. Environ. Sci. Technol. 2005, 39, 3421–3429. [Google Scholar] [CrossRef]
- Auguet, O.; Pijuan, M.; Borrego, C.M.; Rodriguez-Mozaz, S.; Triadó-Margarit, X.; Giustina, S.V.D.; Gutierrez, O. Sewers as Potential Reservoirs of Antibiotic Resistance. Sci. Total Environ. 2017, 605–606, 1047–1054. [Google Scholar] [CrossRef]
- Batt, A.L.; Kim, S.; Aga, D.S. Comparison of the Occurrence of Antibiotics in Four Full-Scale Wastewater Treatment Plants with Varying Designs and Operations. Chemosphere 2007, 68, 428–435. [Google Scholar] [CrossRef]
- Lin, A.Y.-C.; Yu, T.-H.; Lateef, S.K. Removal of Pharmaceuticals in Secondary Wastewater Treatment Processes in Taiwan. J. Hazard. Mater. 2009, 167, 1163–1169. [Google Scholar] [CrossRef]
- Kim, M.; Guerra, P.; Shah, A.; Parsa, M.; Alaee, M.; Smyth, S.A. Removal of Pharmaceuticals and Personal Care Products in a Membrane Bioreactor Wastewater Treatment Plant. Water Sci. Technol. 2014, 69, 2221–2229. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-Q.; Lam, J.C.W.; Li, W.-W.; Yu, H.-Q.; Lam, P.K.S. Spatial Distribution and Removal Performance of Pharmaceuticals in Municipal Wastewater Treatment Plants in China. Sci. Total Environ. 2017, 586, 1162–1169. [Google Scholar] [CrossRef]
- Ofrydopoulou, A.; Nannou, C.; Evgenidou, E.; Christodoulou, A.; Lambropoulou, D. Assessment of a Wide Array of Organic Micropollutants of Emerging Concern in Wastewater Treatment Plants in Greece: Occurrence, Removals, Mass Loading and Potential Risks. Sci. Total Environ. 2022, 802, 149860. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Morales, D.; Masís-Mora, M.; Montiel-Mora, J.R.; Cambronero-Heinrichs, J.C.; Briceño-Guevara, S.; Rojas-Sánchez, C.E.; Méndez-Rivera, M.; Arias-Mora, V.; Tormo-Budowski, R.; Brenes-Alfaro, L.; et al. Occurrence of Pharmaceuticals, Hazard Assessment and Ecotoxicological Evaluation of Wastewater Treatment Plants in Costa Rica. Sci. Total Environ. 2020, 746, 141200. [Google Scholar] [CrossRef] [PubMed]
- Vaudreuil, M.-A.; Vo Duy, S.; Munoz, G.; Sauvé, S. Pharmaceutical Pollution of Hospital Effluents and Municipal Wastewaters of Eastern Canada. Sci. Total Environ. 2022, 846, 157353. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Hu, E.; Liu, S.; Wen, L.; Yang, F.; Li, M. Spatial Distribution and Risk Assessment of Certain Antibiotics in 51 Urban Wastewater Treatment Plants in the Transition Zone between North and South China. J. Hazard. Mater. 2022, 437, 129307. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ge, S.; Shao, P.; Wang, J.; Liu, Y.; Wei, W.; He, C.; Zhang, L. Occurrence and Removal Rate of Typical Pharmaceuticals and Personal Care Products (PPCPs) in an Urban Wastewater Treatment Plant in Beijing, China. Chemosphere 2023, 339, 139644. [Google Scholar] [CrossRef]
- Cairns, J.; Ruokolainen, L.; Hultman, J.; Tamminen, M.; Virta, M.; Hiltunen, T. Ecology Determines How Low Antibiotic Concentration Impacts Community Composition and Horizontal Transfer of Resistance Genes. Commun. Biol. 2018, 1, 35. [Google Scholar] [CrossRef]
- Chow, L.; Waldron, L.; Gillings, M.R. Potential Impacts of Aquatic Pollutants: Sub-Clinical Antibiotic Concentrations Induce Genome Changes and Promote Antibiotic Resistance. Front. Microbiol. 2015, 6, 803. [Google Scholar] [CrossRef]
- Gullberg, E.; Cao, S.; Berg, O.G.; Ilbäck, C.; Sandegren, L.; Hughes, D.; Andersson, D.I. Selection of Resistant Bacteria at Very Low Antibiotic Concentrations. PLoS Pathog. 2011, 7, e1002158. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Palomo, A.; Zhang, H.; Luan, X.; Liu, R.; Awad, M.; Smets, B.F.; Zhang, Y.; Yang, M. Minimum Influent Concentrations of Oxytetracycline, Streptomycin and Spiramycin in Selecting Antibiotic Resistance in Biofilm Type Wastewater Treatment Systems. Sci. Total Environ. 2020, 720, 137531. [Google Scholar] [CrossRef]
- Sanchez-Cid, C.; Guironnet, A.; Keuschnig, C.; Wiest, L.; Vulliet, E.; Vogel, T.M. Gentamicin at Sub-Inhibitory Concentrations Selects for Antibiotic Resistance in the Environment. ISME Commun. 2022, 2, 29. [Google Scholar] [CrossRef]
- Östman, M.; Lindberg, R.H.; Fick, J.; Björn, E.; Tysklind, M. Screening of Biocides, Metals and Antibiotics in Swedish Sewage Sludge and Wastewater. Water Res. 2017, 115, 318–328. [Google Scholar] [CrossRef]
- Xu, Y.-B.; Hou, M.-Y.; Li, Y.-F.; Huang, L.; Ruan, J.-J.; Zheng, L.; Qiao, Q.-X.; Du, Q.-P. Distribution of Tetracycline Resistance Genes and AmpC β-Lactamase Genes in Representative Non-Urban Sewage Plants and Correlations with Treatment Processes and Heavy Metals. Chemosphere 2017, 170, 274–281. [Google Scholar] [CrossRef]
- Cheng, X.; Wei, C.; Ke, X.; Pan, J.; Wei, G.; Chen, Y.; Wei, C.; Li, F.; Preis, S. Nationwide Review of Heavy Metals in Municipal Sludge Wastewater Treatment Plants in China: Sources, Composition, Accumulation and Risk Assessment. J. Hazard. Mater. 2022, 437, 129267. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wu, C.; Zhou, C.; Dong, L.; Liu, B.; Xing, D.; Yang, S.; Fan, J.; Feng, L.; Cao, G.; et al. Fate and Removal of Antibiotic Resistance Genes in Heavy Metals and Dye Co-Contaminated Wastewater Treatment System Amended with β-Cyclodextrin Functionalized Biochar. Sci. Total Environ. 2020, 723, 137991. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, A.Z.; Cen, T.; Li, X.; He, M.; Li, D.; Chen, J. Sub-Inhibitory Concentrations of Heavy Metals Facilitate the Horizontal Transfer of Plasmid-Mediated Antibiotic Resistance Genes in Water Environment. Environ. Pollut. 2018, 237, 74–82. [Google Scholar] [CrossRef]
- Li, X.; Gu, A.Z.; Zhang, Y.; Xie, B.; Li, D.; Chen, J. Sub-Lethal Concentrations of Heavy Metals Induce Antibiotic Resistance via Mutagenesis. J. Hazard. Mater. 2019, 369, 9–16. [Google Scholar] [CrossRef]
- Aendekerk, S.; Ghysels, B.; Cornelis, P.; Baysse, C. Characterization of a New Efflux Pump, MexGHI-OpmD, from Pseudomonas Aeruginosa That Confers Resistance to Vanadium. Microbiology 2002, 148, 2371–2381. [Google Scholar] [CrossRef]
- Hayashi, S.; Abe, M.; Kimoto, M.; Furukawa, S.; Nakazawa, T. The DsbA-DsbB Disulfide Bond Formation System of Burkholderia Cepacia Is Involved in the Production of Protease and Alkaline Phosphatase, Motility, Metal Resistance, and Multi-Drug Resistance. Microbiol. Immunol. 2000, 44, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Hicks, D.B.; Krulwich, T.A. The Purified Bacillus Subtilis Tetracycline Efflux Protein TetA(L) Reconstitutes Both Tetracycline–Cobalt/H+ and Na+(K+)/H+ Exchange. Proc. Natl. Acad. Sci. USA 1996, 93, 14446–14451. [Google Scholar] [CrossRef]
- Mata, M.T.; Baquero, F.; Pérez-Díaz, J.C. A Multidrug Efflux Transporter in Listeria Monocytogenes. FEMS Microbiol. Lett. 2000, 187, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Nishino, K.; Nikaido, E.; Yamaguchi, A. Regulation of Multidrug Efflux Systems Involved in Multidrug and Metal Resistance of Salmonella enterica Serovar Typhimurium. J. Bacteriol. 2007, 189, 9066–9075. [Google Scholar] [CrossRef]
- Li, L.-G.; Xia, Y.; Zhang, T. Co-Occurrence of Antibiotic and Metal Resistance Genes Revealed in Complete Genome Collection. ISME J. 2017, 11, 651–662. [Google Scholar] [CrossRef]
- Perron, K.; Caille, O.; Rossier, C.; Delden, C.; Dumas, J.-L.; Köhler, T. CzcR-CzcS, a Two-Component System Involved in Heavy Metal and Carbapenem Resistance in Pseudomonas aeruginosa. J. Biol. Chem. 2004, 279, 8761–8768. [Google Scholar] [CrossRef] [PubMed]
- Tumah, H.N. Bacterial Biocide Resistance. J. Chemother. 2009, 21, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.; Chakraborty, R.; Mandal, S.M. Biocides and Health-Care Agents Are More than Just Antibiotics: Inducing Cross to Co-Resistance in Microbes. Ecotoxicol. Environ. Saf. 2019, 174, 601–610. [Google Scholar] [CrossRef]
- Chen, J.; Liu, Y.-S.; Deng, W.-J.; Ying, G.-G. Removal of Steroid Hormones and Biocides from Rural Wastewater by an Integrated Constructed Wetland. Sci. Total Environ. 2019, 660, 358–365. [Google Scholar] [CrossRef]
- Juksu, K.; Zhao, J.-L.; Liu, Y.-S.; Yao, L.; Sarin, C.; Sreesai, S.; Klomjek, P.; Jiang, Y.-X.; Ying, G.-G. Occurrence, Fate and Risk Assessment of Biocides in Wastewater Treatment Plants and Aquatic Environments in Thailand. Sci. Total Environ. 2019, 690, 1110–1119. [Google Scholar] [CrossRef]
- Mann, B.C.; Bezuidenhout, J.J.; Bezuidenhout, C.C. Biocide Resistant and Antibiotic Cross-Resistant Potential Pathogens from Sewage and River Water from a Wastewater Treatment Facility in the North-West, Potchefstroom, South Africa. Water Sci. Technol. 2019, 80, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Tran, N.H.; Reinhard, M.; Khan, E.; Chen, H.; Nguyen, V.T.; Li, Y.; Goh, S.G.; Nguyen, Q.B.; Saeidi, N.; Gin, K.Y.-H. Emerging Contaminants in Wastewater, Stormwater Runoff, and Surface Water: Application as Chemical Markers for Diffuse Sources. Sci. Total Environ. 2019, 676, 252–267. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, B. Stochastic Dynamic Mass Spectrometric Quantitative and Structural Analyses of Pharmaceutics and Biocides in Biota and Sewage Sludge. Int. J. Mol. Sci. 2023, 24, 6306. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.; Wu, Z.; Fu, J. Environmental Issues Caused by High-Dose Disinfection Need Urgent Attention. Environ. Health 2023. [Google Scholar] [CrossRef]
- Chen, Z.; Guo, J.; Jiang, Y.; Shao, Y. High Concentration and High Dose of Disinfectants and Antibiotics Used during the COVID-19 Pandemic Threaten Human Health. Environ. Sci. Eur. 2021, 33, 11. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.C.; Oladeinde, A.; Kieran, T.J.; Finger, J.W.; Bayona-Vásquez, N.J.; Cartee, J.C.; Beasley, J.C.; Seaman, J.C.; McArthur, J.V.; Rhodes, O.E.; et al. Co-occurrence of Antibiotic, Biocide, and Heavy Metal Resistance Genes in Bacteria from Metal and Radionuclide Contaminated Soils at the Savannah River Site. Microb. Biotechnol. 2020, 13, 1179–1200. [Google Scholar] [CrossRef]
- Jutkina, J.; Marathe, N.P.; Flach, C.-F.; Larsson, D.G.J. Antibiotics and Common Antibacterial Biocides Stimulate Horizontal Transfer of Resistance at Low Concentrations. Sci. Total Environ. 2018, 616–617, 172–178. [Google Scholar] [CrossRef]
- Lin, W.; Zhang, M.; Zhang, S.; Yu, X. Can Chlorination Co-Select Antibiotic-Resistance Genes? Chemosphere 2016, 156, 412–419. [Google Scholar] [CrossRef]
- Jin, M.; Liu, L.; Wang, D.; Yang, D.; Liu, W.; Yin, J.; Yang, Z.; Wang, H.; Qiu, Z.; Shen, Z.; et al. Chlorine Disinfection Promotes the Exchange of Antibiotic Resistance Genes across Bacterial Genera by Natural Transformation. ISME J. 2020, 14, 1847–1856. [Google Scholar] [CrossRef]
- Lu, J.; Guo, J. Disinfection Spreads Antimicrobial Resistance. Science 2021, 371, 474. [Google Scholar] [CrossRef]
- Rawson, T.M.; Ming, D.; Ahmad, R.; Moore, L.S.P.; Holmes, A.H. Antimicrobial Use, Drug-Resistant Infections and COVID-19. Nat. Rev. Microbiol. 2020, 18, 409–410. [Google Scholar] [CrossRef]
- Carey, D.E.; McNamara, P.J. The Impact of Triclosan on the Spread of Antibiotic Resistance in the Environment. Front. Microbiol. 2015, 5, 780. [Google Scholar] [CrossRef] [PubMed]
- McMurry, L.M.; McDermott, P.F.; Levy, S.B. Genetic Evidence That InhA of Mycobacterium Smegmatis Is a Target for Triclosan. Antimicrob. Agents Chemother. 1999, 43, 711–713. [Google Scholar] [CrossRef] [PubMed]
- Heath, R.J.; Rubin, J.R.; Holland, D.R.; Zhang, E.; Snow, M.E.; Rock, C.O. Mechanism of Triclosan Inhibition of Bacterial Fatty Acid Synthesis. J. Biol. Chem. 1999, 274, 11110–11114. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, H.P. Triclosan: A Widely Used Biocide and Its Link to Antibiotics. FEMS Microbiol. Lett. 2001, 202, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Mcmurry, L.M.; Oethinger, M.; Levy, S.B. Overexpression of MarA, SoxS, or AcrAB Produces Resistance to Triclosan in Laboratory and Clinical Strains of Escherichia coli. FEMS Microbiol. Lett. 1998, 166, 305–309. [Google Scholar] [CrossRef]
- Chuanchuen, R.; Beinlich, K.; Hoang, T.T.; Becher, A.; Karkhoff-Schweizer, R.R.; Schweizer, H.P. Cross-Resistance between Triclosan and Antibiotics in Pseudomonas aeruginosa Is Mediated by Multidrug Efflux Pumps: Exposure of a Susceptible Mutant Strain to Triclosan Selects NfxB Mutants Overexpressing MexCD-OprJ. Antimicrob. Agents Chemother. 2001, 45, 428–432. [Google Scholar] [CrossRef]
- Lu, J.; Yu, Z.; Ding, P.; Guo, J. Triclosan Promotes Conjugative Transfer of Antibiotic Resistance Genes to Opportunistic Pathogens in Environmental Microbiome. Environ. Sci. Technol. 2022, 56, 15108–15119. [Google Scholar] [CrossRef]
- Barber, O.W.; Hartmann, E.M. Benzalkonium Chloride: A Systematic Review of Its Environmental Entry through Wastewater Treatment, Potential Impact, and Mitigation Strategies. Crit. Rev. Environ. Sci. Technol. 2022, 52, 2691–2719. [Google Scholar] [CrossRef]
- Yang, K.; Chen, M.-L.; Zhu, D. Exposure to Benzalkonium Chloride Disinfectants Promotes Antibiotic Resistance in Sewage Sludge Microbiomes. Sci. Total Environ. 2023, 867, 161527. [Google Scholar] [CrossRef]
- Harrison, K.R.; Kappell, A.D.; McNamara, P.J. Benzalkonium Chloride Alters Phenotypic and Genotypic Antibiotic Resistance Profiles in a Source Water Used for Drinking Water Treatment. Environ. Pollut. 2020, 257, 113472. [Google Scholar] [CrossRef] [PubMed]
- Buffet-Bataillon, S.; Tattevin, P.; Bonnaure-Mallet, M.; Jolivet-Gougeon, A. Emergence of Resistance to Antibacterial Agents: The Role of Quaternary Ammonium Compounds—A Critical Review. Int. J. Antimicrob. Agents 2012, 39, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Karatzas, K.A.G.; Randall, L.P.; Webber, M.; Piddock, L.J.V.; Humphrey, T.J.; Woodward, M.J.; Coldham, N.G. Phenotypic and Proteomic Characterization of Multiply Antibiotic-Resistant Variants of Salmonella enterica Serovar Typhimurium Selected Following Exposure to Disinfectants. Appl. Environ. Microbiol. 2008, 74, 1508–1516. [Google Scholar] [CrossRef]
- Laborda, P.; Alcalde-Rico, M.; Blanco, P.; Martínez, J.L.; Hernando-Amado, S. Novel Inducers of the Expression of Multidrug Efflux Pumps That Trigger Pseudomonas aeruginosa Transient Antibiotic Resistance. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef]
- Brown, E.D.; Wright, G.D. Antibacterial Drug Discovery in the Resistance Era. Nature 2016, 529, 336–343. [Google Scholar] [CrossRef]
- Tyers, M.; Wright, G.D. Drug Combinations: A Strategy to Extend the Life of Antibiotics in the 21st Century. Nat. Rev. Microbiol. 2019, 17, 141–155. [Google Scholar] [CrossRef] [PubMed]
- Brown, D. Antibiotic Resistance Breakers: Can Repurposed Drugs Fill the Antibiotic Discovery Void? Nat. Rev. Drug Discov. 2015, 14, 821–832. [Google Scholar] [CrossRef]
- Kristiansen, J.E.; Thomsen, V.F.; Martins, A.; Viveiros, M.; Amaral, L. Non-Antibiotics Reverse Resistance of Bacteria to Antibiotics. In Vivo 2010, 24, 751–754. [Google Scholar]
- Zhang, Y.; Wang, B.; Cagnetta, G.; Duan, L.; Yang, J.; Deng, S.; Huang, J.; Wang, Y.; Yu, G. Typical Pharmaceuticals in Major WWTPs in Beijing, China: Occurrence, Load Pattern and Calculation Reliability. Water Res. 2018, 140, 291–300. [Google Scholar] [CrossRef]
- Archer, E.; Petrie, B.; Kasprzyk-Hordern, B.; Wolfaardt, G.M. The Fate of Pharmaceuticals and Personal Care Products (PPCPs), Endocrine Disrupting Contaminants (EDCs), Metabolites and Illicit Drugs in a WWTW and Environmental Waters. Chemosphere 2017, 174, 437–446. [Google Scholar] [CrossRef]
- Sui, Q.; Huang, J.; Deng, S.; Yu, G.; Fan, Q. Occurrence and Removal of Pharmaceuticals, Caffeine and DEET in Wastewater Treatment Plants of Beijing, China. Water Res. 2010, 44, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Alcalá, I.; Guillén-Navarro, J.M.; Lahora, A. Occurrence and Fate of Pharmaceuticals in a Wastewater Treatment Plant from Southeast of Spain and Risk Assessment. J. Environ. Manag. 2021, 279, 111565. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lu, J.; Engelstädter, J.; Zhang, S.; Ding, P.; Mao, L.; Yuan, Z.; Bond, P.L.; Guo, J. Non-Antibiotic Pharmaceuticals Enhance the Transmission of Exogenous Antibiotic Resistance Genes through Bacterial Transformation. ISME J. 2020, 14, 2179–2196. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lu, J.; Zhang, S.; Li, J.; Mao, L.; Yuan, Z.; Bond, P.L.; Guo, J. Non-Antibiotic Pharmaceuticals Promote the Transmission of Multidrug Resistance Plasmids through Intra- and Intergenera Conjugation. ISME J. 2021, 15, 2493–2508. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yu, Z.; Ding, P.; Lu, J.; Mao, L.; Ngiam, L.; Yuan, Z.; Engelstädter, J.; Schembri, M.A.; Guo, J. Antidepressants Can Induce Mutation and Enhance Persistence toward Multiple Antibiotics. Proc. Natl. Acad. Sci. USA 2023, 120, e2208344120. [Google Scholar] [CrossRef]
- Jia, Y.; Wang, Z.; Fang, D.; Yang, B.; Li, R.; Liu, Y. Acetaminophen Promotes Horizontal Transfer of Plasmid-Borne Multiple Antibiotic Resistance Genes. Sci. Total Environ. 2021, 782, 146916. [Google Scholar] [CrossRef]
- Estahbanati, S.; Fahrenfeld, N.L. Influence of Wastewater Treatment Plant Discharges on Microplastic Concentrations in Surface Water. Chemosphere 2016, 162, 277–284. [Google Scholar] [CrossRef]
- Li, X.; Mei, Q.; Chen, L.; Zhang, H.; Dong, B.; Dai, X.; He, C.; Zhou, J. Enhancement in Adsorption Potential of Microplastics in Sewage Sludge for Metal Pollutants after the Wastewater Treatment Process. Water Res. 2019, 157, 228–237. [Google Scholar] [CrossRef]
- Lv, X.; Dong, Q.; Zuo, Z.; Liu, Y.; Huang, X.; Wu, W.-M. Microplastics in a Municipal Wastewater Treatment Plant: Fate, Dynamic Distribution, Removal Efficiencies, and Control Strategies. J. Clean. Prod. 2019, 225, 579–586. [Google Scholar] [CrossRef]
- Okoffo, E.D.; O’Brien, S.; O’Brien, J.W.; Tscharke, B.J.; Thomas, K.V. Wastewater Treatment Plants as a Source of Plastics in the Environment: A Review of Occurrence, Methods for Identification, Quantification and Fate. Environ. Sci. Water Res. Technol. 2019, 5, 1908–1931. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, J.; Li, J. The Removal of Microplastics in the Wastewater Treatment Process and Their Potential Impact on Anaerobic Digestion Due to Pollutants Association. Chemosphere 2020, 251, 126360. [Google Scholar] [CrossRef] [PubMed]
- Hintersteiner, I.; Himmelsbach, M.; Buchberger, W.W. Characterization and Quantitation of Polyolefin Microplastics in Personal-Care Products Using High-Temperature Gel-Permeation Chromatography. Anal. Bioanal. Chem. 2015, 407, 1253–1259. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lu, J.; Wu, J.; Wang, J.; Luo, Y. Potential Risks of Microplastics Combined with Superbugs: Enrichment of Antibiotic Resistant Bacteria on the Surface of Microplastics in Mariculture System. Ecotoxicol. Environ. Saf. 2020, 187, 109852. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, G.; Song, W.; Ye, C.; Lin, H.; Li, Z.; Liu, W. Plastics in the Marine Environment Are Reservoirs for Antibiotic and Metal Resistance Genes. Environ. Int. 2019, 123, 79–86. [Google Scholar] [CrossRef]
- Wang, J.; Qin, X.; Guo, J.; Jia, W.; Wang, Q.; Zhang, M.; Huang, Y. Evidence of Selective Enrichment of Bacterial Assemblages and Antibiotic Resistant Genes by Microplastics in Urban Rivers. Water Res. 2020, 183, 116113. [Google Scholar] [CrossRef]
- Wang, H.; Xu, K.; Wang, J.; Feng, C.; Chen, Y.; Shi, J.; Ding, Y.; Deng, C.; Liu, X. Microplastic Biofilm: An Important Microniche That May Accelerate the Spread of Antibiotic Resistance Genes via Natural Transformation. J. Hazard. Mater. 2023, 459, 132085. [Google Scholar] [CrossRef]
- Yu, X.; Zhou, Z.-C.; Shuai, X.; Lin, Z.; Liu, Z.; Zhou, J.; Lin, Y.; Zeng, G.; Ge, Z.; Chen, H. Microplastics Exacerbate Co-Occurrence and Horizontal Transfer of Antibiotic Resistance Genes. J. Hazard. Mater. 2023, 451, 131130. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, W.; Zhang, Z.; Grossart, H.-P.; Gadd, G.M. Microplastics Provide New Microbial Niches in Aquatic Environments. Appl. Microbiol. Biotechnol. 2020, 104, 6501–6511. [Google Scholar] [CrossRef]
- Xu, S.; Ma, J.; Ji, R.; Pan, K.; Miao, A.-J. Microplastics in Aquatic Environments: Occurrence, Accumulation, and Biological Effects. Sci. Total Environ. 2020, 703, 134699. [Google Scholar] [CrossRef]
- Wu, X.; Pan, J.; Li, M.; Li, Y.; Bartlam, M.; Wang, Y. Selective Enrichment of Bacterial Pathogens by Microplastic Biofilm. Water Res. 2019, 165, 114979. [Google Scholar] [CrossRef]
- Caruso, G. Microplastics as Vectors of Contaminants. Mar. Pollut. Bull. 2019, 146, 921–924. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Han, Y.; Tang, Y.; Shi, W.; Du, X.; Sun, S.; Liu, G. Microplastics Aggravate the Bioaccumulation of Two Waterborne Veterinary Antibiotics in an Edible Bivalve Species: Potential Mechanisms and Implications for Human Health. Environ. Sci. Technol. 2020, 54, 8115–8122. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Li, Y.; Huang, G.; Yang, C.; Chen, C.; Zhou, T.; Zhao, Y.; Ma, J. Adsorption Behavior of the Antibiotic Levofloxacin on Microplastics in the Presence of Different Heavy Metals in an Aqueous Solution. Chemosphere 2020, 260, 127650. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gao, J.; Li, D.; Dai, H.; Zhao, Y. Co-Occurrence of Microplastics and Triclosan Inhibited Nitrification Function and Enriched Antibiotic Resistance Genes in Nitrifying Sludge. J. Hazard. Mater. 2020, 399, 123049. [Google Scholar] [CrossRef]
- Zhao, Y.; Gao, J.; Wang, Z.; Cui, Y.; Zhang, Y.; Dai, H.; Li, D. Distinct Bacterial Communities and Resistance Genes Enriched by Triclocarban-Contaminated Polyethylene Microplastics in Antibiotics and Heavy Metals Polluted Sewage Environment. Sci. Total Environ. 2022, 839, 156330. [Google Scholar] [CrossRef]
- Nies, D.H. Efflux-Mediated Heavy Metal Resistance in Prokaryotes. FEMS Microbiol. Rev. 2003, 27, 313–339. [Google Scholar] [CrossRef]
- Li, A.-D.; Li, L.-G.; Zhang, T. Exploring Antibiotic Resistance Genes and Metal Resistance Genes in Plasmid Metagenomes from Wastewater Treatment Plants. Front. Microbiol. 2015, 6, 1025. [Google Scholar] [CrossRef]
- Wei, Z.; Wei, Y.; Li, H.; Shi, D.; Yang, D.; Yin, J.; Zhou, S.; Chen, T.; Li, J.; Jin, M. Emerging Pollutant Metformin in Water Promotes the Development of Multiple-Antibiotic Resistance in Escherichia coli via Chromosome Mutagenesis. J. Hazard. Mater. 2022, 430, 128474. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, F.; Yu, Q.; Zhang, X.-X. A Mini-Review of Antibiotic Resistance Drivers in Urban Wastewater Treatment Plants: Environmental Concentrations, Mechanism and Perspectives. Water 2023, 15, 3165. https://doi.org/10.3390/w15173165
Zhao F, Yu Q, Zhang X-X. A Mini-Review of Antibiotic Resistance Drivers in Urban Wastewater Treatment Plants: Environmental Concentrations, Mechanism and Perspectives. Water. 2023; 15(17):3165. https://doi.org/10.3390/w15173165
Chicago/Turabian StyleZhao, Fuzheng, Qingmiao Yu, and Xu-Xiang Zhang. 2023. "A Mini-Review of Antibiotic Resistance Drivers in Urban Wastewater Treatment Plants: Environmental Concentrations, Mechanism and Perspectives" Water 15, no. 17: 3165. https://doi.org/10.3390/w15173165
APA StyleZhao, F., Yu, Q., & Zhang, X.-X. (2023). A Mini-Review of Antibiotic Resistance Drivers in Urban Wastewater Treatment Plants: Environmental Concentrations, Mechanism and Perspectives. Water, 15(17), 3165. https://doi.org/10.3390/w15173165






