Low Concentrations of Antibiotics Alter Microbial Communities and Induce High Abundances of Antibiotic-Resistant Genes in Ornamental Water
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Sites and Sample Collection
2.2. ARGs Mutation Experiments
2.2.1. Determining Inhibitory Concentration
2.2.2. Construction of Experimental Systems
2.3. Sequencing of Bacterial 16S rRNA Genes
2.4. Quantification of ARGs and Integrase Genes
2.5. Statistical Analysis
3. Results and Discussion
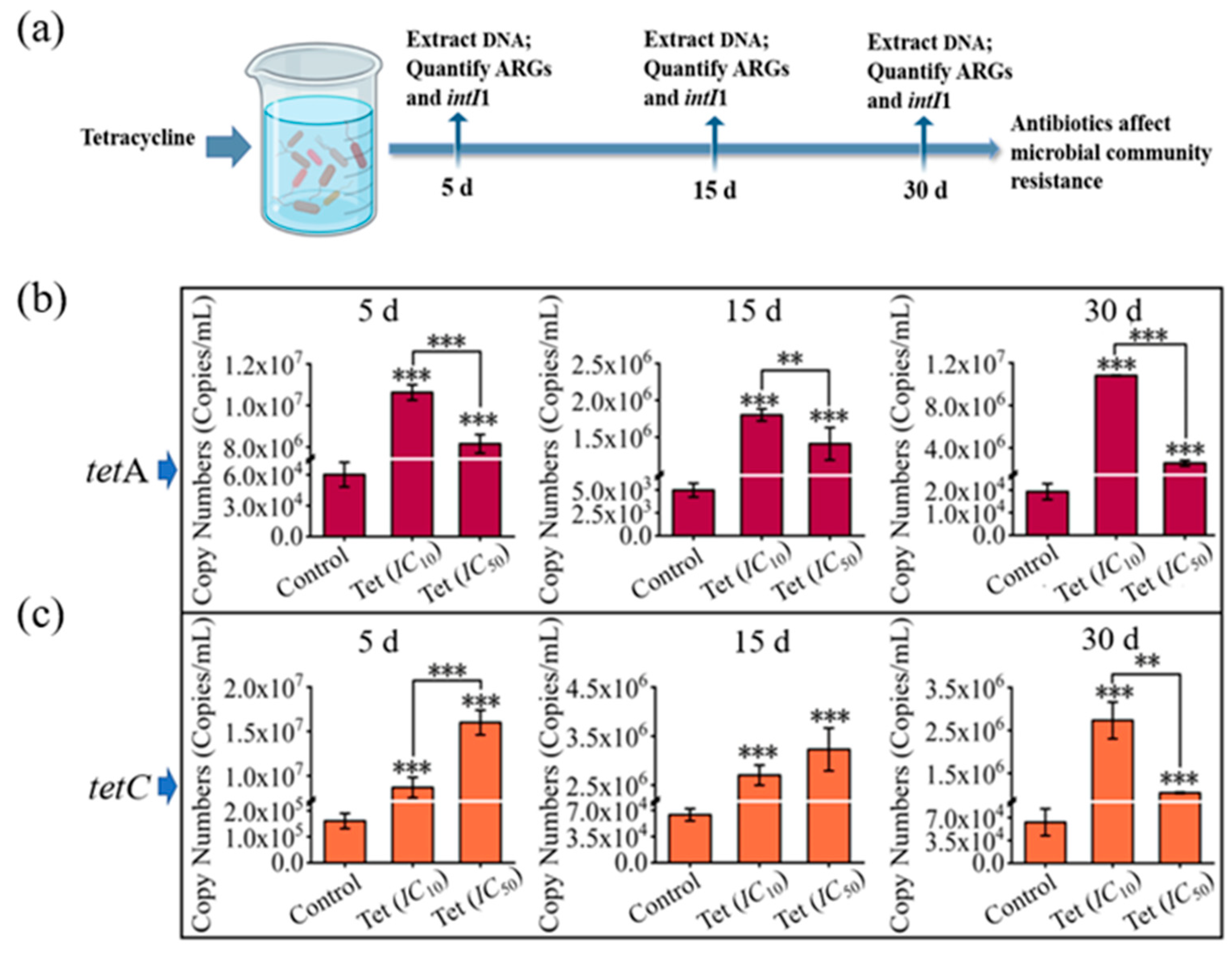
3.1. Effects of Tetracycline on Microbial Community Composition in Pond Water
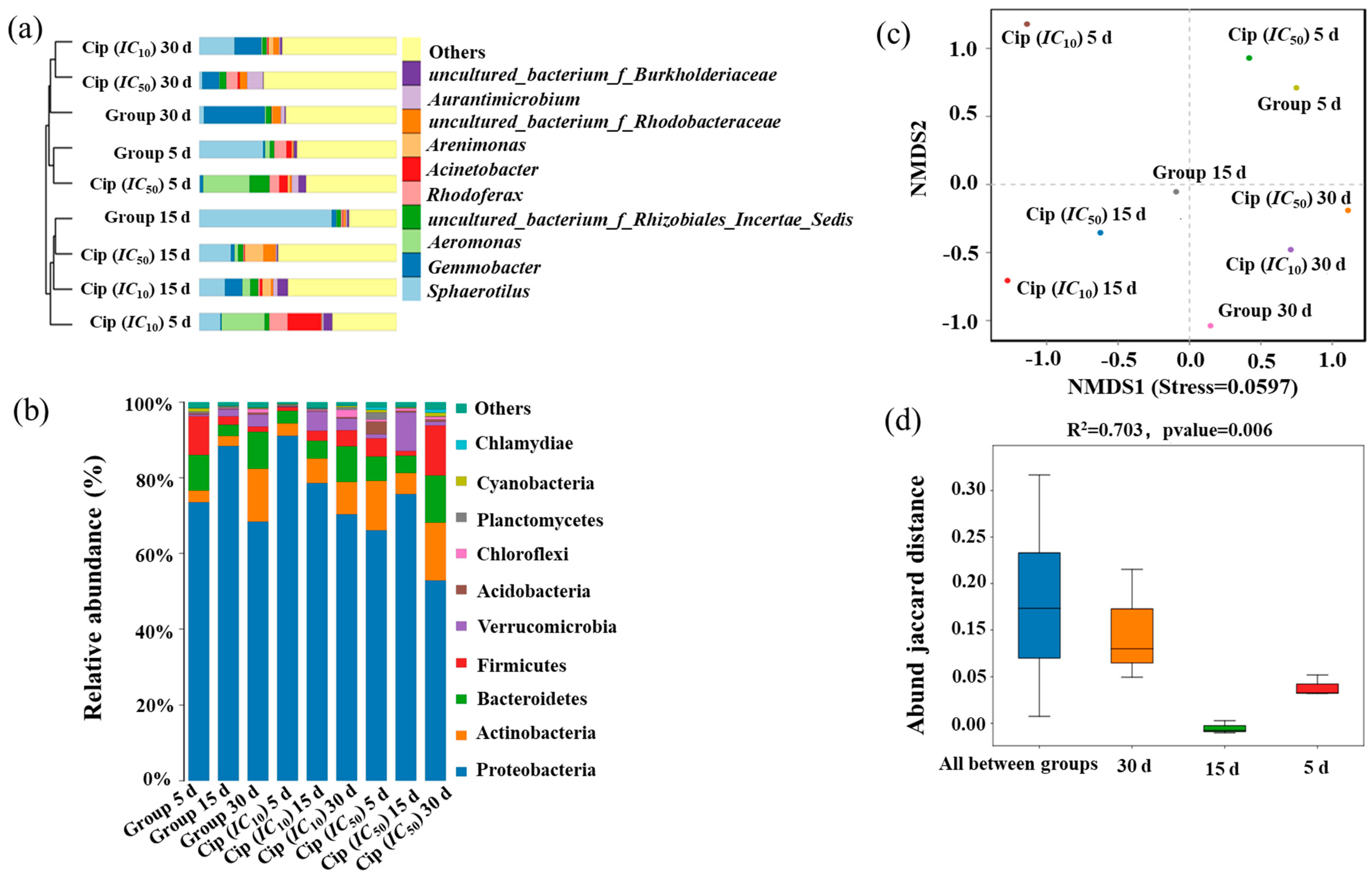
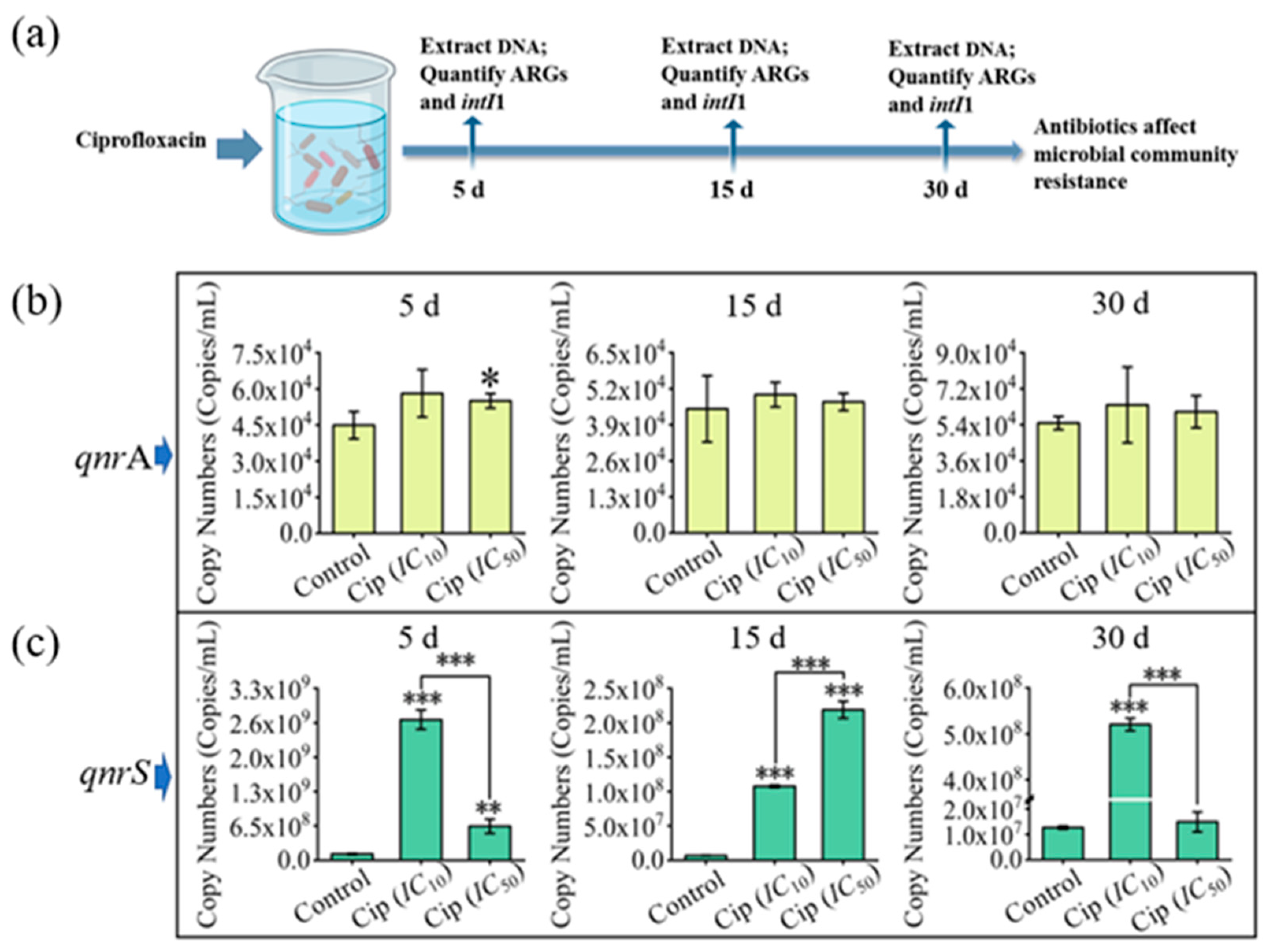
3.2. Impact of Ciprofloxacin on Microbial Community Composition in Pond Water
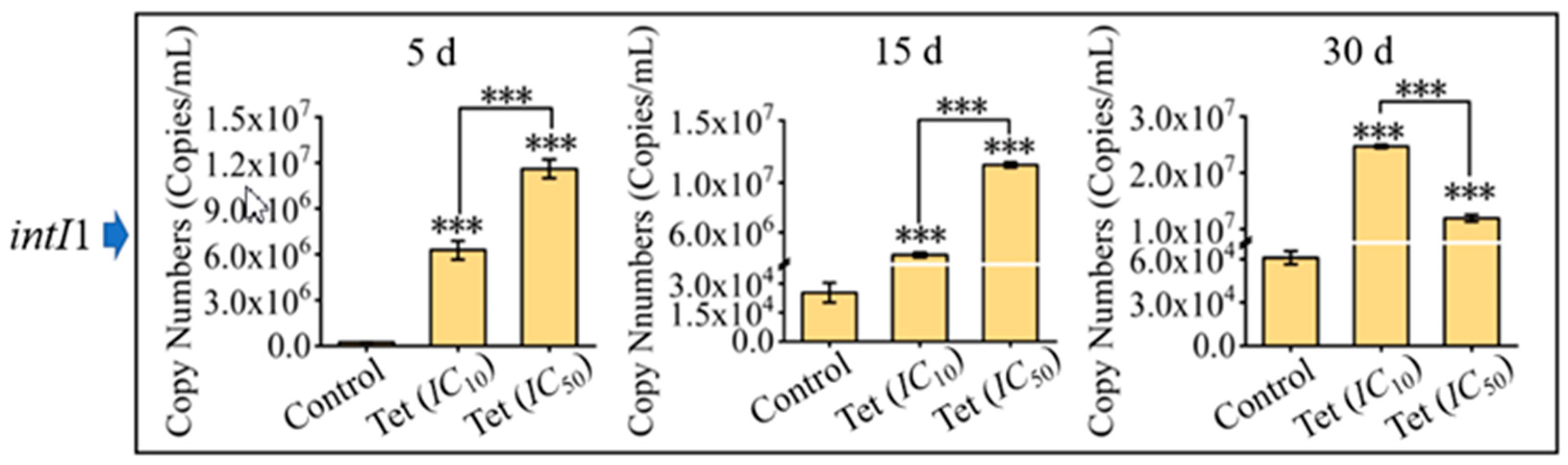
3.3. Impacts of Antibiotics on Microbial Community Resistance in Pond Water
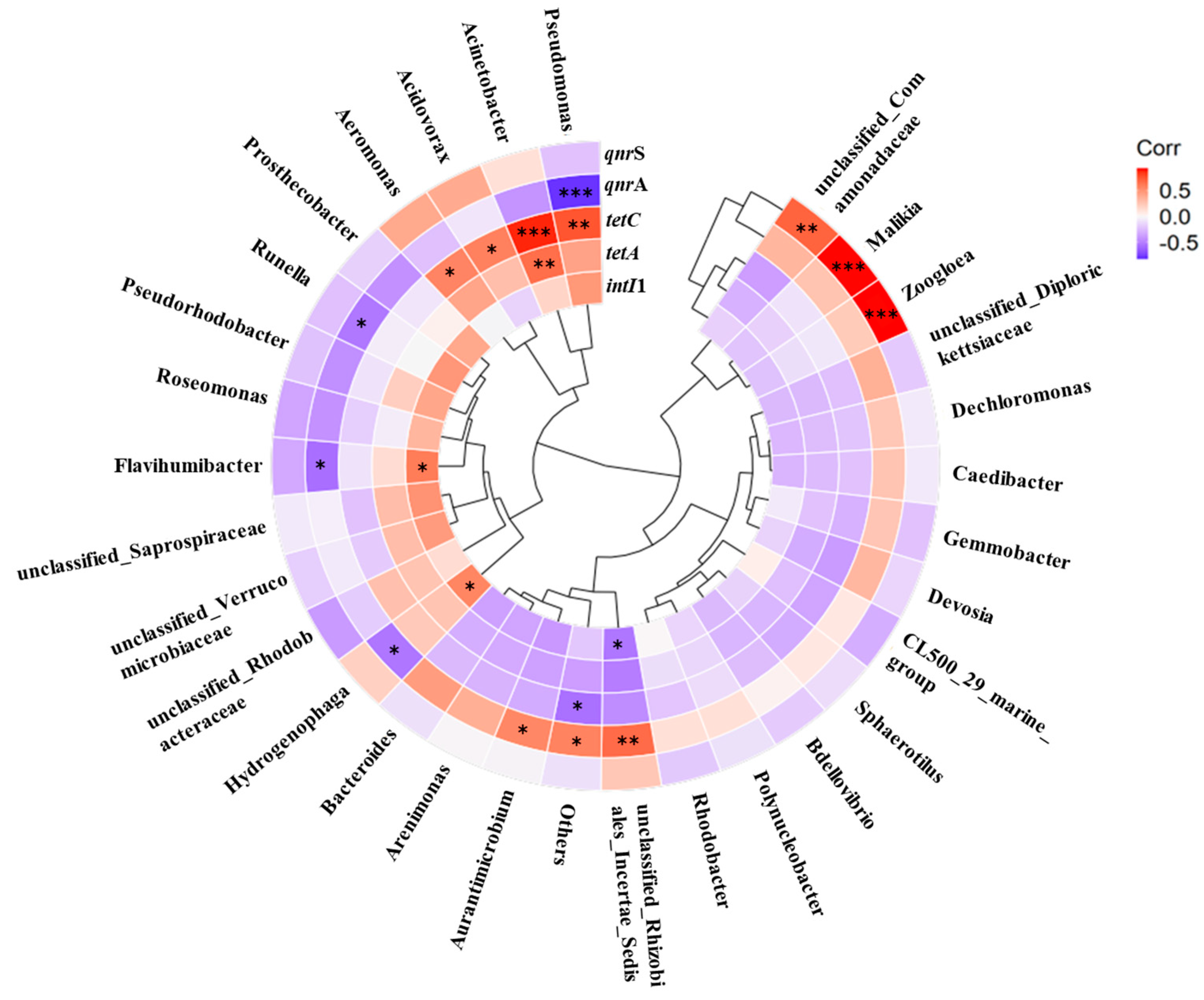
3.4. Interrelationship among Microbial Community, intI1, and ARGs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Halling-Sorensen, B.; Nors Nielsen, S.; Lanzky, P.F.; Ingerslev, F.; Holten Lutzhoft, H.C.; Jorgensen, S.E. Occurrence, fate and effects of pharmaceutical substances in the environment: A review. Chemosphere 1998, 36, 357–393. [Google Scholar] [CrossRef] [PubMed]
- Qiao, M.; Ying, G.G.; Singer, A.C.; Zhu, Y.G. Review of antibiotic resistance in China and its environment. Environ. Int. 2018, 110, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lin, Y.; Zheng, Y.; Meng, F. Antibiotics in mariculture systems: A review of occurrence, environmental behavior, and ecological effects. Environ. Pollut. 2022, 293, 118541. [Google Scholar] [CrossRef]
- Kemper, N. Veterinary antibiotics in the aquatic and terrestrial environment. Ecol. Indic. 2008, 8, 1–13. [Google Scholar] [CrossRef]
- Nappier, S.P.; Liguori, K.; Ichida, A.M.; Stewart, J.R.; Jones, K.R. Antibiotic resistance in recreational waters: State of the science. Int. J. Environ. Res. Public Health 2020, 17, 8034. [Google Scholar] [CrossRef]
- Liu, L.; Wang, Q.; Lin, H.; Das, R.; Wang, S.; Qi, H.; Yang, J.; Xue, Y.; Mao, D.; Luo, Y. Amoxicillin increased functional pathway genes and beta-lactam resistance genes by pathogens bloomed in intestinal microbiota using a simulator of the human intestinal microbial ecosystem. Front. Microbiol. 2020, 11, 1213. [Google Scholar] [CrossRef]
- Qin, K.N.; Wei, L.L.; Li, J.J.; Lai, B.; Zhu, F.Y.; Yu, H.; Zhao, Q.L.; Wang, K. A review of ARGs in WWTPs: Sources, stressors, and elimination. Chin. Chem. Lett. 2020, 31, 2603–2613. [Google Scholar] [CrossRef]
- Chopra, I.; Roberts, M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef]
- Pruden, A.; Pei, R.; Storteboom, H.; Carlson, K.H. Antibiotic resistance genes as emerging contaminants: Studies in northern Colorado. Environ. Sci. Technol. 2006, 40, 7445–7450. [Google Scholar] [CrossRef]
- Li, S.N.; Zhang, C.F.; Li, F.X.; Hua, T.; Zhou, Q.X.; Ho, S.H. Technologies towards antibiotic resistance genes (ARGs) removal from aquatic environment: A critical review. J. Hazard. Mater. 2021, 411, 125148. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, Y.; Lu, J.; Yu, Z.; Song, H.; Bond, P.L.; Guo, J. Chlorine disinfection facilitates natural transformation through ROS-mediated oxidative stress. ISME J. 2021, 15, 2969–2985. [Google Scholar] [CrossRef]
- Zainab, S.M.; Junaid, M.; Xu, N.; Malik, R.N. Antibiotics and antibiotic resistant genes (ARGs) in groundwater: A global review on dissemination, sources, interactions, environmental and human health risks. Water Res. 2020, 187, 116455. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.W.; Wang, J.T.; Li, J.; Shi, X.Z.; Ma, Y.B.; Chen, D.; He, J.Z. Long-term nickel contamination increases the occurrence of antibiotic resistance genes in agricultural soils. Environ. Sci. Technol. 2017, 51, 790–800. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liu, Q.; Han, Y.N. Assessing the fishery resource status of China’s coastal waters using surplus production models. J. Ocean Univ. China 2021, 20, 1236–1244. [Google Scholar] [CrossRef]
- Santos, L.; Ramos, F. Analytical strategies for the detection and quantification of antibiotic residues in aquaculture fishes: A review. Trends Food Sci. Technol. 2016, 52, 16–30. [Google Scholar] [CrossRef]
- Ning, K.; Ji, L.; Zhang, L.; Zhu, X.; Wei, H.; Han, M.; Wang, Z. Is rice-crayfish co-culture a better aquaculture model: From the perspective of antibiotic resistome profiles. Environ. Pollut. 2022, 292, 118450. [Google Scholar] [CrossRef]
- Wang, C.; Liu, X.; Yang, Y.; Wang, Z. Antibiotic and antibiotic resistance genes in freshwater aquaculture ponds in China: A meta-analysis and assessment. J. Clean. Prod. 2021, 329, 129719. [Google Scholar] [CrossRef]
- Xu, J.Y.; Sangthong, R.; McNeil, E.; Tang, R.; Chongsuvivatwong, V. Antibiotic use in chicken farms in northwestern China. Antimicrob. Resist. Infect. Control 2020, 9, 10. [Google Scholar] [CrossRef]
- Zinia, N.J.; McShane, P. Urban ecosystems and ecosystem services in megacity Dhaka: Mapping and inventory analysis. Urban Ecosyst. 2021, 24, 915–928. [Google Scholar] [CrossRef]
- Wang, H.; Mei, C.; Liu, J.; Shao, W. A new strategy for integrated urban water management in China: Sponge city. Sci. China Technol. Sci. 2018, 61, 317–329. [Google Scholar] [CrossRef]
- Chen, S.; van de Ven, F.H.M.; Zevenbergen, C.; Verbeeck, S.; Ye, Q.; Zhang, W.; Wei, L. Revisiting China’s sponge city planning approach: Lessons from a case study on qinhuai district, Nanjing. Front. Environ. Sci. 2021, 9, 748231. [Google Scholar] [CrossRef]
- Luis Martinez, J. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 2009, 157, 2893–2902. [Google Scholar] [CrossRef]
- Mo, W.Y.; Chen, Z.; Leung, H.M.; Leung, A.O.W. Application of veterinary antibiotics in China’s aquaculture industry and their potential human health risks. Environ. Sci. Pollut. Res. 2017, 24, 8978–8989. [Google Scholar] [CrossRef]
- Bunnajirakul, S.; Pavasutthipaisit, S.; Steinhagen, D. Pathological alterations due to motile Aeromonas infection in red swordtail fish. Tieraerztl. Prax. K. H. 2015, 43, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Cabello, F.C.; Godfrey, H.P.; Tomova, A.; Ivanova, L.; Doelz, H.; Millanao, A.; Buschmann, A.H. Antimicrobial use in aquaculture re-examined: Its relevance to antimicrobial resistance and to animal and human health. Environ. Microbiol. 2013, 15, 1917–1942. [Google Scholar] [CrossRef]
- Pham, T.D.M.; Ziora, Z.M.; Blaskovich, M.A.T. Quinolone antibiotics. MedChemComm 2019, 10, 1719–1739. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, J.; Mao, L.; Li, J.; Yuan, Z.; Bond, P.L.; Guo, J. Antiepileptic drug carbamazepine promotes horizontal transfer of plasmid-borne multi-antibiotic resistance genes within and across bacterial genera. ISME J. 2019, 13, 509–522. [Google Scholar] [CrossRef]
- Du, R.J.; Qu, Y.J.; Zhao, M.; Liu, Y.H.; Qi, P.X.; Sun, X.B. Logistic modeling to predict the minimum inhibitory concentration (MIC) of olive leaf extract (OLE) against Listeria monocytogenes. PLoS ONE 2022, 17, e0263359. [Google Scholar] [CrossRef]
- Hou, W.; Sun, S.; Wang, M.; Gu, B.; Li, X.; Zhang, C.; Jia, R. Variations in stable carbon and nitrogen isotopes of particulate organic matter in surface waters of water-receiving area of Eastern Route of South-to-North Water Transfer Project, China. Environ. Sci. Pollut. Res. 2020, 27, 2805–2818. [Google Scholar] [CrossRef]
- Liu, X.W.; Lv, K.; Deng, C.X.; Yu, Z.M.; Shi, J.H.; Johnson, A.C. Persistence and migration of tetracycline, sulfonamide, fluoroquinolone, and macrolide antibiotics in streams using a simulated hydrodynamic system. Environ. Pollut. 2019, 252, 1532–1538. [Google Scholar] [CrossRef]
- Sun, R.; He, L.; Li, T.; Dai, Z.; Sun, S.; Ren, L.; Liang, Y.Q.; Zhang, Y.; Li, C. Impact of the surrounding environment on antibiotic resistance genes carried by microplastics in mangroves. Sci. Total Environ. 2022, 837, 155771. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Ren, S.; Niu, T.; Guo, Y.; Qi, S.; Han, X.; Liu, D.; Pan, F. Distribution of antibiotic-resistant bacteria in chicken manure and manure-fertilized vegetables. Environ. Sci. Pollut. Res. 2014, 21, 1231–1241. [Google Scholar] [CrossRef]
- Gillings, M.R.; Gaze, W.H.; Pruden, A.; Smalla, K.; Tiedje, J.M.; Zhu, Y.G. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 2015, 9, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; He, J. Effect of antibiotics in the environment on microbial populations. Appl. Microbiol. Biotechnol. 2010, 87, 925–941. [Google Scholar] [CrossRef] [PubMed]
- Lundstrom, S.V.; Ostman, M.; Bengtsson-Palme, J.; Rutgersson, C.; Thoudal, M.; Sircar, T.; Blanck, H.; Eriksson, K.M.; Tysklind, M.; Flach, C.F.; et al. Minimal selective concentrations of tetracycline in complex aquatic bacterial biofilms. Sci. Total Environ. 2016, 553, 587–595. [Google Scholar] [CrossRef]
- Henderson, M.; Ergas, S.J.; Ghebremichael, K.; Gross, A.; Ronen, Z. Occurrence of antibiotic-resistant genes and bacteria in household greywater treated in constructed wetlands. Water 2022, 14, 758. [Google Scholar] [CrossRef]
- Hou, Y.; Li, B.; Feng, G.; Zhang, C.; He, J.; Li, H.; Zhu, J. Responses of bacterial communities and organic matter degradation in surface sediment to macrobrachium nipponense bioturbation. Sci. Total Environ. 2021, 759, 143534. [Google Scholar] [CrossRef]
- Gibson, M.K.; Forsberg, K.J.; Dantas, G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 2015, 9, 207–216. [Google Scholar] [CrossRef]
- Somerfield, P.J.; Clarke, K.R.; Gorley, R.N. Analysis of similarities (ANOSIM) for 2-way layouts using a generalised ANOSIM statistic, with comparative notes on permutational multivariate analysis of variance (PERMANOVA). Austral Ecol. 2021, 46, 911–926. [Google Scholar] [CrossRef]
- He, H.; Xie, Y. Effect of different hemodialysis methods on microbiota in uremic patients. Biomed. Res. Int. 2020, 2020, 6739762. [Google Scholar] [CrossRef]
- Tong, C.; Hu, H.; Chen, G.; Li, Z.; Li, A.; Zhang, J. Chlorine disinfectants promote microbial resistance in Pseudomonas sp. Environ. Res. 2021, 199, 111296. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Jin, D.; Freitag, T.E.; Sun, W.; Yu, Q.; Fu, J.; Ma, J. A compositional shift in the soil microbiome induced by tetracycline, sulfamonomethoxine and ciprofloxacin entering a plant-soil system. Environ. Pollut. 2016, 212, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.B.; Li, B.X.; Zou, R.S.; Dai, Y.; Xie, S.G.; Yuan, B.L. Biodegradation of antibiotic ciprofloxacin: Pathways, influential factors, and bacterial community structure. Environ. Sci. Pollut. Res. 2016, 23, 7911–7918. [Google Scholar] [CrossRef]
- Kang, J.; Zhang, Z.; Chen, Y.; Zhou, Z.; Zhang, J.; Xu, N.; Zhang, Q.; Lu, T.; Peijnenburg, W.J.G.M.; Qian, H. Machine learning predicts the impact of antibiotic properties on the composition and functioning of bacterial community in aquatic habitats. Sci. Total Environ. 2022, 828, 154412. [Google Scholar] [CrossRef]
- Liu, F.; Wu, J.; Ying, G.G.; Luo, Z.; Feng, H. Changes in functional diversity of soil microbial community with addition of antibiotics sulfamethoxazole and chlortetracycline. Appl. Microbiol. Biotechnol. 2012, 95, 1615–1623. [Google Scholar] [CrossRef]
- Du, B.; Yang, Q.; Li, X.; Yuan, W.; Chen, Y.; Wang, R. Impacts of long-term exposure to tetracycline and sulfamethoxazole on the sludge granules in an anoxic-aerobic wastewater treatment system. Sci. Total Environ. 2019, 684, 67–77. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Y.; Wu, N.; Li, W.; Song, X.; Ma, Y.; Niu, Z. Colonization characteristics of bacterial communities on plastic debris: The localization of immigrant bacterial communities. Water Res. 2021, 19, 116883. [Google Scholar] [CrossRef]
- Grenni, P.; Ancona, V.; Caracciolo, A.B. Ecological effects of antibiotics on natural ecosystems: A review. Microchem. J. 2018, 136, 25–39. [Google Scholar] [CrossRef]
- Zhang, Y.; Pei, M.; Zhang, B.; He, Y.; Zhong, Y. Changes of antibiotic resistance genes and bacterial communities in the advanced biological wastewater treatment system under low selective pressure of tetracycline. Water Res. 2021, 207, 117834. [Google Scholar] [CrossRef]
- Gao, P.; Xu, W.; Ruan, X.; Qian, Y.; Xue, G.; Jia, H. Long-term impact of a tetracycline concentration gradient on the bacterial resistance in anaerobic-aerobic sequential bioreactors. Chemosphere 2018, 205, 308–316. [Google Scholar] [CrossRef]
- Kohanski, M.A.; DePristo, M.A.; Collins, J.J. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol. Cell 2010, 37, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Jutkina, J.; Marathe, N.P.; Flach, C.F.; Larsson, D.G.J. Antibiotics and common antibacterial biocides stimulate horizontal transfer of resistance at low concentrations. Sci. Total Environ. 2018, 616, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Huang, C.; Xi, B.; Tang, Z.; Tan, W.; Li, W.; Zhang, Y.; Li, W. The maturity period is the main stage of antibiotic resistance genes reduction in aerobic composting process of swine manure in sub-scale farms. Bioresour. Technol. 2021, 319, 124139. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Ye, N.; Huang, K.; Yu, J.; Zhang, S. Mobile genetic elements drive the antibiotic resistome alteration in freshwater shrimp aquaculture. Water 2021, 13, 1461. [Google Scholar] [CrossRef]
- Chen, J.; Yang, Y.; Jiang, X.; Ke, Y.; He, T.; Xie, S. Metagenomic insights into the profile of antibiotic resistomes in sediments of aquaculture wastewater treatment system. J. Environ. Sci. 2022, 113, 345–355. [Google Scholar] [CrossRef]
- Roberts, M.C.; Schwarz, S. Tetracycline and phenicol resistance genes and mechanisms: Importance for agriculture, the environment, and humans. J. Environ. Qual. 2016, 45, 576–592. [Google Scholar] [CrossRef]
- Yuan, Q.B.; Huang, Y.M.; Wu, W.B.; Zuo, P.; Hu, N.; Zhou, Y.Z.; Alvarez, P.J.J. Redistribution of intracellular and extracellular free & adsorbed antibiotic resistance genes through a wastewater treatment plant by an enhanced extracellular DNA extraction method with magnetic beads. Environ. Int. 2019, 131, 104986. [Google Scholar]
- Wang, Z.; Yuan, S.; Deng, Z.; Wang, Y.; Deng, S.; Song, Y.; Sun, C.; Bu, N.; Wang, X. Evaluating responses of nitrification and denitrification to the co-selective pressure of divalent zinc and tetracycline based on resistance genes changes. Bioresour. Technol. 2020, 314, 123769. [Google Scholar] [CrossRef]
- Zhu, N.; Jin, H.; Ye, X.; Liu, W.; Li, D.; Shah, G.M.; Zhu, Y. Fate and driving factors of antibiotic resistance genes in an integrated swine wastewater treatment system: From wastewater to soil. Sci. Total Environ. 2020, 721, 137654. [Google Scholar] [CrossRef]
- Ng, L.K.; Martin, I.; Alfa, M.; Mulvey, M. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probe. 2001, 15, 209–215. [Google Scholar] [CrossRef]
- Aminov, R.I.; Chee-Sanford, J.C.; Garrigues, N.; Teferedegne, B.; Krapac, I.J.; White, B.A.; Mackie, R.I. Development, validation, and application of PCR primers for detection of tetracycline efflux genes of gram-negative bacteria. Appl. Environ. Microb. 2002, 68, 1786–1793. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Huang, Z.; Yang, K.; Graham, D.; Xie, B. Relationships between Antibiotics and Antibiotic Resistance Gene Levels in Municipal Solid Waste Leachates in Shanghai, China. Environ. Sci. Technol. 2015, 49, 4122–4128. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.G.; Johnson, T.A.; Su, J.Q.; Qiao, M.; Guo, G.X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef] [PubMed]
- Marti, E.; Luis Balcazar, J. Real-Time PCR Assays for Quantification of qnr Genes in Environmental Water Samples and Chicken Feces. Appl. Environ. Microb. 2013, 79, 1743–1745. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, L.; Fang, H.; Mao, Q.; Bai, Y.; Ye, Z.; Hu, D.; Wang, X.; Hou, Y.; Ye, N.; Zhang, S.; et al. Low Concentrations of Antibiotics Alter Microbial Communities and Induce High Abundances of Antibiotic-Resistant Genes in Ornamental Water. Water 2023, 15, 3047. https://doi.org/10.3390/w15173047
Tian L, Fang H, Mao Q, Bai Y, Ye Z, Hu D, Wang X, Hou Y, Ye N, Zhang S, et al. Low Concentrations of Antibiotics Alter Microbial Communities and Induce High Abundances of Antibiotic-Resistant Genes in Ornamental Water. Water. 2023; 15(17):3047. https://doi.org/10.3390/w15173047
Chicago/Turabian StyleTian, Lingyun, Hao Fang, Qianbo Mao, Yi Bai, Zirui Ye, Dingjun Hu, Xiaoheng Wang, Yiyu Hou, Nan Ye, Shuai Zhang, and et al. 2023. "Low Concentrations of Antibiotics Alter Microbial Communities and Induce High Abundances of Antibiotic-Resistant Genes in Ornamental Water" Water 15, no. 17: 3047. https://doi.org/10.3390/w15173047
APA StyleTian, L., Fang, H., Mao, Q., Bai, Y., Ye, Z., Hu, D., Wang, X., Hou, Y., Ye, N., Zhang, S., & Ma, Y. (2023). Low Concentrations of Antibiotics Alter Microbial Communities and Induce High Abundances of Antibiotic-Resistant Genes in Ornamental Water. Water, 15(17), 3047. https://doi.org/10.3390/w15173047








