Bioaugmentation Potential Investigation Using a Phenol Affinity Analysis of Three Acinetobacter Strains in a Multi-Carbon-Source Condition
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture Preparation
2.2. Analytical Methods
2.3. Phenol Degradation in Culture Media
2.4. Inoculum Preparation
2.5. Phenol Degradation in Synthetic Wastewater
3. Results and Discussion
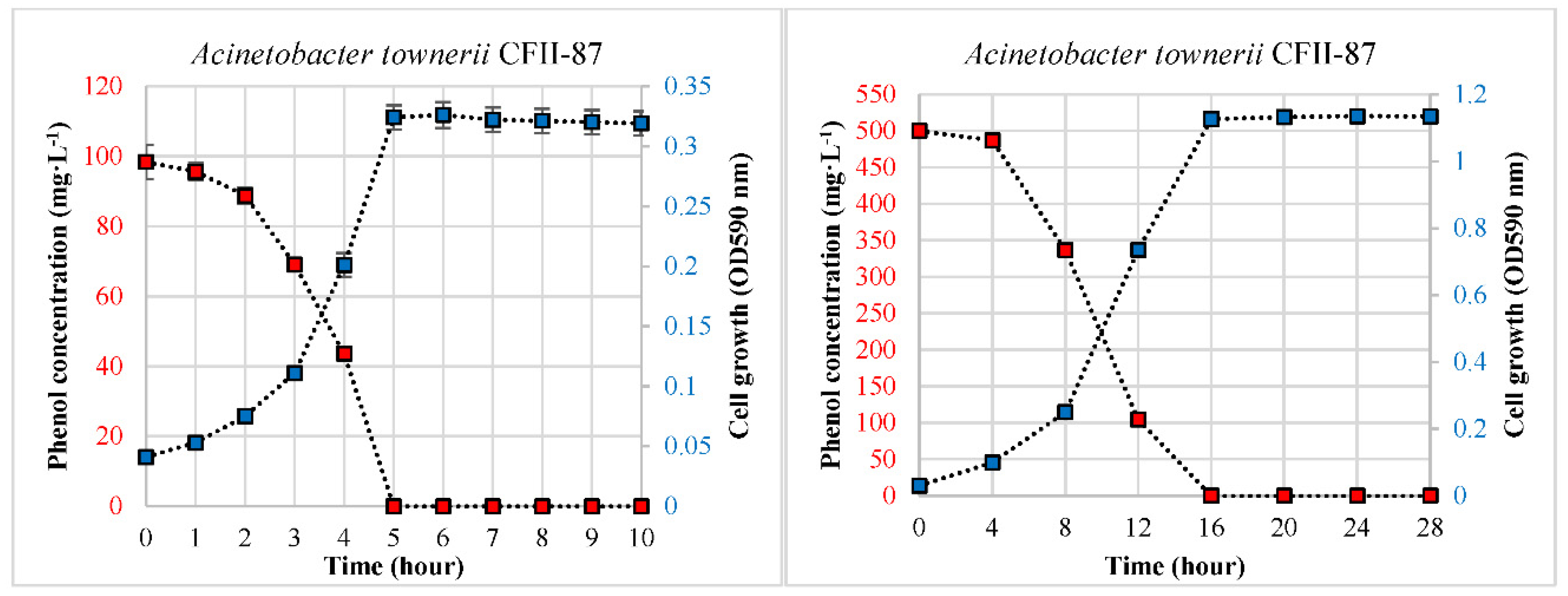
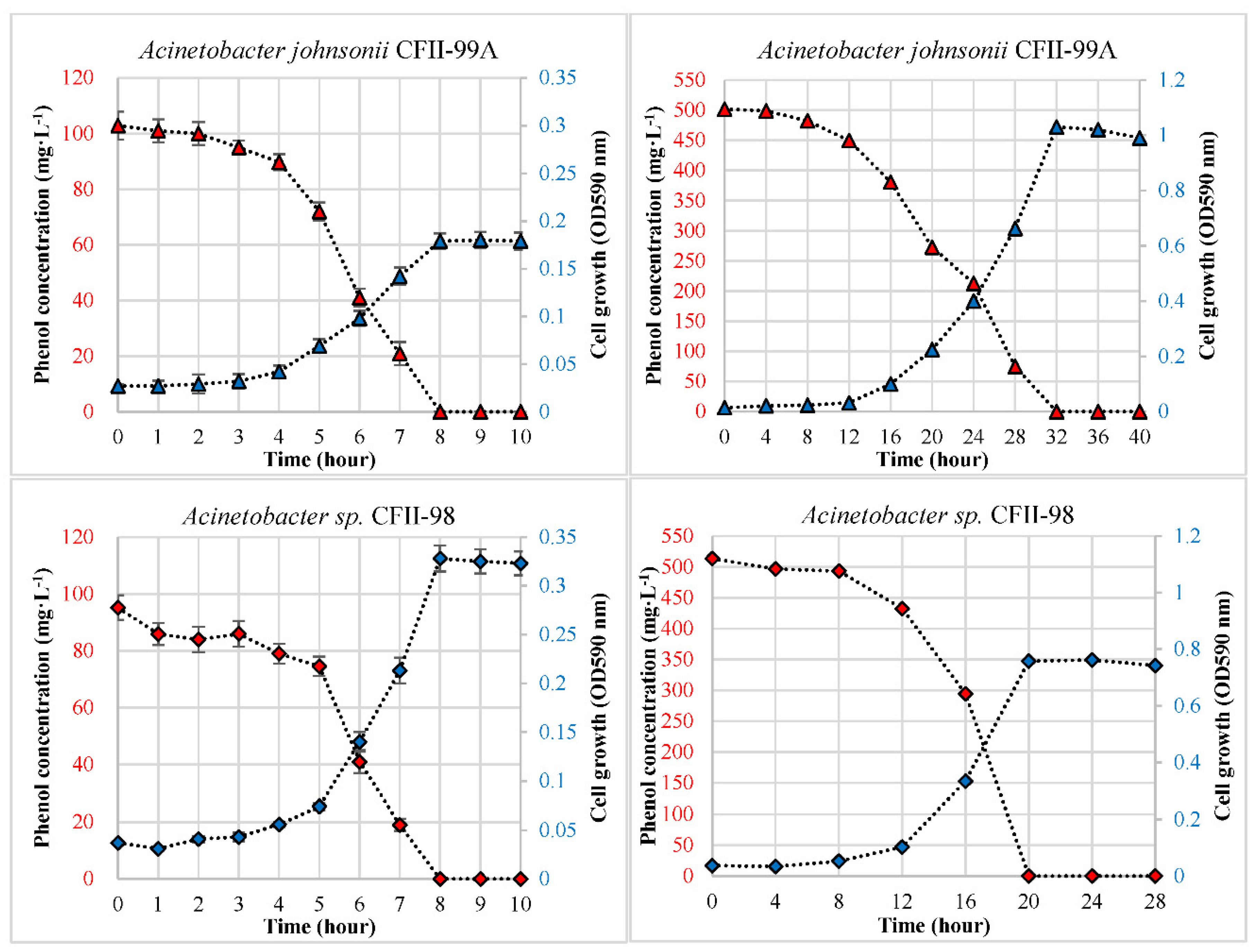
3.1. Phenol Degradation in the Single-Carbon-Source Condition
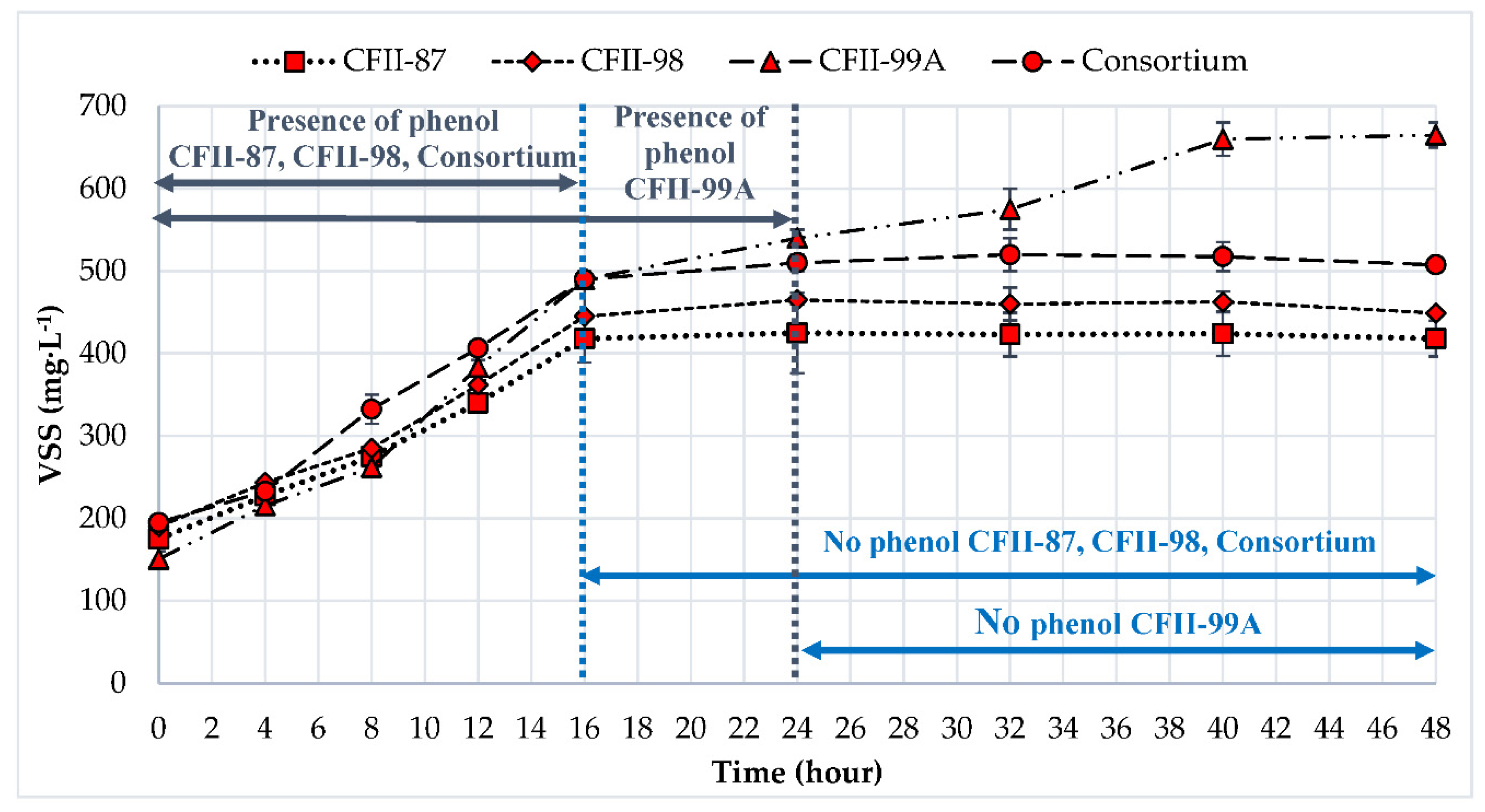
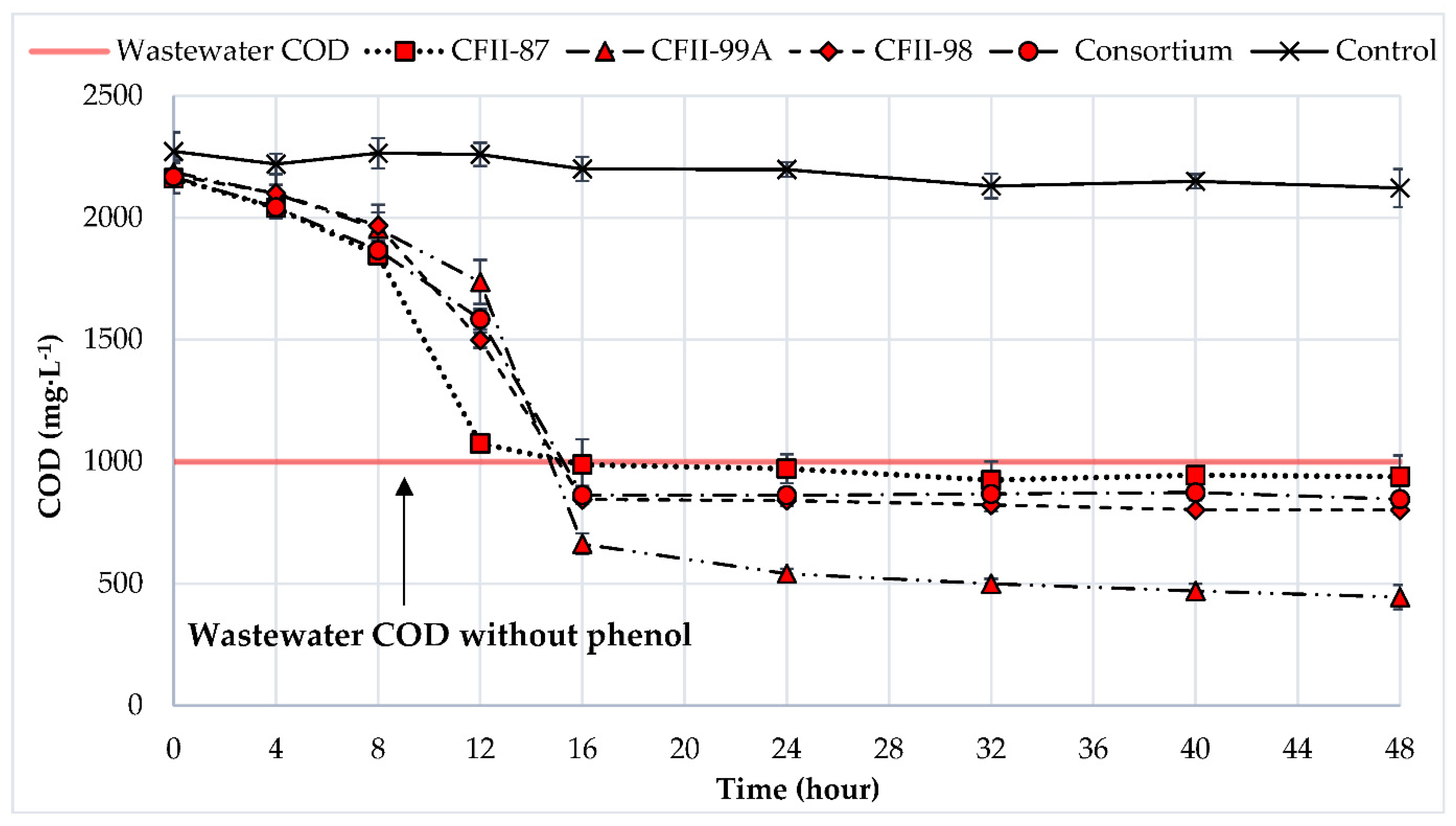
3.2. Phenol Degradation in a Multi-Carbon-Source Condition
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nuhoglu, A.; Yalcin, B. Modelling of phenol removal in a batch reactor. Process. Biochem. 2005, 40, 1233–1239. [Google Scholar] [CrossRef]
- Duraisamy, P.; Sekar, J.; Arunkumar, A.D.; Ramalingam, P.V. Kinetics of Phenol Biodegradation by Heavy Metal Tolerant Rhizobacteria Glutamicibacter nicotianae MSSRFPD35 from Distillery Effluent Contaminated Soils. Front. Microbiol. 2020, 11, 1573. [Google Scholar] [CrossRef] [PubMed]
- Radziff, S.B.M.; Ahmad, S.A.; Shaharuddin, N.A.; Merican, F.; Kok, Y.-Y.; Zulkharnain, A.; Gomez-Fuentes, C.; Wong, C.-Y. Potential Application of Algae in Biodegradation of Phenol: A Review and Bibliometric Study. Plants 2021, 10, 2677. [Google Scholar] [CrossRef] [PubMed]
- Guerin, T.F. The effect of interactions between soil compaction and phenol contamination on plant growth characteristics: Implications for scaling bioremediation at industrial sites. J. Environ. Manag. 2022, 302, 114017. [Google Scholar] [CrossRef]
- Li, H.; Zhang, G.C.; Xie, H.C.; Li, K.; Zhang, S.Y. The effects of the phenol concentrations on photosynthetic parameters of Salix babylonica L. Photosynthetica 2015, 53, 430–435. [Google Scholar] [CrossRef]
- GESAMP. Revised GESAMP: Hazard Evaluation Procedure for Chemical Substances Carried by Ships, 2nd ed.; International Maritime Organization: London, UK.
- Duan, W.; Meng, F.; Cui, H.; Lin, Y.; Wang, G.; Wu, J. Ecotoxicity of phenol and cresols to aquatic organisms: A review. Ecotoxicol. Environ. Saf. 2018, 157, 441–456. [Google Scholar] [CrossRef]
- Canadian Council of Ministers of the Environment. Canadian water quality guidelines for the protection of aquatic life: Phenols—Mono- and dihydric phenols. In Canadian Environmental Quality Guidelines; Canadian Council of Ministers of the Environment: Winnipeg, MB, Canda, 1999. [Google Scholar]
- Public Health England. Phenol General Information; PHE Publications: Letchworth, UK, 2016; pp. 1–4.
- Hannaford, A.M.; Kuek, C. Aerobic batch degradation of phenol using immobilized Pseudomonas putida. J. Ind. Microbiol. Biotechnol. 1999, 22, 121–126. [Google Scholar] [CrossRef]
- Arutchelvan, V.; Kanakasabai, V.; Elangovan, R.; Nagarajan, S.; Muralikrishnan, V. Kinetics of high strength phenol degradation using Bacillus brevis. J. Hazard. Mater. 2006, 29, 216–222. [Google Scholar] [CrossRef]
- World Meteorological Organization (WMO). 2021 State of Climate Services: Water; World Meteorological Organization (WMO): Geneva, Switzerland, 2021. [Google Scholar]
- Said, K.A.M.; Ismail, A.F.; Karim, Z.A.; Abdullah, M.S.; Hafeez, A. A review of technologies for the phenolic compounds recovery and phenol removal from wastewater. Process. Saf. Environ. Prot. 2021, 151, 257–289. [Google Scholar] [CrossRef]
- Nakhli, S.A.A.; Ahmadizadeh, K.; Fereshtehnejad, M.; Rostami, M.H.; Safari, M.; Borghei, S.M. Biological removal of phenol from saline wastewater using a moving bed biofilm reactor containing acclimated mixed consortia. Springerplus 2014, 3, 112. [Google Scholar] [CrossRef] [Green Version]
- Pradeep, N.V.; Anupama, S.; Navya, K.; Shalini, H.N.; Idris, M.; Hampannavar, U.S. Biological removal of phenol from wastewaters: A mini review. Appl. Water Sci. 2015, 5, 105–112. [Google Scholar] [CrossRef]
- Inglezakis, V.; Malamis, S.; Omirkhan, A.; Nauruzbayeva, J.; Makhtayeva, Z.; Seidakhmetov, T.; Kudarova, A. Investigating the inhibitory effect of cyanide, phenol and 4-nitrophenol on the activated sludge process employed for the treatment of petroleum wastewater. J. Environ. Manag. 2017, 203, 825–830. [Google Scholar] [CrossRef]
- Rongsayamanont, C.; Khongkhaem, P.; Luepromchai, E.; Khan, E. Inhibitory effect of phenol on wastewater ammonification. Bioresour. Technol. 2020, 309, 123312. [Google Scholar] [CrossRef] [PubMed]
- Herrero, M.; Stuckey, D. Bioaugmentation and its application in wastewater treatment: A review. Chemosphere 2015, 140, 119–128. [Google Scholar] [CrossRef]
- Bajaj, M.; Gallert, C.; Winter, J. Biodegradation of high phenol containing synthetic wastewater by an aerobic fixed bed reactor. Bioresour. Technol. 2008, 99, 8376–8381. [Google Scholar] [CrossRef]
- Sivasubramanian, S.; Namasivayam, S.K.R. Phenol degradation studies using microbial consortium isolated from environmental sources. J. Environ. Chem. Eng. 2015, 3, 243–252. [Google Scholar] [CrossRef]
- Felföldi, T.; Nagymáté, Z.; Székely, A.J.; Jurecska, L.; Márialigeti, K. Biological treatment of coke plant effluents: From a microbiological perspective. Biol. Futur. 2020, 71, 359–370. [Google Scholar] [CrossRef]
- Hao, O.J.; Kim, M.H.; Seagren, E.A.; Kim, H. Kinetics of phenol and chlorophenol utilization by Acinetobacter species. Chemosphere 2002, 46, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Poi, G.; Aburto-Medina, A.; Mok, P.C.; Ball, A.S.; Shahsavari, E. Bioremediation of Phenol-Contaminated Industrial Wastewater Using a Bacterial Consortium—From Laboratory to Field. Water Air Soil Pollut. 2017, 228, 89. [Google Scholar] [CrossRef]
- Silva, C.C.; Hayden, H.; Sawbridge, T.; Mele, P.; De Paula, S.O.; Silva, L.C.F.; Vidigal, P.M.P.; Vicentini, R.; Sousa, M.P.; Torres, A.P.R.; et al. Identification of Genes and Pathways Related to Phenol Degradation in Metagenomic Libraries from Petroleum Refinery Wastewater. PLoS ONE 2013, 8, e61811. [Google Scholar] [CrossRef] [Green Version]
- Van Schie, P.M.; Young, L.Y. Biodegradation of Phenol: Mechanisms and Applications. Bioremediation J. 2000, 4, 1–18. [Google Scholar] [CrossRef]
- Zhang, X.; Gao, P.; Chao, Q.; Wang, L.; Senior, E.; Zhao, L. Microdiversity of phenol hydroxylase genes among phenol-degrading isolates of Alcaligenes sp. from an activated sludge system. FEMS Microbiol. Lett. 2004, 237, 369–375. [Google Scholar] [CrossRef] [Green Version]
- Szilveszter, S.; Fikó, D.-R.; Máthé, I.; Felföldi, T.; Ráduly, B. Kinetic characterization of a new phenol degrading Acinetobacter towneri strain isolated from landfill leachate treating bioreactor. World J. Microbiol. Biotechnol. 2023, 39, 1–11. [Google Scholar] [CrossRef] [PubMed]
- APHA. Standard Methods for the Examination of Water and Wastewater; American Public Health Association (APHA): Washington, DC, USA, 2005.
- Watanabe, K.; Teramoto, M.; Futamata, H.; Harayama, S. Molecular Detection, Isolation, and Physiological Characterization of Functionally Dominant Phenol-Degrading Bacteria in Activated Sludge. Appl. Environ. Microbiol. 1998, 64, 4396–4402. [Google Scholar] [CrossRef]
- Nopens, I.; Capalozza, C.; Vanrolleghem, P.A. Stability Analysis of a Synthetic Municipal Wastewater (Technical Report); Department of Applied Mathematics Biometrics and Process Control, University of Gent: Gent, Belgium, 2001. [Google Scholar]
- Xu, N.; Qiu, C.; Yang, Q.; Zhang, Y.; Wang, M.; Ye, C.; Guo, M. Analysis of Phenol Biodegradation in Antibiotic and Heavy Metal Resistant Acinetobacter lwoffii NL1. Front. Microbiol. 2021, 12, 725755. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Du, D.; Zhou, W.; Zeng, X.; Cheng, G. Phenol degradation and genotypic analysis of dioxygenase genes in bacteria isolated from sediments. Braz. J. Microbiol. 2017, 48, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xie, W.; Li, D.; Peng, Y.; Li, Z.; Liu, S. Biodegradation of Phenol by Bacteria Strain Acinetobacter Calcoaceticus PA Isolated from Phenolic Wastewater. Int. J. Environ. Res. Public Health 2016, 13, 300. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tian, Y.; Han, B.; Zhao, H.-B.; Bi, J.-N.; Cai, B.-L. Biodegradation of phenol by free and immobilized Acinetobacter sp. strain PD12. J. Environ. Sci. 2007, 19, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Adav, S.S.; Chen, M.-Y.; Lee, D.-J.; Ren, N.-Q. Degradation of phenol by Acinetobacter strain isolated from aerobic granules. Chemosphere 2007, 67, 1566–1572. [Google Scholar] [CrossRef]
- Gracioso, L.H.; Vieira, P.B.; Baltazar, M.P.G.; Avanzi, I.R.; Karolski, B.; Nascimento, C.A.O.; Perpetuo, E.A. Removal of phenolic compounds from raw industrial wastewater by Achromobacter sp. isolated from a hydrocarbon-contaminated area. Water Environ. J. 2018, 33, 40–50. [Google Scholar] [CrossRef] [Green Version]
- Alexievaa, Z.; Gerginova, M.; Zlateva, P.; Peneva, N. Comparison of growth kinetics and phenol metabolizing enzymes of Trichosporon cutaneum R57 and mutants with modified degradation abilities. Enzym. Microb. Technol. 2004, 34, 242–247. [Google Scholar] [CrossRef]
- Adav, S.S.; Chen, M.-Y.; Lee, D.-J.; Ren, N.-Q. Degradation of phenol by aerobic granules and isolated yeast Candida tropicalis. Biotechnol. Bioeng. 2006, 96, 844–852. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-H.; Cheng, Y.-S. Phenol Degradation Kinetics by Free and Immobilized Pseudomonas putida BCRC 14365 in Batch and Continuous-Flow Bioreactors. Processes 2020, 8, 721. [Google Scholar] [CrossRef]
- Kamali, M.; Gameiro, T.; Costa, M.E.; Capela, I.; Aminabhavi, T.M. Enhanced biodegradation of phenolic wastewaters with acclimatized activated sludge—A kinetic study. Chem. Eng. J. 2019, 378, 122186. [Google Scholar] [CrossRef]
- Lin, H.; Chen, G.; Zhang, Z.; Ren, Y. Exploration of the mechanism of 2-CP degradation by Acinetobacter sp. stimulated by Lactobacillus plantarum fermentation waste: A bio-waste reuse. Environ. Res. 2023, 231, 116119. [Google Scholar] [CrossRef]
- Wang, P.; Wei, H.; Ke, T.; Fu, Y.; Zeng, Y.; Chen, C.; Chen, L. Characterization and genome analysis of Acinetobacter oleivorans S4 as an efficient hydrocarbon-degrading and plant-growth-promoting rhizobacterium. Chemosphere 2023, 331, 138732. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Li, S.; Rene, E.R.; Lun, X.; Zhang, P.; Ma, W. Detoxification of fluoroglucocorticoid by Acinetobacter pittii C3 via a novel defluorination pathway with hydrolysis, oxidation and reduction: Performance, genomic characteristics, and mechanism. J. Hazard. Mater. 2023, 452, 131302. [Google Scholar] [CrossRef]
- Tang, L.; Zhao, X.; Chen, X.; Jiang, Y.; Gudda, F.; Wang, Y.; Ling, W. Distribution of bound-PAH residues and their correlations with the bacterial community at different depths of soil from an abandoned chemical plant site. J. Hazard. Mater. 2023, 453, 131328. [Google Scholar] [CrossRef] [PubMed]
- Zhan, H.; Wang, H.; Liao, L.; Feng, Y.; Fan, X.; Zhang, L.; Chen, S. Kinetics and Novel Degradation Pathway of Permethrin in Acinetobacter baumannii ZH-14. Front. Microbiol. 2018, 9, 98. [Google Scholar] [CrossRef] [Green Version]
- Ye, T.; Zhou, T.; Fan, X.; Bhatt, P.; Zhang, L.; Chen, S. Acinetobacter lactucae Strain QL-1, a Novel Quorum Quenching Candidate Against Bacterial Pathogen Xanthomonas campestris pv. campestris. Front. Microbiol. 2019, 10, 2867. [Google Scholar] [CrossRef] [Green Version]
- Ye, T.; Zhou, T.; Xu, X.; Zhang, W.; Fan, X.; Mishra, S.; Zhang, L.; Zhou, X.; Chen, S. Whole-Genome Sequencing Analysis of Quorum Quenching Bacterial Strain Acinetobacter lactucae QL-1 Identifies the FadY Enzyme for Degradation of the Diffusible Signal Factor. Int. J. Mol. Sci. 2020, 21, 6729. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Luo, Q.; Zhang, Y.; Fan, X.; Ye, T.; Mishra, S.; Bhatt, P.; Zhang, L.; Chen, S. Quorum Quenching in a Novel Acinetobacter sp. XN-10 Bacterial Strain against Pectobacterium carotovorum subsp. carotovorum. Microorganisms 2020, 8, 1100. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Liu, R.; Liu, C.; Chen, L. Bioaugmentation with isolated strains for the removal of toxic and refractory organics from coking wastewater in a membrane bioreactor. Biodegradation 2015, 26, 465–474. [Google Scholar] [CrossRef]
- Fuller, M.E.; Hatzinger, P.B.; Condee, C.W.; Andaya, C.; Vainberg, S.; Michalsen, M.M.; Crocker, F.H.; Indest, K.J.; Jung, C.M.; Eaton, H.; et al. Laboratory evaluation of bioaugmentation for aerobic treatment of RDX in groundwater. Biodegradation 2015, 26, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Tchelet, R.; Meckenstock, R.; Steinle, P.; van der Meer, J.R. Population dynamics of an introduced bacterium degrading chlorinated benzenes in a soil column and in sewage sludge. Biodegradation 1999, 10, 113–125. [Google Scholar] [CrossRef]


| mg·L−1 | COD, mg·L−1 | N, mg·L−1 | P, mg·L−1 | g·2 L−1 conc. = 240 L | |
|---|---|---|---|---|---|
| Chemical Compounds | |||||
| Urea | 91.74 | 23.22 | 42.81 | 0 | 22.01 |
| NH4Cl | 12.75 | 0 | 3.52 | 0 | 3.06 |
| Na-acetate 3H2O | 131.64 | 79.37 | 0 | 0 | 31.59 |
| Peptone | 17.41 | 17.41 | 0.67 | 0 | 4.17 |
| MgHPO4·3H2O | 29.02 | 0 | 0 | 5.14 | 6.96 |
| KH2PO4 | 23.40 | 0 | 0 | 3.14 | 5.61 |
| FeSO4·7H2O | 5.80 | 0 | 0 | 0 | 1.39 |
| Main carbon sources | |||||
| Starch | 122.00 | 122.00 | 0 | 0 | 29.28 |
| Milk powder | 116.19 | 116.19 | 6.95 | 1.14 | 27.88 |
| Yeast extract | 52.24 | 52.24 | 6.28 | 0 | 12.53 |
| Soy oil | 29.02 | 29.02 | 0 | 0 | 6.96 |
| Total | 631.21 | 439.45 | 60.23 | 9.42 | 151.44 |
| Trace metals | mg metal/L | ||||
| Cr(NO3)3·9H2O | 0.770 | 0.100 | 0.1848 | ||
| CuCl2·2H2O | 0.536 | 0.200 | 0.1286 | ||
| MnSO4·H2O | 0.108 | 0.035 | 0.0259 | ||
| NiSO4·6H2O | 0.336 | 0.075 | 0.0806 | ||
| PbCl2 | 0.100 | 0.075 | 0.0240 | ||
| ZnCl2 | 0.208 | 0.100 | 0.0499 | ||
| Total | 2.058 | 0.585 | 0.4938 | ||
| Bacterial Strain/Phenol Concentration | 100 mg·L−1 | 500 mg·L−1 | ||||||
|---|---|---|---|---|---|---|---|---|
| Lag Time (h) | µ | µmax | mg Phenol·L−1·h−1 | Lag Time (h) | µ | µmax | mg Phenol·L−1·h−1 | |
| A. towneri CFII-87 | 0 | 0.42 | 0.59 | 19.66 | 0 | 0.23 | 0.30 | 31.25 |
| A. johnsonii CFII-99A | 3 | 0.36 | 0.50 | 12.86 | 12 | 0.14 | 0.20 | 15.68 |
| A. sp. CFII-98 | 4 | 0.46 | 0.64 | 11.90 | 8 | 0.21 | 0.29 | 25.68 |
| Time Interval (Hour)/ Sample | mg Phenol·g VSS h−1 | |||
|---|---|---|---|---|
| A. towneri CFII-87 | A. johnsonii CFII-99A | A. sp. CFII-98 | Consortium | |
| 1–4 | 76.07 ± 11.79 | 63.08 ± 18.08 | 42.76 ± 3.29 | 69.54 ± 15.70 |
| 4–8 | 100.82 ± 7.42 | 72.78 ± 15.39 | 59.26 ± 2.34 | 81.26 ± 0.72 |
| 8–12 | 282.11 ± 13.34 | 90.41 ± 9.78 | 178.84 ± 20.95 | 93.20 ± 5.48 |
| 12–16 | 30.46 ± 0.94 | 141.01 ± 11.94 | 133.06 ± 7.82 | 146.76 ± 3.33 |
| 16–24 | 21.75 ± 0.95 | |||
| Average | 122.37 ± 1.83 | 77.81 ± 1.41 | 103.48 ± 1.87 | 97.69 ± 3.69 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fikó, D.-R.; Ráduly, B.; Máthé, I.; Felföldi, T.; Lányi, S.; Szilveszter, S. Bioaugmentation Potential Investigation Using a Phenol Affinity Analysis of Three Acinetobacter Strains in a Multi-Carbon-Source Condition. Water 2023, 15, 2815. https://doi.org/10.3390/w15152815
Fikó D-R, Ráduly B, Máthé I, Felföldi T, Lányi S, Szilveszter S. Bioaugmentation Potential Investigation Using a Phenol Affinity Analysis of Three Acinetobacter Strains in a Multi-Carbon-Source Condition. Water. 2023; 15(15):2815. https://doi.org/10.3390/w15152815
Chicago/Turabian StyleFikó, Dezső-Róbert, Botond Ráduly, István Máthé, Tamás Felföldi, Szabolcs Lányi, and Szabolcs Szilveszter. 2023. "Bioaugmentation Potential Investigation Using a Phenol Affinity Analysis of Three Acinetobacter Strains in a Multi-Carbon-Source Condition" Water 15, no. 15: 2815. https://doi.org/10.3390/w15152815
APA StyleFikó, D.-R., Ráduly, B., Máthé, I., Felföldi, T., Lányi, S., & Szilveszter, S. (2023). Bioaugmentation Potential Investigation Using a Phenol Affinity Analysis of Three Acinetobacter Strains in a Multi-Carbon-Source Condition. Water, 15(15), 2815. https://doi.org/10.3390/w15152815






