Enhancement of Microbial and Metabolic Mechanisms in an Aerobic Bioreactor with Immobilized Microflora by Simple and Complex Electron Donors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Screening and Isolation of N Removal Microbes (MZ-5, S2, and S9)
2.2. Preparation of Polymer Complex Beads Carrying Microflora
2.3. Assessment of N Removal Potential of the Microflora Using Different Electron Donors
2.4. Establishment of the Aerobic Reactor Device and Operation Conditions
2.5. Metagenomic DNA Extraction and 16S rRNA Sequencing
2.6. Analytical Methods
3. Results and Discussions
3.1. Selection of Simple Electron Donor for the Microflora
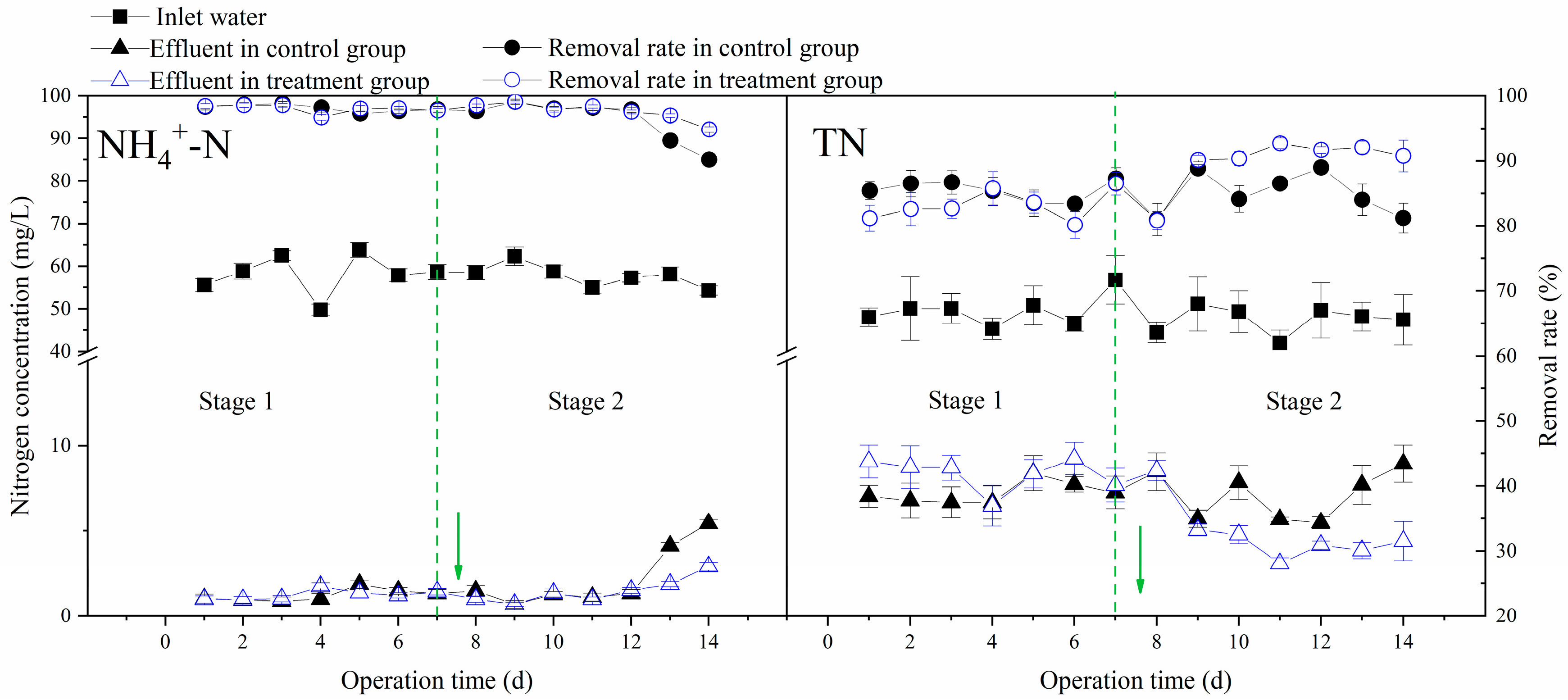
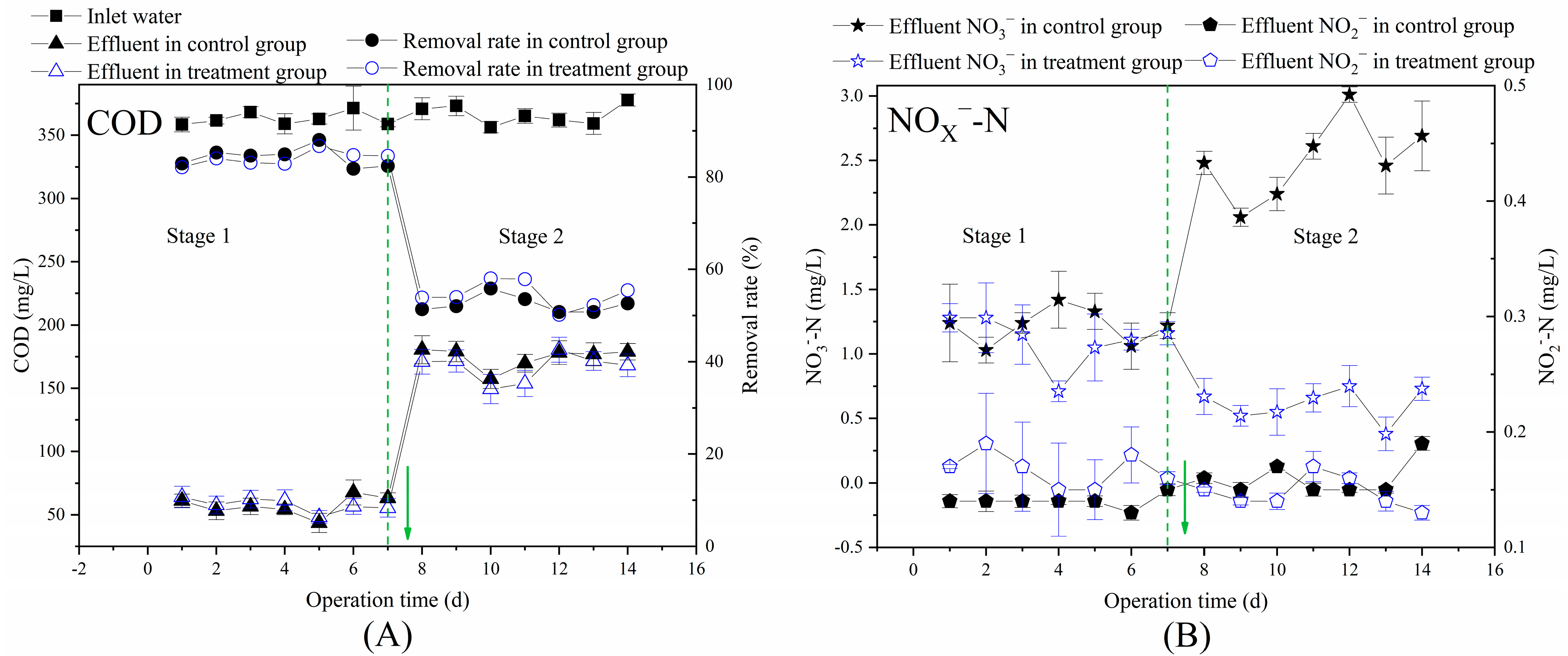
3.2. Effects of Multi-Electron Donors on the Performance of Aerobic Bioreactors
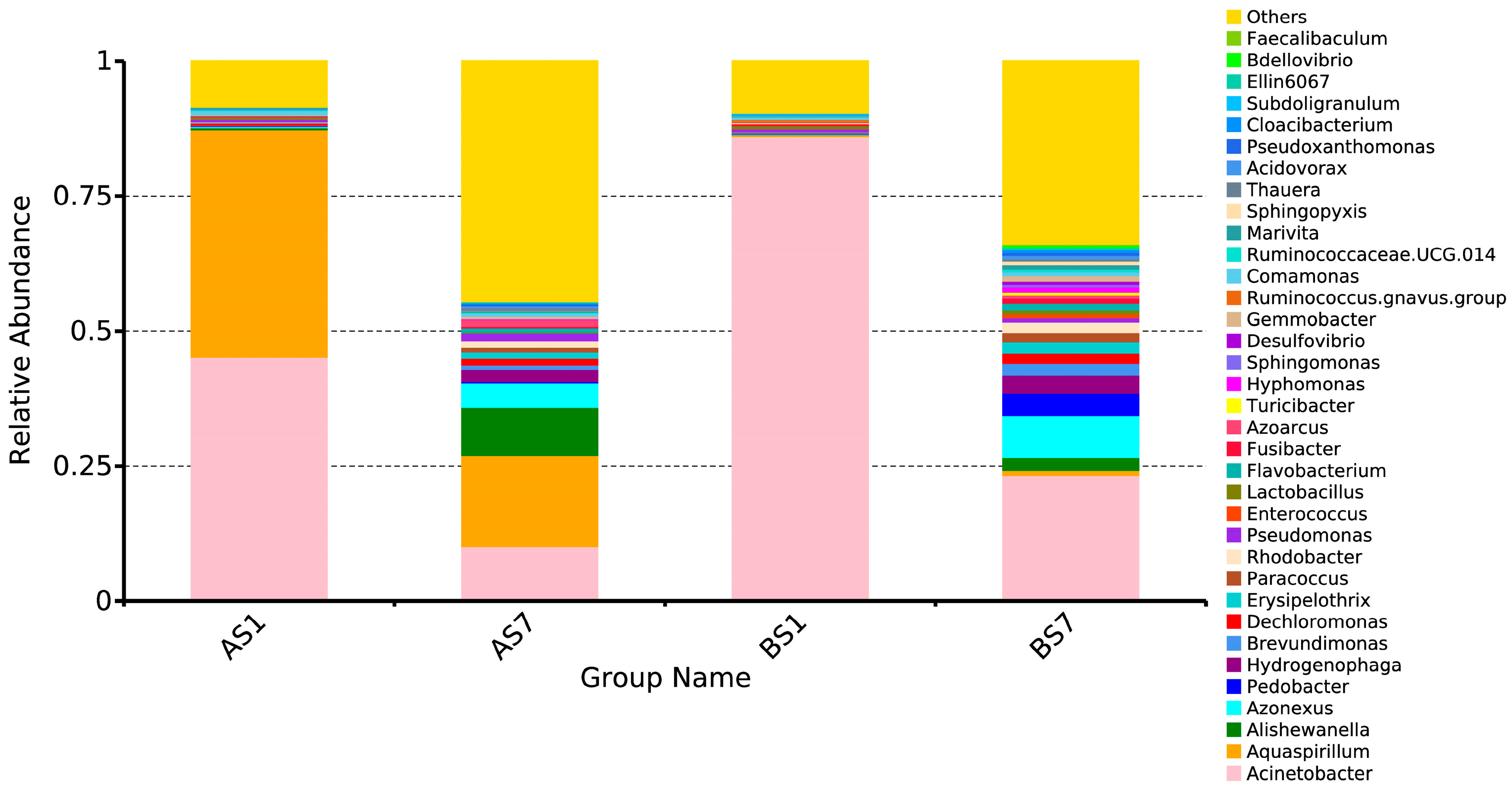
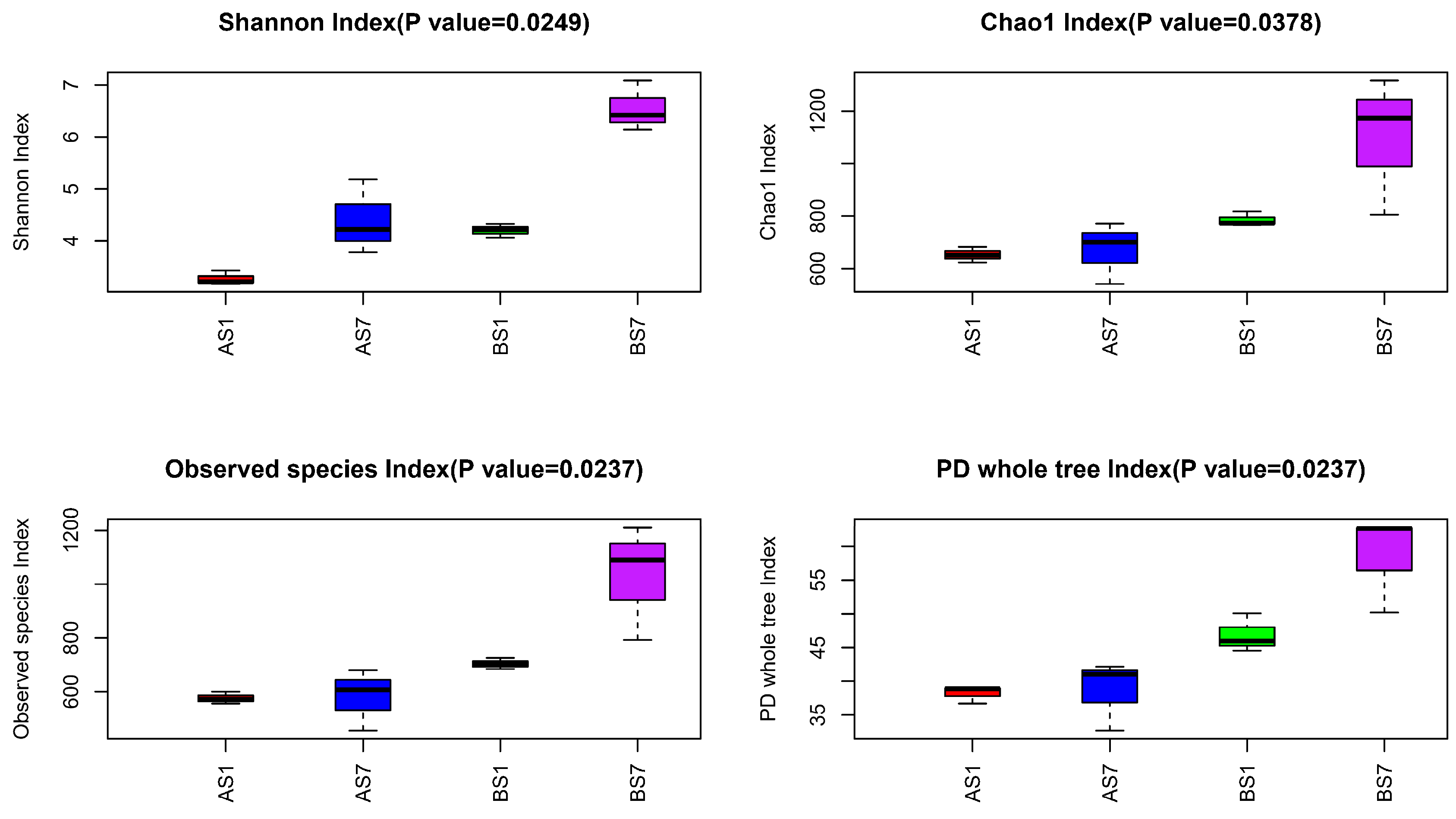
3.3. The Relationships between the Microbial Diversity and Electron Donor
3.4. Role of Non-Dominant Bacteria and The Characteristic Genus of the Treatment Groups
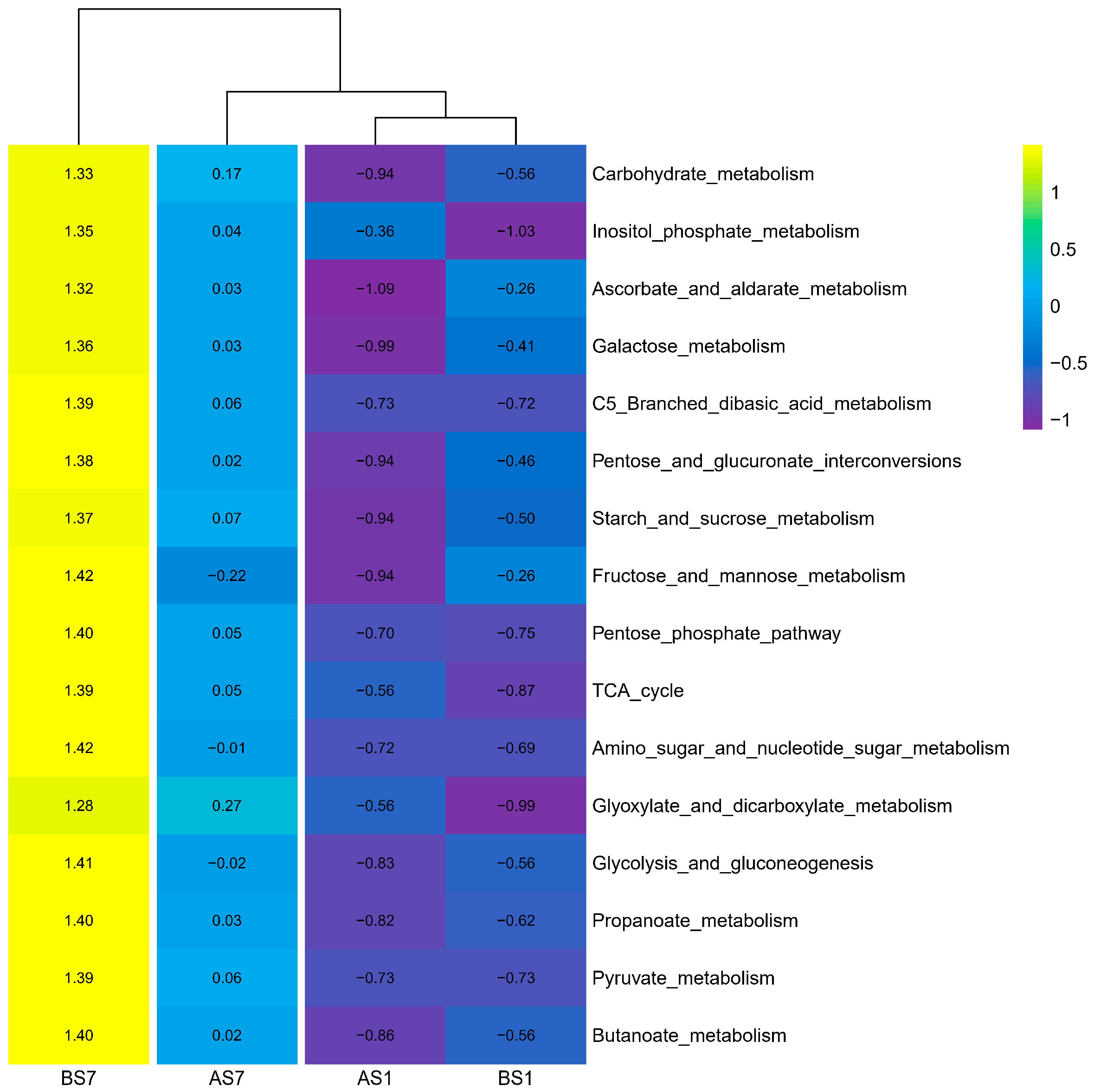
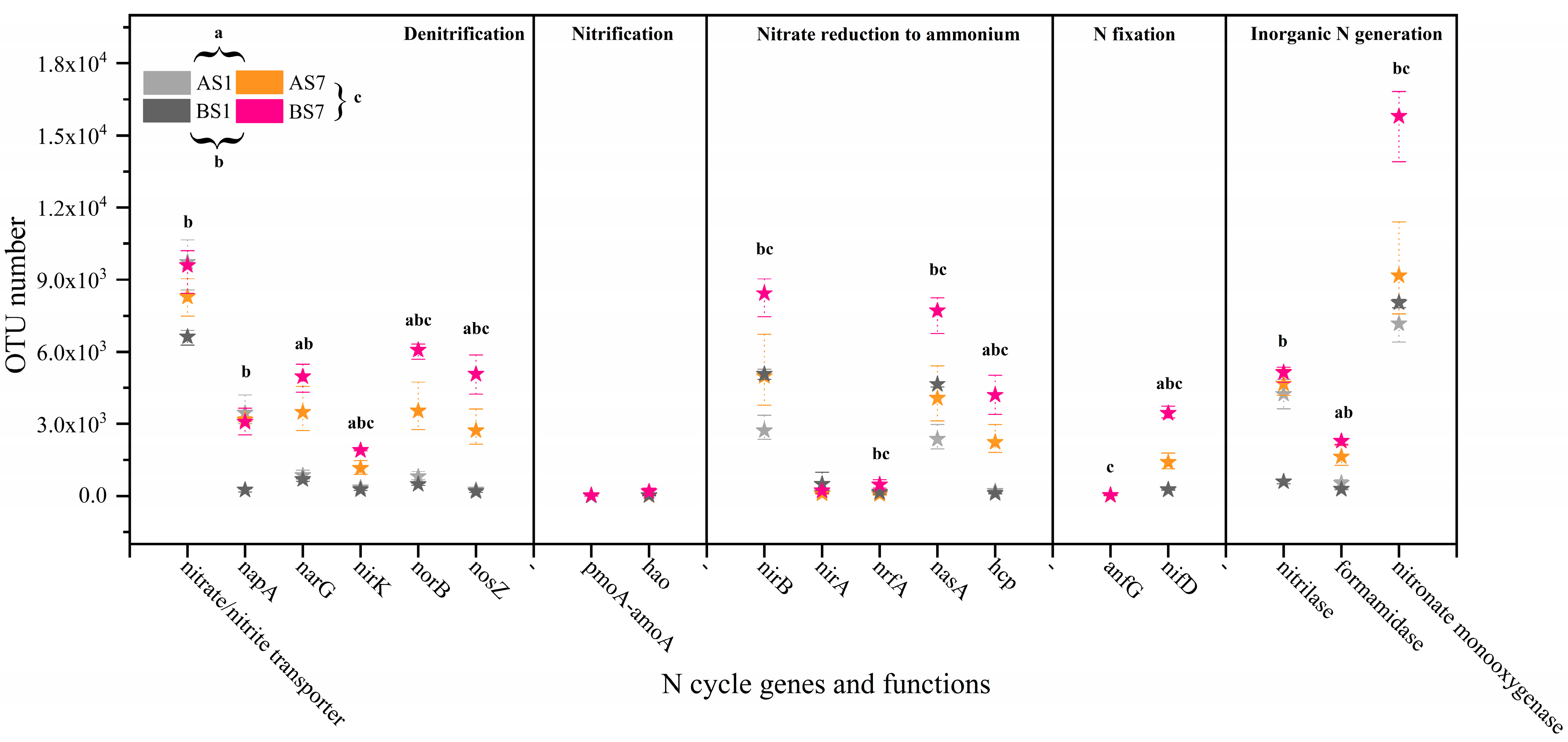
3.5. C Metabolism and N Cycle Genes of Activated Sludge
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wang, X.; Xia, S.; Chen, L.; Zhao, J.; Renault, N.; Chovelon, J. Nutrients removal from municipal wastewater by chemical precipitation in a moving bed biofilm reactor. Process. Biochem. 2006, 41, 824–828. [Google Scholar] [CrossRef]
- Kuai, L.P.; Verstraete, W. Ammonium removal by the oxygen-limited autotrophic nitrification-denitrification system. Appl. Environ. Microb. 1998, 64, 4500–4506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Peng, Y.; Liu, X.; Zeng, W.; Mino, T.; Satoh, H. Nitrogen Removal via Nitrite from Municipal Wastewater at Low Temperatures Using Real-Time Control to Optimize Nitrifying Communities. Environ. Sci. Technol. 2007, 41, 8159–8164. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Li, H.; Wang, S.; Wang, Y.; Li, A.; Feng, X.; Li, J. Enhanced proteins and amino acids production based on ammonia nitrogen assimilation and sludge increment by the integration of bioadsorption with anaerobic-anoxic-oxic (AAO) process. Chemosphere 2021, 280, 130721. [Google Scholar] [CrossRef]
- Park, J.Y.; Yoo, Y.J. Biological nitrate removal in industrial wastewater treatment: Which electron donor we can choose. Appl. Microbiol. Biotechnol. 2009, 82, 415–429. [Google Scholar] [CrossRef]
- Isaacs, S.; Henze, M.; Søeberg, H.; Kümmel, M. External carbon source addition as a means to control an activated sludge nutrient removal process. Water Res. 1994, 28, 511–520. [Google Scholar] [CrossRef]
- Hu, W.; Tian, J.; Li, X.; Chen, L. Wastewater treatment system optimization with an industrial symbiosis model: A case study of a Chinese eco-industrial park. J. Ind. Ecol. 2020, 24, 1338–1351. [Google Scholar] [CrossRef]
- Pang, Y.; Wang, J. Various electron donors for biological nitrate removal: A review. Sci. Total Environ. 2021, 794, 148699. [Google Scholar] [CrossRef]
- Castellar, J.; Formosa, J.; Fernandez, A.I.; Jové, P.; Bosch, M.G.; Morató, J.; Brix, H.; Arias, C.A. Cork as a sustainable carbon source for nature-based solutions treating hydroponic wastewaters—Preliminary batch studies. Sci. Total Environ. 2019, 650, 267–276. [Google Scholar] [CrossRef]
- Fowdar, H.S.; Hatt, B.E.; Breen, P.; Cook, P.L.; Deletic, A. Evaluation of sustainable electron donors for nitrate removal in different water media. Water Res. 2015, 85, 487–496. [Google Scholar] [CrossRef]
- Hu, R.; Zheng, X.; Xin, J.; Sun, Z.; Zheng, T. Selective enhancement and verification of woody biomass digestibility as a denitrification carbon source. Bioresour. Technol. 2017, 244, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Si, Z.; Song, X.; Wang, Y.; Cao, X.; Zhao, Y.; Wang, B.; Chen, Y.; Arefe, A. Intensified heterotrophic denitrification in constructed wetlands using four solid carbon sources: Denitrification efficiency and bacterial community structure. Bioresour. Technol. 2018, 267, 416–425. [Google Scholar] [CrossRef]
- Deng, Q.; Wan, L.; Li, X.; Cao, X.; Zhou, Y.; Song, C. Metagenomic evidence reveals denitrifying community diversity rather than abundance drives nitrate removal in stormwater biofilters amended with different organic and inorganic electron donors. Chemosphere 2020, 257, 127269. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Deng, Q.; Li, X.; Tang, M.; Ma, X.; Cao, X.; Zhou, Y.; Song, C. Organic carbon quantity and quality jointly triggered the switch between dissimilatory nitrate reduction to ammonium (DNRA) and denitrification in biofilters. Chemosphere 2021, 280, 130917. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Ma, C.; Huang, X.; Liu, J.; Lu, L.; Peng, K.; Li, S. Research progress in solid carbon source–based denitrification technologies for different target water bodies. Sci. Total Environ. 2021, 782, 146669. [Google Scholar] [CrossRef]
- Ling, Y.; Yan, G.; Wang, H.; Dong, W.; Wang, H.; Chang, Y.; Chang, M.; Li, C. Release Mechanism, Secondary Pollutants and Denitrification Performance Comparison of Six Kinds of Agricultural Wastes as Solid Carbon Sources for Nitrate Removal. Int. J. Environ. Res. Public Health 2021, 18, 1232. [Google Scholar] [CrossRef]
- Tao, M.; Jing, Z.; Tao, Z.; Luo, H.; Zuo, S. Improvements of nitrogen removal and electricity generation in microbial fuel cell-constructed wetland with extra corncob for carbon-limited wastewater treatment. J. Clean. Prod. 2021, 297, 126639. [Google Scholar] [CrossRef]
- Zhong, H.; Cheng, Y.; Ahmad, Z.; Shao, Y.; Zhang, H.; Lu, Q.; Shim, H. Solid-phase denitrification for water remediation: Processes, limitations, and new aspects. Crit. Rev. Biotechnol. 2020, 40, 1113–1130. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.-X.; Shao, L.; Yin, H.-L.; Chu, H.-Q.; Yao, Y.-J. Biological Denitrification Using Corncobs as a Carbon Source and Biofilm Carrier. Water Environ. Res. 2009, 81, 242–247. [Google Scholar] [CrossRef]
- Li, G.; Chen, J.; Yang, T.; Sun, J.; Yu, S. Denitrification with corncob as carbon source and biofilm carriers. Water Sci. Technol. 2012, 65, 1238–1243. [Google Scholar] [CrossRef]
- Li, C.; Wang, H.; Yan, G.; Dong, W.; Chu, Z.; Wang, H.; Chang, Y.; Ling, Y.; Zhang, Y. Initial carbon release characteristics, mechanisms and denitrification performance of a novel slow release carbon source. J. Environ. Sci. 2022, 118, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Xiong, R.; Yu, X.; Zhang, Y.; Peng, Z.; Yu, L.; Cheng, L.; Li, T. Comparison of agricultural wastes and synthetic macromolecules as solid carbon source in treating low carbon nitrogen wastewater. Sci. Total Environ. 2020, 739, 139885. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Tan, H. Alginate-Based Biomaterials for Regenerative Medicine Applications. Materials 2013, 6, 1285–1309. [Google Scholar] [CrossRef] [PubMed]
- Chang, Q.; Ali, A.; Su, J.; Wen, Q.; Bai, Y.; Gao, Z.; Xiong, R. Efficient removal of nitrate, manganese, and tetracycline by a polyvinyl alcohol/sodium alginate with sponge cube immobilized bioreactor. Bioresour. Technol. 2021, 331, 125065. [Google Scholar] [CrossRef]
- Warneke, S.; Schipper, L.A.; Matiasek, M.G.; Scow, K.M.; Cameron, S.; Bruesewitz, D.A.; McDonald, I.R. Nitrate removal, communities of denitrifiers and adverse effects in different carbon substrates for use in denitrification beds. Water Res. 2011, 45, 5463–5475. [Google Scholar] [CrossRef] [Green Version]
- Khin, T.; Annachhatre, A.P. Novel microbial nitrogen removal processes. Biotechnol. Adv. 2004, 22, 519–532. [Google Scholar] [CrossRef]
- Joo, H.-S.; Hirai, M.; Shoda, M. Characteristics of ammonium removal by heterotrophic nitrification-aerobic denitrification by Alcaligenes faecalis No. 4. J. Biosci. Bioeng. 2005, 100, 184–191. [Google Scholar] [CrossRef]
- Xu, N.; Liao, M.; Liang, Y.; Guo, J.; Zhang, Y.; Xie, X.; Fan, Q.; Zhu, Y. Biological nitrogen removal capability and pathways analysis of a novel low C/N ratio heterotrophic nitrifying and aerobic denitrifying bacterium (Bacillus thuringiensis strain WXN-23). Environ. Res. 2021, 195, 110797. [Google Scholar] [CrossRef]
- Ouyang, L.; Wang, K.; Liu, X.; Wong, M.H.; Hu, Z.; Chen, H.; Yang, X.; Li, S. A study on the nitrogen removal efficacy of bacterium Acinetobacter tandoii MZ-5 from a contaminated river of Shenzhen, Guangdong Province, China. Bioresour. Technol. 2020, 315, 123888. [Google Scholar] [CrossRef]
- Xie, N.; Zhong, L.; Ouyang, L.; Xu, W.; Zeng, Q.; Wang, K.; Zaynab, M.; Chen, H.; Xu, F.; Li, S. Community Composition and Function of Bacteria in Activated Sludge of Municipal Wastewater Treatment Plants. Water 2021, 13, 852. [Google Scholar] [CrossRef]
- APHA. Standard Methods for the Examination of Water and Wastewater, 22nd ed.; American Public Health Association (APHA): Washington, DC, USA, 2012. [Google Scholar]
- Jiang, F.; Qi, Y.; Shi, X. Effect of liquid carbon sources on nitrate removal, characteristics of soluble microbial products and microbial community in denitrification biofilters. J. Clean. Prod. 2022, 339, 130776. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Z.; Li, J.; Liu, D.; Yuan, Y.; Chen, Z.; Wang, G. Cooperation between two strains of Enterobacter and Klebsiella in the simultaneous nitrogen removal and phosphate accumulation processes. Bioresour. Technol. 2019, 291, 121854. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, Z.; Zhao, M.; Yuan, B.; Yao, J.; Chen, J.; Hrynshpan, D.; Savitskaya, T. A fungus–bacterium co-culture synergistically promoted nitrogen removal by enhancing enzyme activity and electron transfer. Sci. Total Environ. 2021, 754, 142109. [Google Scholar] [CrossRef] [PubMed]
- Van Hofwegen, D.J.; Hovde, C.J.; Minnich, S.A. Rapid Evolution of Citrate Utilization by Escherichia coli by Direct Selection Requires citT and dctA. J. Bacteriol. 2016, 198, 1022–1034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiu, B.; Liang, S.-K.; He, X.-L.; Wang, X.-K.; Cui, Z.-G.; Jiang, Z.-J. Bioavailability of dissolved organic nitrogen and its uptake by Ulva prolifera: Implications in the outbreak of a green bloom off the coast of Qingdao, China. Mar. Pollut. Bull. 2019, 140, 563–572. [Google Scholar] [CrossRef]
- Sun, C.; Zhang, B.; Ning, D.; Zhang, Y.; Dai, T.; Wu, L.; Li, T.; Liu, W.; Zhou, J.; Wen, X. Seasonal dynamics of the microbial community in two full-scale wastewater treatment plants: Diversity, composition, phylogenetic group based assembly and co-occurrence pattern. Water Res. 2021, 200, 117295. [Google Scholar] [CrossRef]
- Grießmeier, V.; Gescher, J. Influence of the Potential Carbon Sources for Field Denitrification Beds on Their Microbial Diversity and the Fate of Carbon and Nitrate. Front. Microbiol. 2018, 9, 1313. [Google Scholar] [CrossRef]
- Zhou, Z.; Wang, C.; Luo, Y. Meta-analysis of the impacts of global change factors on soil microbial diversity and functionality. Nat. Commun. 2020, 11, 3072. [Google Scholar] [CrossRef]
- Gong, X.; Li, Q.; Li, T.; Li, C.; Huang, J.; Zhou, N.; Jia, X. Chemical composition and monolignin in alkali and acid treated corncob affect sugar release. Ind. Crop. Prod. 2022, 176, 114317. [Google Scholar] [CrossRef]
- Gao, H.; Mao, Y.; Zhao, X.; Liu, W.-T.; Zhang, T.; Wells, G. Genome-centric metagenomics resolves microbial diversity and prevalent truncated denitrification pathways in a denitrifying PAO-enriched bioprocess. Water Res. 2019, 155, 275–287. [Google Scholar] [CrossRef]
- Fesefeldt, A.; Kloos, K.; Bothe, H.; Lemmer, H.; Gliesche, C. Distribution of denitrification and nitrogen fixation genes in Hyphomicrobium spp. and other budding bacteria. Can. J. Microbiol. 1998, 44, 181–186. [Google Scholar] [CrossRef]
- Harter, J.; Weigold, P.; El-Hadidi, M.; Huson, D.H.; Kappler, A.; Behrens, S. Soil biochar amendment shapes the composition of N2O-reducing microbial communities. Sci. Total Environ. 2016, 562, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Li, X.; Zhang, M.; Liu, Z.; Han, Y.; Li, Q.; Zhou, W. Solid-phase denitrification in high salinity and low-temperature wastewater treatment. Bioresour. Technol. 2021, 341, 125801. [Google Scholar] [CrossRef]
- Aranda, C.; Godoy, F.; Becerra, J.; Barra, R.O.; Martinez, M. Aerobic secondary utilization of a non-growth and inhibitory substrate 2,4,6-trichlorophenol by Sphingopyxis chilensis S37 and Sphingopyxis-like strain S32. Biodegradation 2003, 14, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Mamoto, R.; Tang, M.; Kawai, F. Polyvinyl alcohol degradation by Sphingopyxis sp. strain 113P3. J. Biotechnol. 2008, 136, S308. [Google Scholar] [CrossRef]
- Kaminski, M.A.; Sobczak, A.; Dziembowski, A.; Lipinski, L. Genomic Analysis of γ-Hexachlorocyclohexane-Degrading Sphingopyxis lindanitolerans WS5A3p Strain in the Context of the Pangenome of Sphingopyxis. Genes 2019, 10, 688. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Duan, C.; Yu, J.; Dong, W. Transforming heterotrophic to autotrophic denitrification process: Insights into microbial community, interspecific interaction and nitrogen metabolism. Bioresour. Technol. 2022, 345, 126471. [Google Scholar] [CrossRef]
- Lin, Z.; Zhou, J.; He, L.; He, X.; Pan, Z.; Wang, Y.; He, Q. High-temperature biofilm system based on heterotrophic nitrification and aerobic denitrification treating high-strength ammonia wastewater: Nitrogen removal performances and temperature-regulated metabolic pathways. Bioresour. Technol. 2022, 344, 126184. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Z.; Chen, Y. Biochar Mitigates N2O Emission of Microbial Denitrification through Modulating Carbon Metabolism and Allocation of Reducing Power. Environ. Sci. Technol. 2021, 55, 8068–8078. [Google Scholar] [CrossRef]

| Component | Concentration (g/L) | Component | Concentration (g/L) |
|---|---|---|---|
| CH3COONa | 3.13 | MgSO4·7H2O | 0.05 |
| (NH4)2SO4 | 0.24 | MnSO4·4H2O | 0.01 |
| K2HPO4 | 0.25 | FeSO4·7H2O | 0.01 |
| NaCl | 0.125 | C:N | 18 |
| Strains | NH4+-N | NO2−-N | NO3−-N | TN |
|---|---|---|---|---|
| MZ-5 | 98.97 ± 0.09% | 67.44 ± 0.12% | 65.21 ± 0.12% | 68.52 ± 0.26% |
| S2 | 99.26 ± 0.13% | 7.74 ± 0.17% | 93.06 ± 0.16% | 69.69 ± 0.09% |
| S9 | 99.15 ± 0.03% | 17.51 ± 0.22% | 55.10 ± 0.15% | 60.05 ± 0.09% |
| Carbon Source | NH4+-N | NO2−-N | NO3−-N | TN |
|---|---|---|---|---|
| Disodium succinate | 99.58 ± 0.23% | 4.10 ± 0.08% | 81.14 ± 0.12% | 65.91 ± 0.22% |
| Sodium acetate | 99.34 ± 0.31% | 46.06 ± 0.15% | 98.58 ± 0.32% | 84.64 ± 0.16% |
| Trisodium citrate | 3.51 ± 0.11% | −4.15 ± 0.26% | 2.14 ± 0.12% | 10.24 ± 0.17% |
| Glucose | 56.41 ± 0.21% | 3.60 ± 0.23% | −4.79 ± 0.22% | 7.89 ± 0.13% |
| Sucrose | 41.84 ± 0.27% | 4.18 ± 0.14% | −9.43 ± 0.26% | 8.47 ± 0.22% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, Q.; Wang, K.; Xu, W.; Yu, X.; Feng, J.; Li, S.; Chen, H. Enhancement of Microbial and Metabolic Mechanisms in an Aerobic Bioreactor with Immobilized Microflora by Simple and Complex Electron Donors. Water 2023, 15, 2548. https://doi.org/10.3390/w15142548
Deng Q, Wang K, Xu W, Yu X, Feng J, Li S, Chen H. Enhancement of Microbial and Metabolic Mechanisms in an Aerobic Bioreactor with Immobilized Microflora by Simple and Complex Electron Donors. Water. 2023; 15(14):2548. https://doi.org/10.3390/w15142548
Chicago/Turabian StyleDeng, Qinghui, Keju Wang, Wang Xu, Xinfan Yu, Jie Feng, Shuangfei Li, and Huirong Chen. 2023. "Enhancement of Microbial and Metabolic Mechanisms in an Aerobic Bioreactor with Immobilized Microflora by Simple and Complex Electron Donors" Water 15, no. 14: 2548. https://doi.org/10.3390/w15142548
APA StyleDeng, Q., Wang, K., Xu, W., Yu, X., Feng, J., Li, S., & Chen, H. (2023). Enhancement of Microbial and Metabolic Mechanisms in an Aerobic Bioreactor with Immobilized Microflora by Simple and Complex Electron Donors. Water, 15(14), 2548. https://doi.org/10.3390/w15142548







