1. Introduction
Invasive plant species present a major threat to native diversity. After all, they can grow in every habitat [
1]. Globalization and human exchange over the last twenty years facilitated the introduction of several plant species to novel terrestrial regions [
2]. After introduction into new habitats, some plant species become invasive plants in these novel environments, and destructively affect the ecosystem, economy, and culture [
3]. Invasive plant species can capture resources from above and below ground [
4]. These abilities enable them to overcome the growth of their native competitor under mixed culture [
5]. Understanding the mechanisms by which invasive plant species sustain their growth under different environmental conditions is crucial for their management [
6].
The role of functional traits is very important for the successful invasion of invasive plant species under different habitat conditions [
7]. Functional traits also assist invasive plant species in boosting their growth under competition, and reduce the growth of their native competitor [
8]. Functional trait responses under different habitat conditions, such as nutrient availability, water fluctuation, and temperature variation, greatly impact invasive plant species’ effective invasion and the growth of native plants [
9]. Different functional traits play a different role in the effective invasion of these invasive plant species; for example, root and shoot functional traits are indicators of growth development [
10]. Additionally, photosynthesis, transpiration, and evaporation are the main indicators of leaf growth development [
11]. Therefore, understanding the responses of functional traits under different environmental conditions is very fruitful for managing invasive plant species.
Considering globalization, many environmental factors, i.e., water, nutrients, CO
2, light, and temperature, affect the growth of native plant species, but assist invasive plant species in sustaining their development because they prefer to grow in disturbed habitats [
6]. In wetland or riparian zones, the main issues are submergence and eutrophication. The former, called submergence, is a type of flooding during which the shoot of a plant is under water [
12]. Expulsion of water from riversides, dams, and canals will create submergence close to these areas, and vegetation will face submergence. Submergence imposes considerable stress by decreasing energy and carbohydrate values [
13]. Functional traits, such as shoot elongation and leaves, assist both invasive and native plant species in sustaining their growth under submergence, by enabling exposure to sunlight for photosynthesis [
5,
14]. Eutrophication is another environmental factor that negatively affects the aquatic ecosystem [
15]. Eutrophication decreases the growth of native plant species. It boosts the growth of invasive plant species because invasive plant species prefer to grow in an environment with rich resources [
16]. Eutrophication helps to overcome the stress of submergence by enhancing the shoot of invasive species because these plants can obtain more CO
2, light for photosynthesis, and oxygen for transpiration [
14]. Meanwhile, invasive plant species enhance their root length in order to capture resources below ground, and overcome the growth of their native competitor by enhancing their root length [
17]. Therefore, for managing invasive plant species under submergence and eutrophication habitats, the traits of different growth parameters assist us in understanding their invasion.
Growth parameters play the main role in successfully invading invasive plant species under different habitats, especially under competition. In the agriculture sector, many growth prediction models have been developed with the help of varying growth parameters, under additional irrigation and planting methods [
18,
19,
20]. Most modeling for the prediction of growth was done with the help of different statistical techniques such as correlation, path analysis, multiple linear regression (MLR), stepwise regression (SWR), factor analysis, and principal component analysis (PCA) [
21,
22,
23]. All of these methods presume to follow the linear relationship of input and output. These methods could not explain the complex relationship among the input variables and the output [
24]. These complex relationships required non-linear methods, such as Bayesian classification (BC), artificial neural network (ANN) models, genetic expression programming (GEP), and adaptive neuro-fuzzy inference systems (ANFIS) in order to overcome the drawback of these linear models and provide more accurate results [
25,
26,
27]. The ANN model is most commonly used for modeling crop yield in agriculture [
28]. It is a mathematical tool that attempts to represent low-level intelligence in normal creatures. The construction of ANN models is fairly basic, and it may create a non-linear relationship between the input and output variables [
29]. The neuron types, training technique, transfer function, and hidden layer of ANN models are used to classify them [
30]. Multi-layer perceptron (MLP) networks are commonly used for ANN modeling in the agriculture sector [
23,
31].
These non-linear complex models were created in order to forecast the growth prediction of different crops by using their growth parameters as input variables [
21,
22,
28]. In the ecological sector, there is a lack of research with the help of these non-linear complex models being used to forecast the growth of invasive plants. Furthermore, there is no study using the ANN model to predict the growth of invasive plants under different environmental conditions, in China or all over the world, even though invasive plant species are major threat to native biodiversity. The ANN model must be constructed in order to predict invasive plant species responses under different environmental conditions.
Furthermore, the hypothesis of this study that these predicting models describe to us would help to control the growth of these invasive species under native biodiversity. As a result, this work aimed to develop ANN and MLR models that could predict which growth parameters help Wedelia trilobata and its native species, Wedelia chinensis, to survive under submergence and eutrophication. Furthermore, predicting which growth parameters of Wedelia trilobata would reveal the important factors that allow it to overcome the growth of its native competitor under competition within submergence and eutrophication. These prediction models could be very helpful for managing invasive plant species under submergence and eutrophication habitats.
2. Materials and Methods
Wedelia trilobata (WT) is among the top most invasive plants in China [
6]; WT and
Wedelia chinensis (WC) both come from the Asteraceae family [
3]. It was mostly found worldwide in arid, semiarid, and humid regions [
9]. It is familiar as a groundcover plant species in China, but over time, it moved speedily from gardens to the roadside and finally into agricultural fields. It is also found in the wetland areas near the riverside [
1]. This likely indicates that WT can sustain its growth under submergence and eutrophication.
2.1. Study Site and Material Preparation
Plant material (Ramets) of WT and WC were collected around the Yangtze River bank (29.7204° N, 112.6501° E), in Yangzhou, Jiangsu, China. Ramets of both plant species were harvested after obtaining necessary approval, and all processes were conducted in compliance with applicable standards and laws. It is considered a cultivation study because ramets of both plant species have been used. This experiment was conducted in the greenhouse of Jiangsu University (32.1993° N, 119.5143° E), Zhenjiang, Jiangsu China, from the start of September to 20 November 2019, as presented in
Figure 1. In the seedling tray, ramets of WT and WC were prepared with sandy soil and peat moss (1:1) as growth media for the experimental investigation. These seedling trays were kept in a greenhouse with a temperature of 30 ± 5 °C and a relative humidity of 60%. Seedlings were watered every day, and Hoagland solution was applied once a week.
When the prepared seedlings had two completely grown leaves, then these seedlings were shifted into plastic pots (12.7 cm height and 17.78 cm diameter) filled with sandy soil as a growth medium. Pots of these seedlings were located in a bin (80 × 40 × 20 cm) for mesocosm investigation within the greenhouse. Both species were grown together in mixed culture, with 15 replicates of every treatment. For one week, normal water was provided to the seedlings to allow them to sustain in their new habitat condition. After 7 days, these transplanted seedlings were divided into 2 submergence and 3 eutrophication levels in order to simulate the naturally existing submergence and eutrophication under the wetland environment. There were two submergence stages (S1 = 7 cm and S2 = 14 cm) and three nutrient groups—N1 containing nitrogen (0.45 mg/L) and phosphorus (0.097 mg/L); N2 containing nitrogen (4.5 mg/L) and phosphorus (0.97 mg/L); and N3 containing nitrogen (45 mg/L) and phosphorus (9.7 mg/L),—using KNO
3, NH
4Cl, and KH
2PO
4 to prepare each of these treatments, respectively [
5]. There were 90 pots in total. Every day, tap water was added to each treatment bin to maintain the submergence level, and the nutrient solution was renewed one time after seven days. Plants were harvested after 30 days.
2.2. Morphological Trait Measurement
Plant height and root length of the plants from each treatment were measured with a ruler at the time of harvesting. ImageJ software was used to measure leaf area; the number of leaves and nodes were counted at the time of harvesting for the plants from every treatment. The dry weight (DW) of each plant per treatment was measured after drying at ≤80 °C for 48 h. The specific leaf area was calculated using the formula of leaf area to dry mass.
A portable LI-6400XT photosynthesis measurement instrument was used to measure the photosynthesis, transpiration, and stomatal conductance of the plants from each treatment. A fully expanded leaf was selected for measurement. Measurement was taken from 10.00 to 11.00 am under full sunshine. Using LI-6400XT, the following settings were noted during the measurement of data: atmospheric pressure 98.9 kpa, air molar flow 402.5 mmol m−2s−1, photosynthetically active radiation up to 1000 μmol m−2 s−1, CO2 concentration 402 μmol mol−1, vapor pressure 7.0 to 8.8 mbar, and ambient temperature 30.2 to 34.8 °C.
The water potential of each plant that underwent treatment was calculated using Psypro, Wescor, USA before harvesting of plants. Details of the growth parameters, along with their units and abbreviations, are presented in Abbreviations Part.
2.3. Processing of Data and Statistical Analysis
2.3.1. Criteria for Input Variables
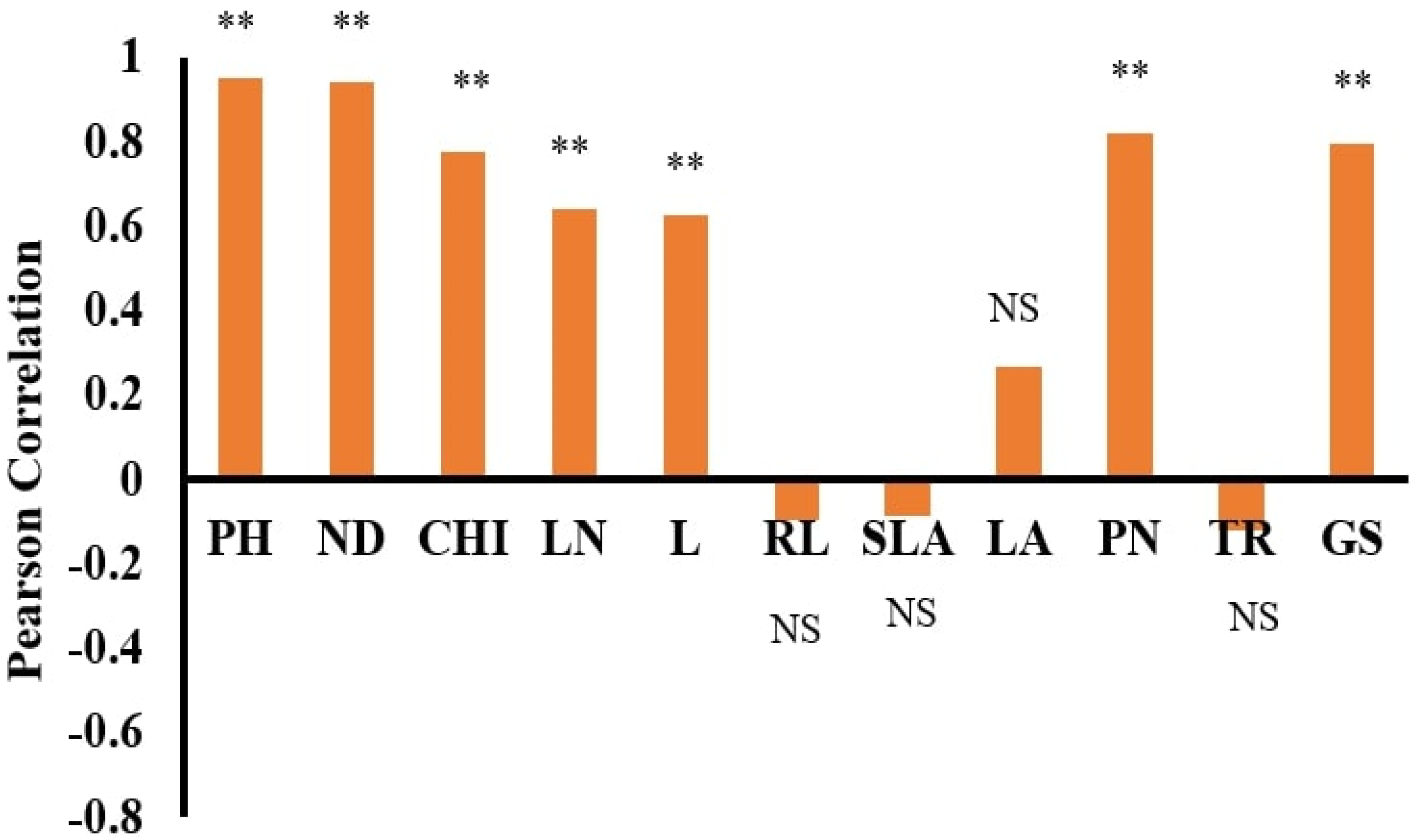
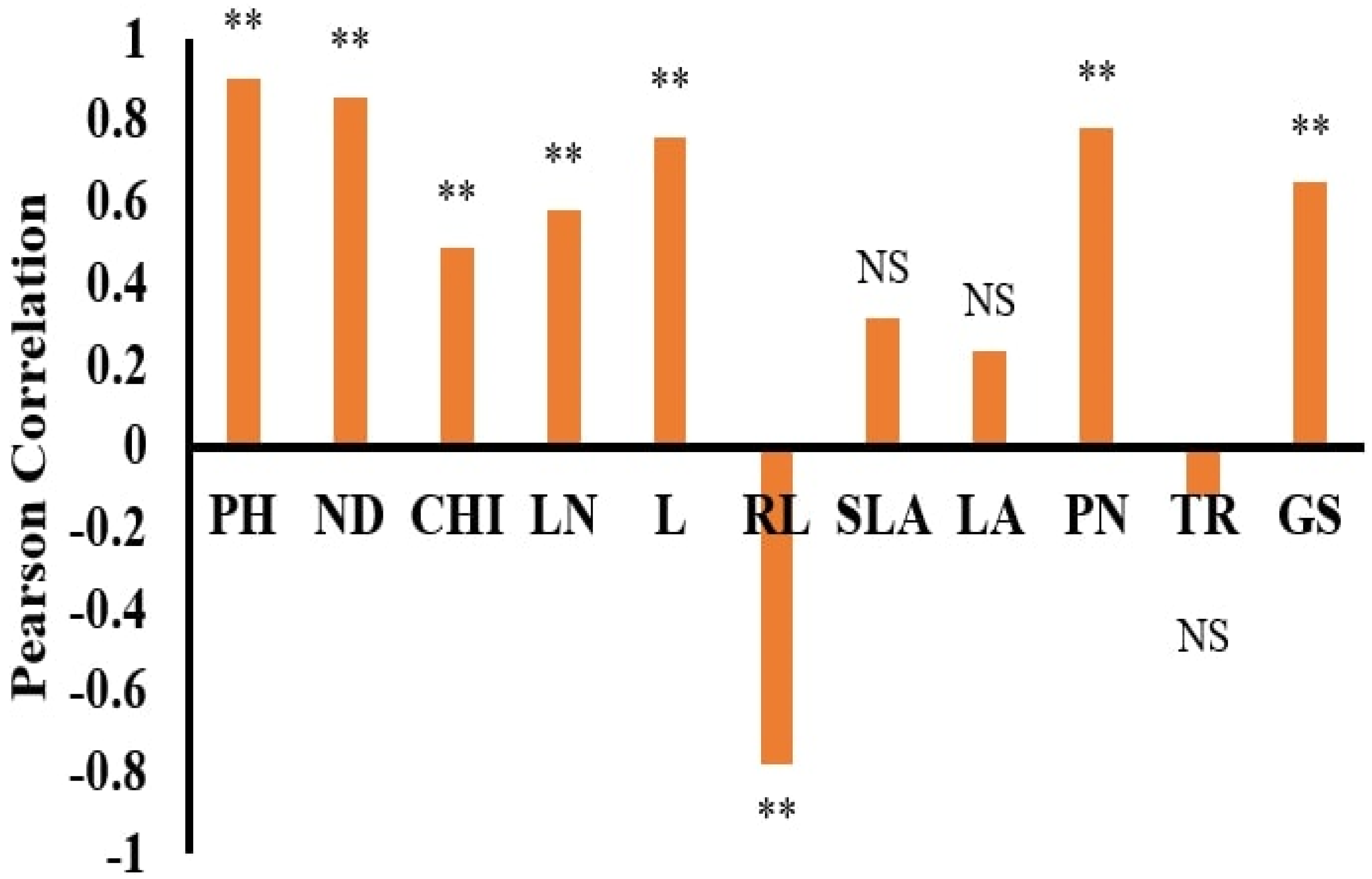
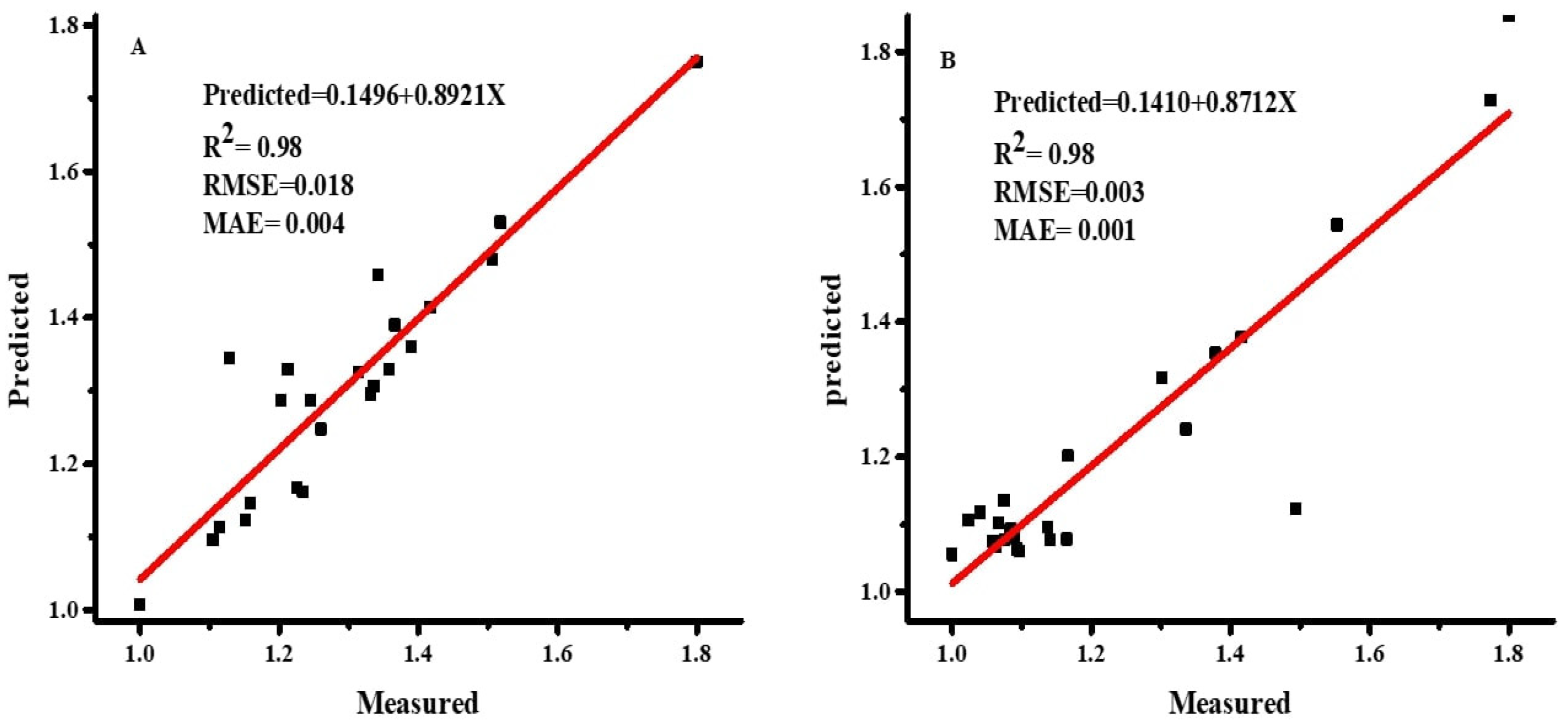
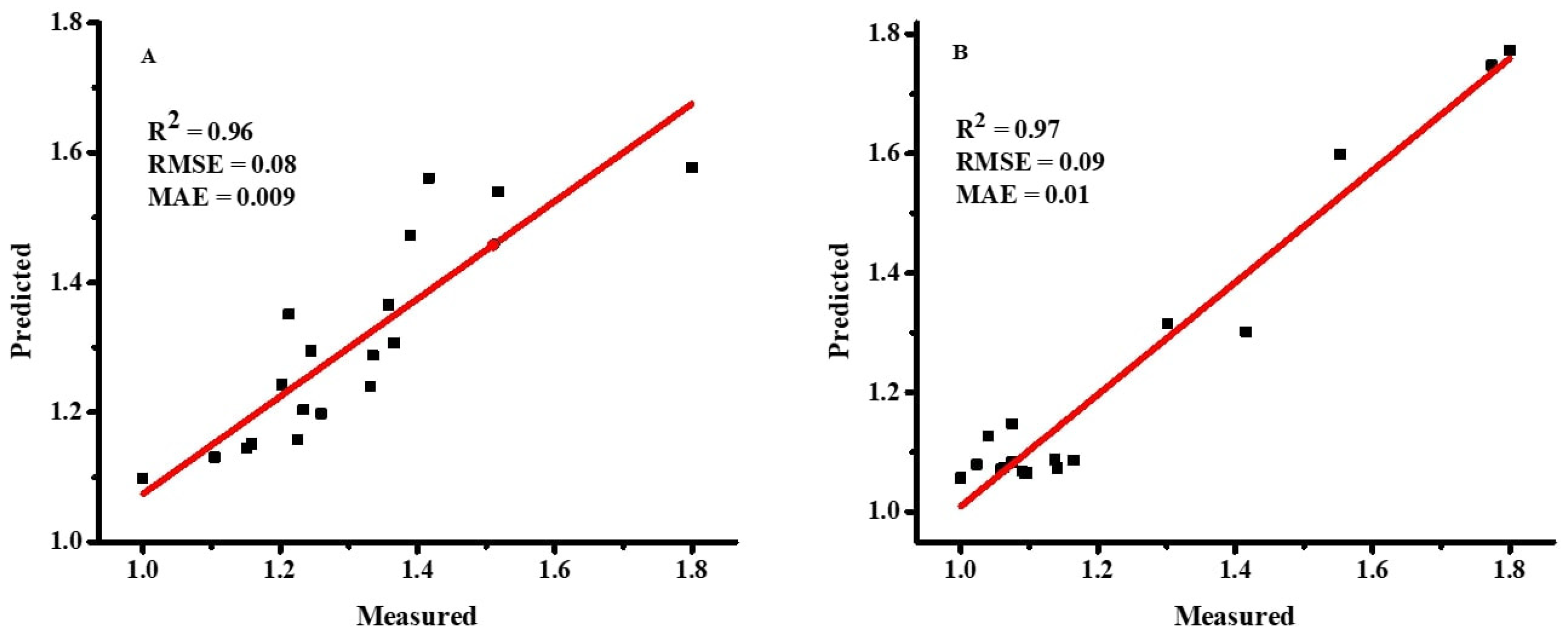
The dataset’s normality was checked with the Anderson–Darling test using SPSS 22 statistical software. Pearson correlation coefficients and stepwise regression (SWR) were used to assess the association between the morphological features and DW using SPSS 22 statistical software. Statistica software was used to run the ANN models, which used DW as the dependent and other features as independent parameters [
22]. Nightly samples were used to train, test and validate the ANN model. The descriptive statistics of the trait variables of both plant species are presented in
Table 1 and
Table 2.
2.3.2. Artificial Neural Network (ANN)
The dry weights (DWs) of both plant species, WT and WC, were used as output variables. In contrast, the remaining parameters were used as input variables. ANN training and testing were done based on the morphological traits of the invasive plant species under the submergence and eutrophication experiments. All datasets were divided into training, testing, and validation by a 70:15:15% ratio. This selection was done according to the previous literature [
24].
All data have been normalized and placed within the data ranges, where the Tanh [–1, 1] and Sigmoid [0, 1] are activation functions [
32]. For normalization, Equation (1) was utilized.
where X
norm is the normalized value of an independent or dependent variable. X
min and X
max are the variable’s minimum and maximum values, and Xi’s are the original data.
The optimal neural network structure contains three primary layers: input, hidden, and output. The output of the network is presumed via Equation (2).
Yf represents the model output (DW), n and m represent the hidden layers and input nodes, and the transfer function is donated by f. Cij i = 1, 2, …, m; j = 0, 1, …, n is the weight from the input through the hidden node, and dj j = 0, 1,…, n are the weight vectors extending from the hidden layers to the output nodes. The weight of leading arcs from bias terms, denoted by a
0 and b
0j, are always equal to 1. Based on earlier research [
21,
22,
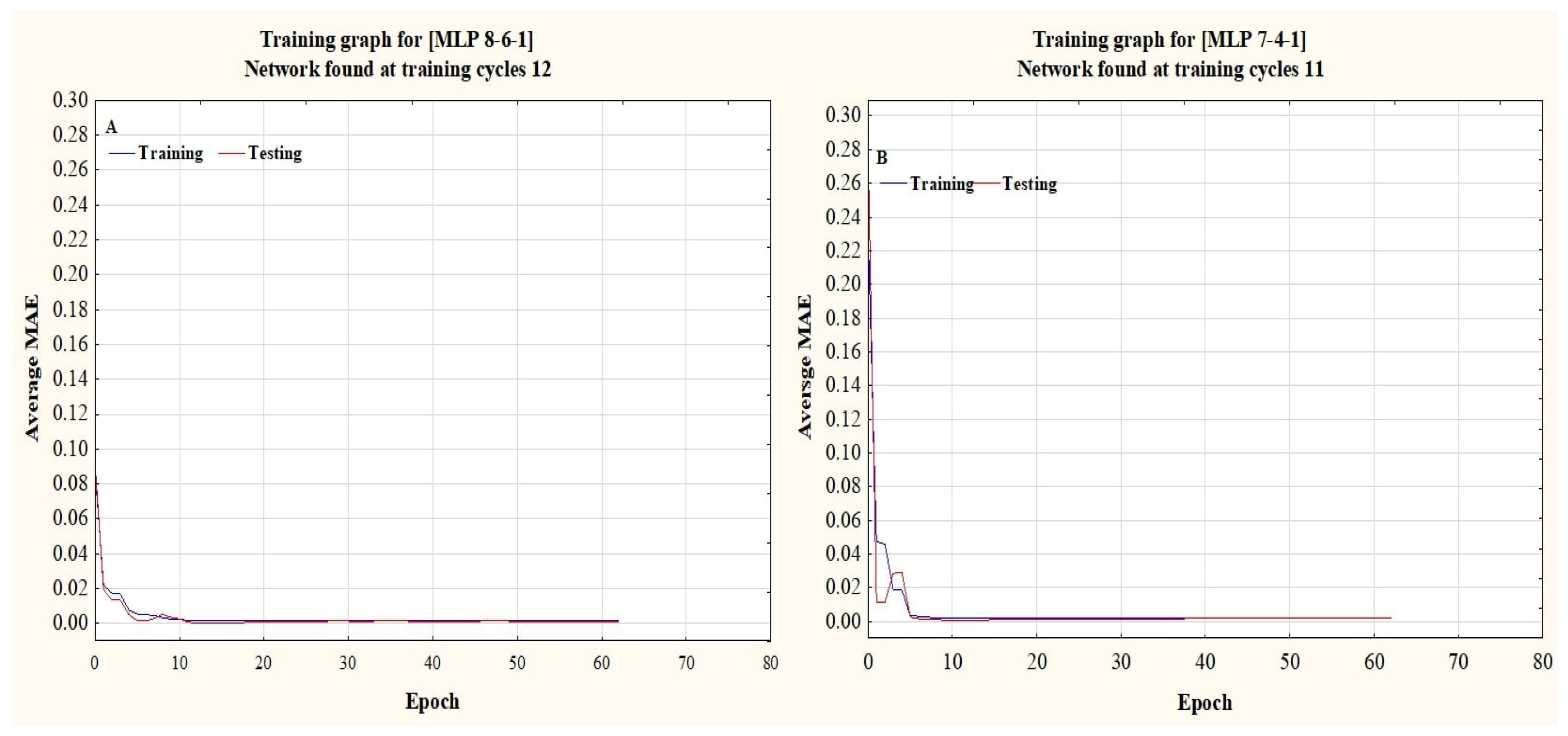
32], the current study used a feed-forward multi-layer perceptron (MLP) topology with three layers, and the back propagation (BP) training technique, along with the Levenberg–Marquardt, Momentum, and Conjugate Gradient learning algorithms. Trial-and-error testing was used to identify hidden layers (1–3) and neurons [
25]. Sigmoid Axon, Tangent Hyperbolic Axon, Linear Sigmoid Axon, and Linear Tangent Hyperbolic Axon activation functions were used to determine the best equation with high accuracy within the hidden and output layers [
33,
34].
2.3.3. Multiple Linear Regression (MLR)
Stepwise regression analysis (SWR) was conducted in order to determine the relative contributions of independent variables and to create a prediction model for the DW of WT and WC [
35]. Equation (3) was used to generate the SWR model using the same data as the ANN model. Independence of error was determined using the Durbin–Watson test. The tolerance value and variance inflation factor (VIF) were also tested in order to check the occurrence of multicollinearity for predictor variables. Higher collinearity described a smaller tolerance value (<0.1) or a high VIF (>10) value.
where a
0 + a
n is the regression coefficient, X
1 − X
n are independent variables, and € is the error of the nth observation.
2.3.4. Performance and Sensitivity Analysis
As demonstrated in Equations (4)–(6) [
36], the accuracy of the ANN and MLR models was evaluated using the root mean square error (RMSE), coefficient of determination (R
2), and mean absolute error (MAE).
where n represents the number of data, Y
i denotes the actual value, and Y
j denotes the predicted value. Y
io and Y
jo are the mean values of observed and predicted values, respectively. A model with lower values of RMSE and MAE and a higher value of R
2 is considered the best prediction model.
Sensitivity analysis was performed by choosing the most appropriate input parameters that control the dry weight of WT and WC within submergence and eutrophication, after determining the best ANN model. The sensitivity analysis was performed by running a dataset without any input variables, and the values of R
2, RMSE, and MAE determined the accuracy of the model [
37].