Optimization of White-Rot Fungi Mycelial Culture Components for Bioremediation of Pharmaceutical-Derived Pollutants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Precultivation
2.2. Screening Test
2.3. Selection of the Most-Effective Media Variants
2.4. Determination of Growth Curves
2.5. Enzymatic Activity
2.6. Removal of Pharmaceuticals
2.7. Calculations
2.7.1. Percentage of C and N in the Investigated Substrates
2.7.2. The Efficiency Coefficient of a Single Component in a Two-Component Medium
2.7.3. Composition of One-Component Media
2.7.4. Enzymatic Activity
2.7.5. Estimation of Growth Curves Parameters
2.7.6. Specific Growth Rate
2.7.7. Growth Model for One-Component Media Containing DDGS and CSL
2.7.8. Removal Degree
2.7.9. Statistics
3. Results
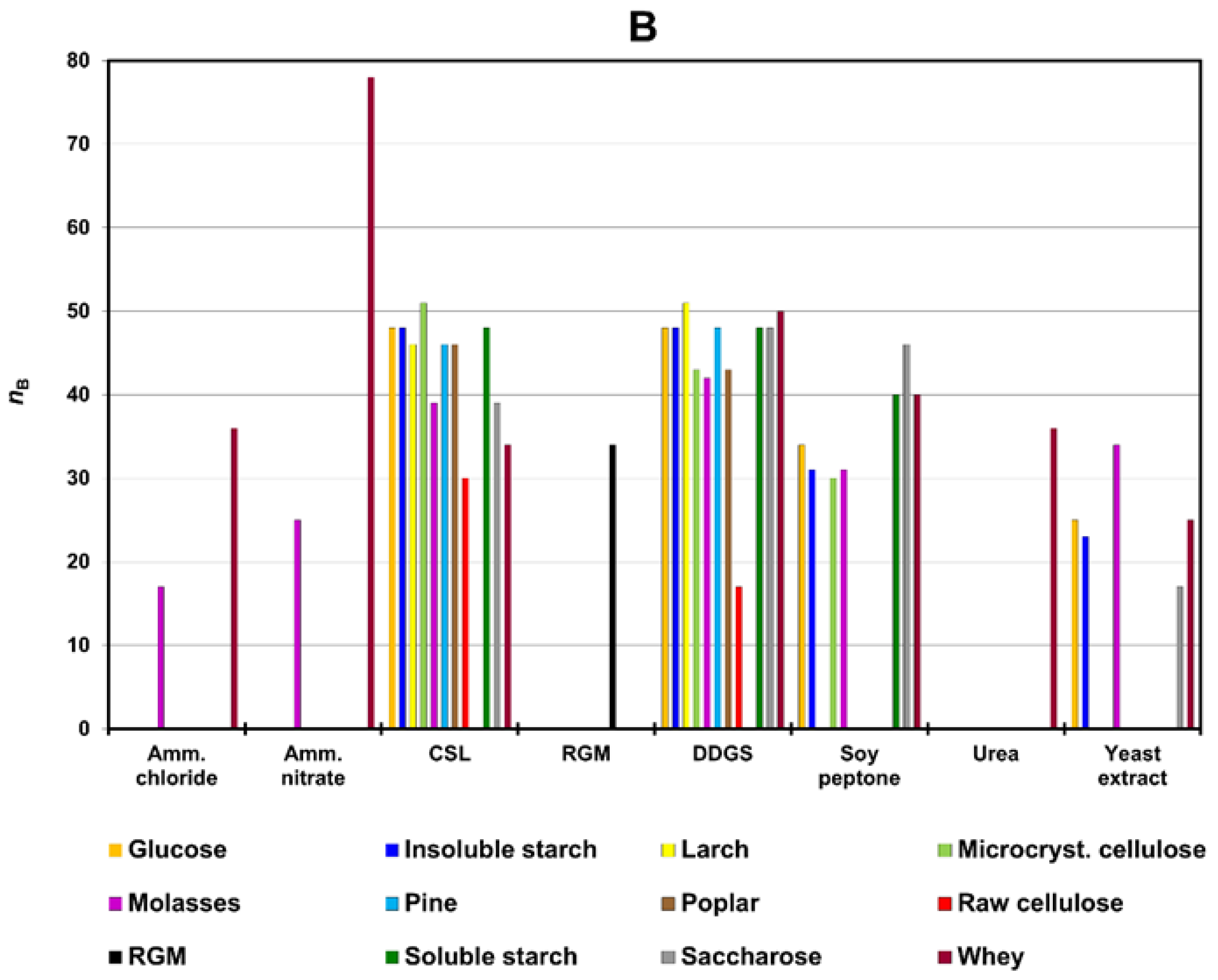
3.1. Screening Test
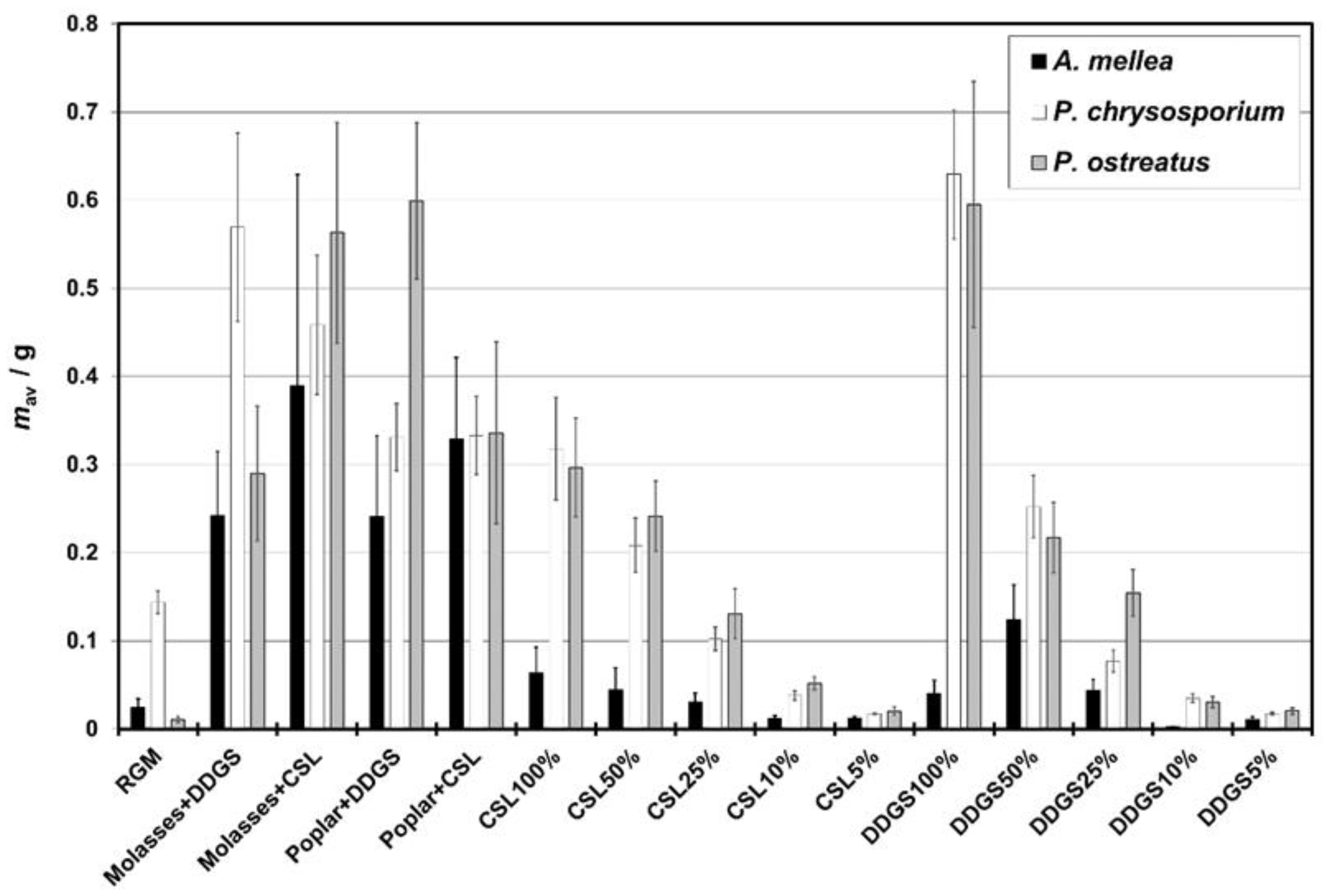
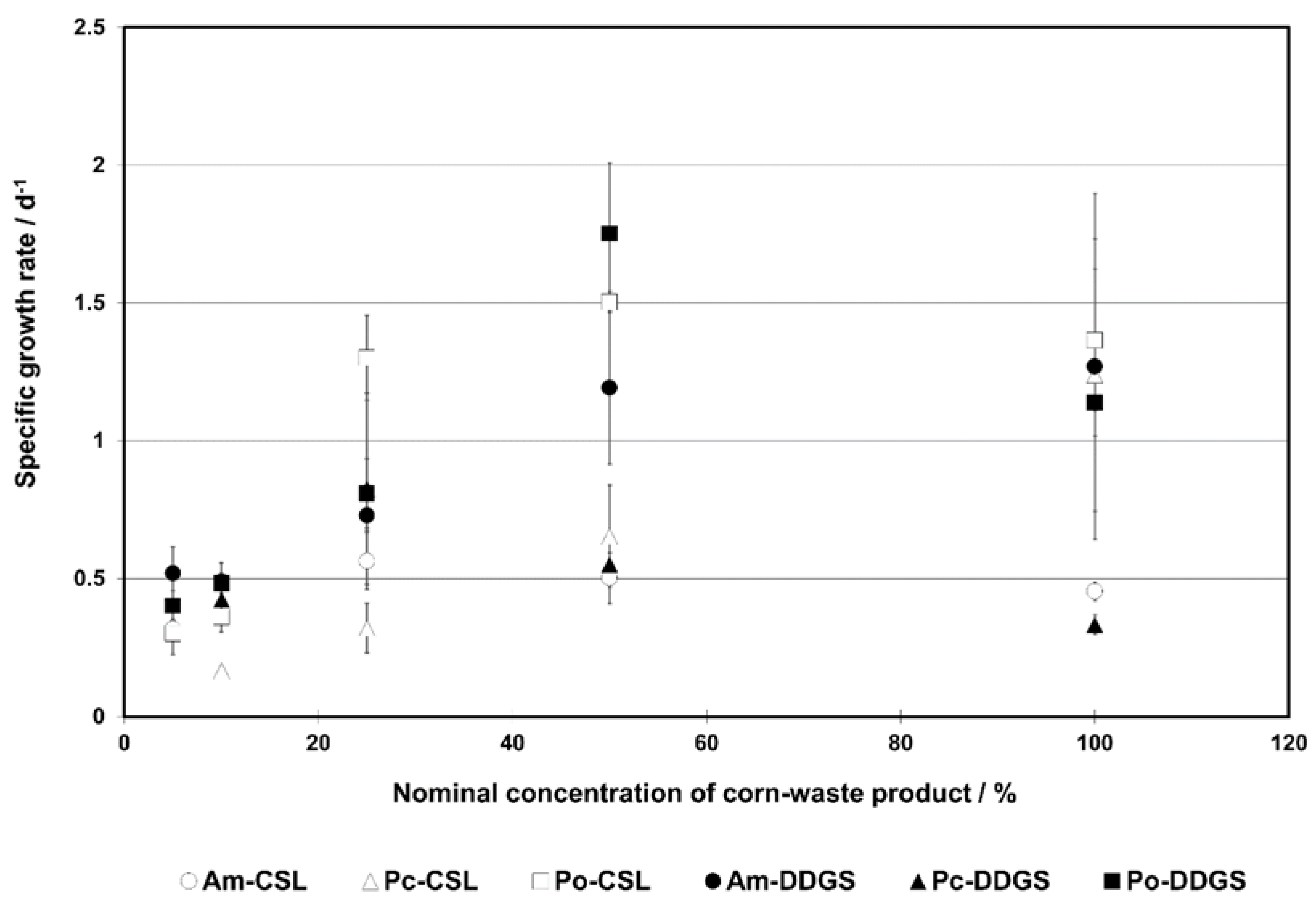
3.2. Growth Curves—Parameters, Enzymatic Activity, and Results Description
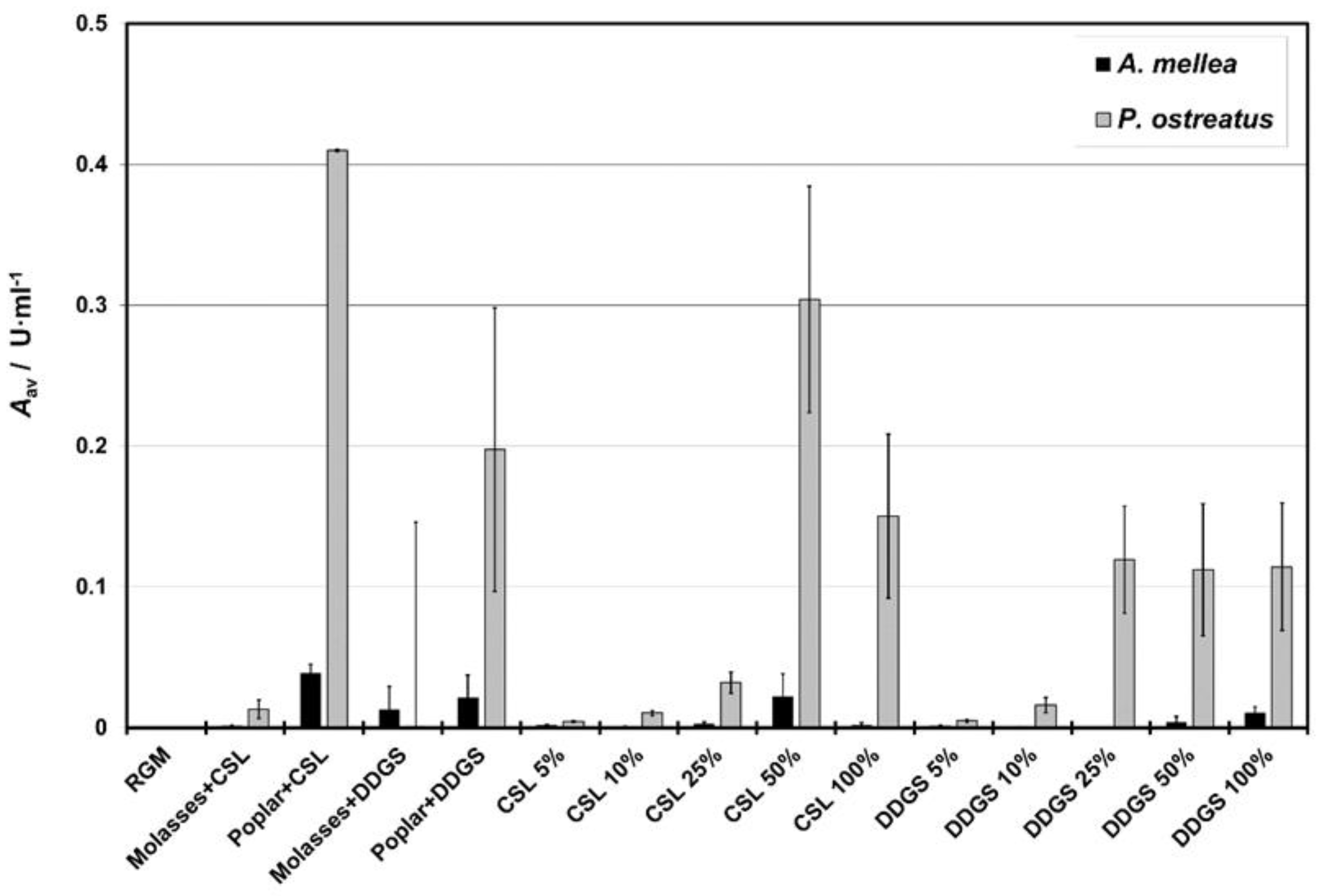
3.3. Laccase
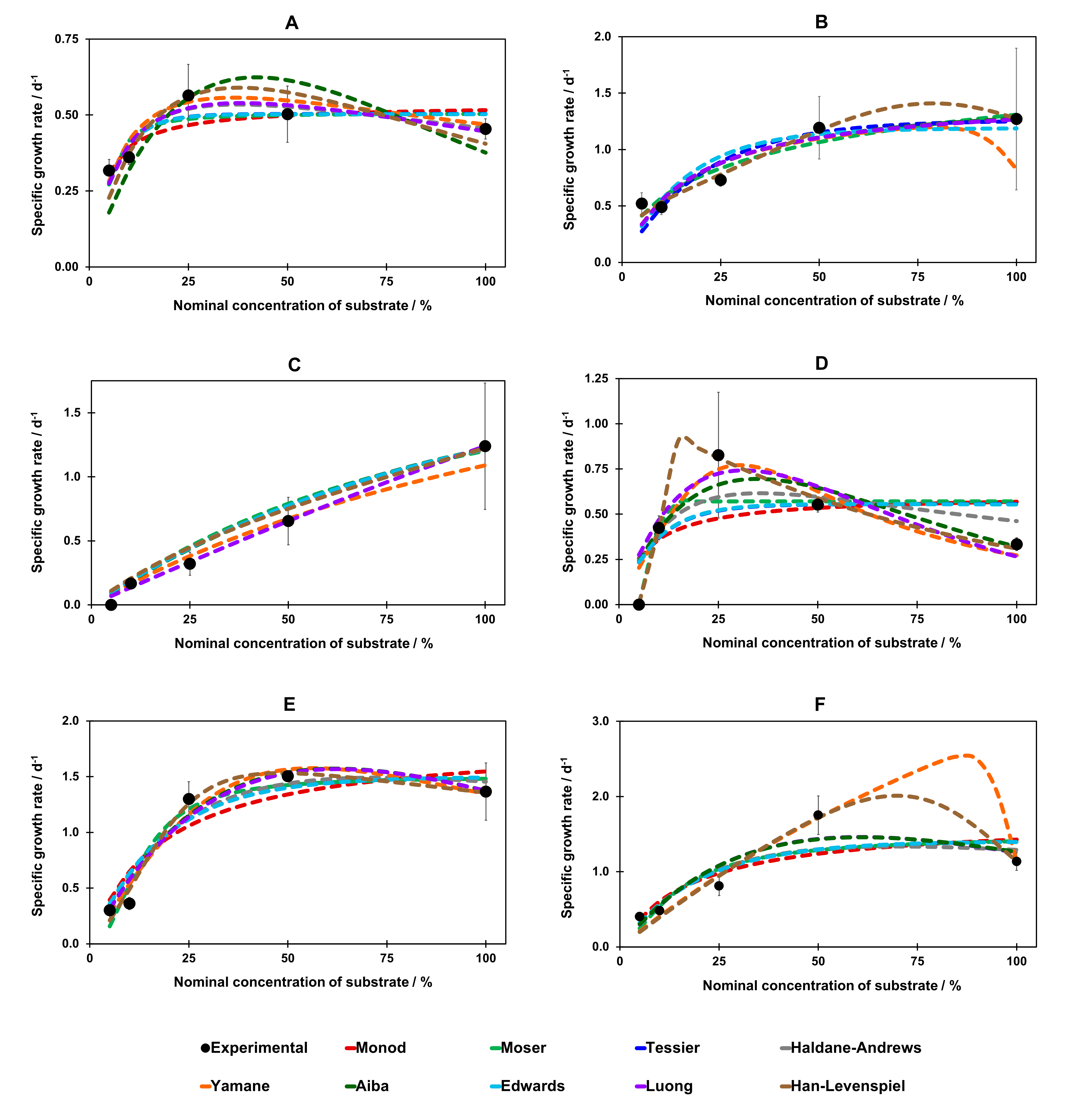
3.4. Growth Model on a One-Component Medium
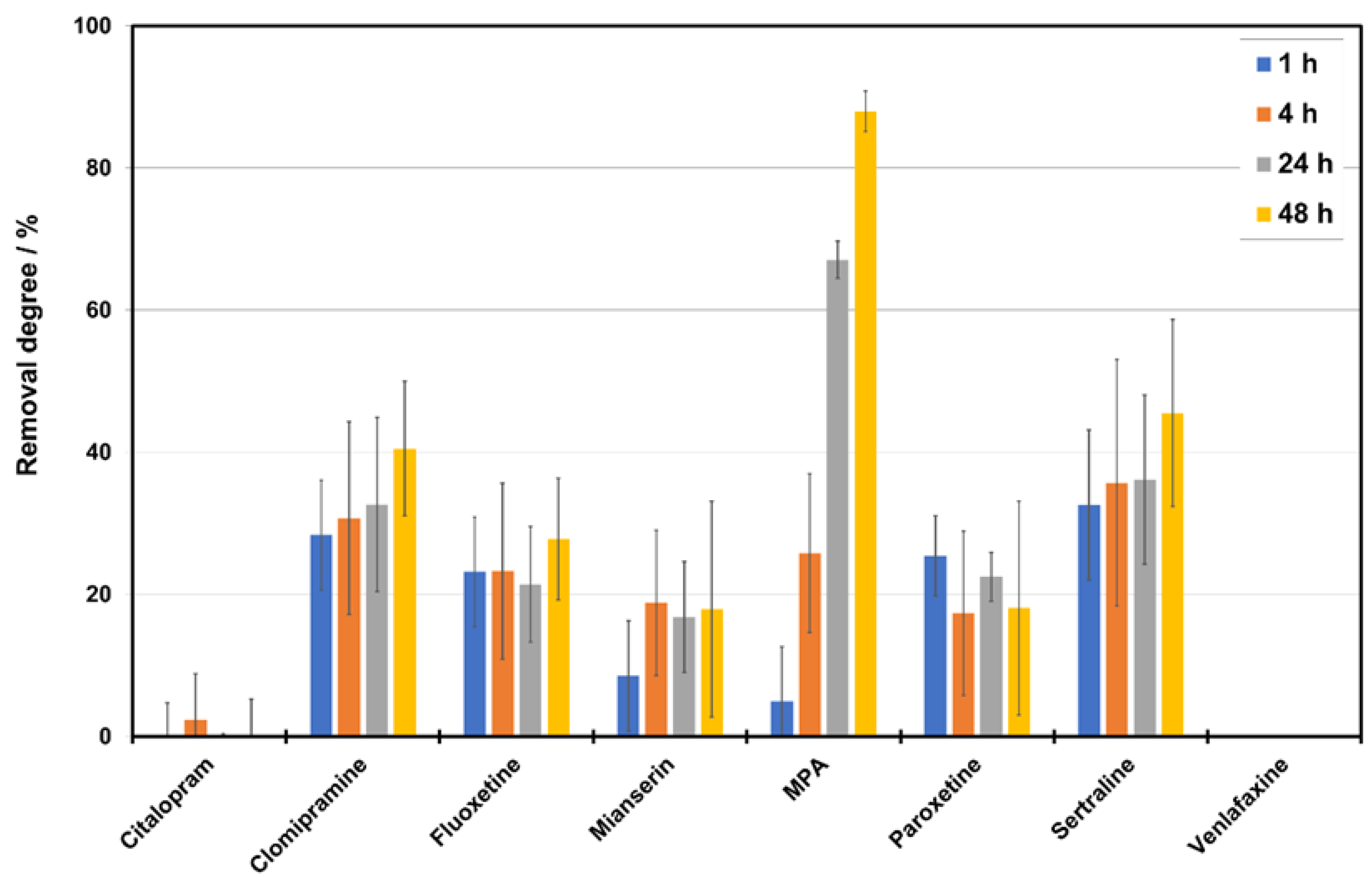
3.5. Removal of Pharmaceuticals
4. Discussion
4.1. Screening Test
4.2. Growth Curve
4.3. Enzymatic Activity
4.4. Growth Model
4.5. Removal Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kulshreshtha, S.; Mathur, N.; Bhatnagar, P. Mycoremediation of paper, pulp and cardboard industrial wastes and pollutants. In Fungi as Bioremediators; Goltapeh, E.M., Danesh, Y.R., Varma, A., Eds.; Springer: Berlin, Heidelberg, Germany, 2013; Volume 32, pp. 77–116. [Google Scholar]
- Asemoloye, M.D.; Ahmad, R.; Jonathan, S.G. Synergistic rhizosphere degradation of γ-hexachlorocyclohexane (lindane) through the combinatorial plant-fungal action. PLoS ONE 2017, 12, e0183373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, N.; Singhvi, R. Effects of chemical fertilizers and pesticides on human health and environment: A review. Int. J. Agric. Environ. Biotechnol. 2017, 10, 675–680. [Google Scholar] [CrossRef]
- Castellet-Rovira, F.; Lucas, D.; Villagrasa, M.; Rodríguez-Mozaz, S.; Barceló, D.; Sarrà, M. Stropharia rugosoannulata and Gymnopilus luteofolius: Promising fungal species for pharmaceutical biodegradation in contaminated water. J. Environ. Manag. 2018, 207, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Couto, C.F.; Lange, L.C.; Amaral, M.C.S. Occurrence, fate and removal of pharmaceutically active compounds (PhACs) in water and wastewater treatment plants—A review. J. Water Process Eng. 2019, 32, 100927. [Google Scholar] [CrossRef]
- Asif, M.B.; Hai, F.I.; Singh, L.; Price, W.E.; Nghiem, L.D. Degradation of Pharmaceuticals and Personal Care Products by White-Rot Fungi-a Critical Review. Curr. Pollut. Rep. 2017, 3, 88–103. [Google Scholar] [CrossRef] [Green Version]
- Kaleniecka, A.; Zarzycki, P.K. Pharmaceuticals in the aquatic environment: Sources, effects, treatment methods. Arch. Physiother. Glob. Res. 2015, 19, 39–52. [Google Scholar] [CrossRef]
- Zwiener, C.; Frimmel, F. Oxidative treatment of pharmaceuticals in water. Water Res. 2000, 34, 1881–1885. [Google Scholar] [CrossRef]
- Ternes, T.A.; Meisenheimer, M.; McDowell, D.; Sacher, F.; Brauch, H.-J.; Haist-Gulde, B.; Preuss, G.; Wilme, U.; Zulei-Seibert, N. Removal of pharmaceuticals during drinking water treatment. Environ. Sci. Technol. 2002, 36, 3855–3863. [Google Scholar] [CrossRef]
- Xia, Y.; Wen, X.; Zhang, B.; Yang, Y. Diversity and assembly patterns of activated sludge microbial communities: A review. Biotechnol. Adv. 2018, 36, 1038–1047. [Google Scholar] [CrossRef]
- Olivieri, G.; Russo, M.; Giardina, P.; Marzocchella, A.; Sannia, G.; Salatino, P. Strategies for dephenolization of raw olive mill wastewater by means of Pleurotus ostreatus. J. Ind. Microbiol. Biotechnol. 2012, 39, 719–729. [Google Scholar] [CrossRef]
- Asamudo, N.; Daba, A.; Ezeronye, O. Bioremediation of textile effluent using Phanerochaete chrysosporium. Afr. J. Biotechnol. 2005, 4, 1548–1553. [Google Scholar] [CrossRef]
- Harms, H.; Schlosser, D.; Wick, L.Y. Untapped potential: Exploiting fungi in bioremediation of hazardous chemicals. Nat. Rev. Microbiol. 2011, 9, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Grandclément, C.; Seyssiecq, I.; Piram, A.; Wong-Wah-Chung, P.; Vanot, G.; Tiliacos, N.; Roche, N.; Doumenq, P. From the conventional biological wastewater treatment to hybrid processes, the evaluation of organic micropollutant removal: A review. Water Res. 2017, 111, 297–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mir-Tutusaus, J.A.; Baccar, R.; Caminal, G.; Sarrà, M. Can white-rot fungi be a real wastewater treatment alternative for organic micropollutants removal? A review. Water Res. 2018, 138, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Jebapriya, G.R.; Gnanadoss, J.J. Bioremediation of textile dye using white rot fungi: A review. Int. J. Curr. Res. Rev. 2013, 5, 1. [Google Scholar]
- Mir-Tutusaus, J.A.; Masís-Mora, M.; Corcellas, C.; Eljarrat, E.; Barceló, D.; Sarrà, M.; Caminal, G.; Vicent, T.; Rodríguez-Rodríguez, C.E. Degradation of selected agrochemicals by the white rot fungus Trametes versicolor. Sci. Total Environ. 2014, 500, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Hasanin, M.S.; Hashem, A.H. Eco-friendly, economic fungal universal medium from watermelon peel waste. J. Microbiol. Methods 2020, 168, 105802. [Google Scholar] [CrossRef]
- Costa, S.; Dedola, D.G.; Pellizzari, S.; Blo, R.; Rugiero, I.; Pedrini, P.; Tamburini, E.J.W. Lignin biodegradation in pulp-and-paper mill wastewater by selected white rot fungi. Water 2017, 9, 935. [Google Scholar] [CrossRef] [Green Version]
- Das, A.; Bhattacharya, S.; Panchanan, G.; Navya, B.S.; Nambiar, P. Production, characterization and Congo red dye decolourizing efficiency of a laccase from Pleurotus ostreatus MTCC 142 cultivated on co-substrates of paddy straw and corn husk. J. Genet. Eng. Biotechnol. 2016, 14, 281–288. [Google Scholar] [CrossRef] [Green Version]
- Gern, R.M.M.; Wisbeck, E.; Rampinelli, J.R.; Ninow, J.L.; Furlan, S.A. Alternative medium for production of Pleurotus ostreatus biomass and potential antitumor polysaccharides. Bioresour. Technol. 2008, 99, 76–82. [Google Scholar] [CrossRef]
- Papaspyridi, L.; Katapodis, P.; Gonou-Zagou, Z.; Kapsanaki-Gotsi, E.; Christakopoulos, P. Optimization of biomass production with enhanced glucan and dietary fibres content by Pleurotus ostreatus ATHUM 4438 under submerged culture. Biochem. Eng. J. 2010, 50, 131–138. [Google Scholar] [CrossRef]
- Fahy, V.; FitzGibbon, F.J.; McMullan, G.; Singh, D.; Marchant, R. Decolourisation of molasses spent wash by Phanerochaete chrysosporium. Biotechnol. Lett. 1997, 19, 97–99. [Google Scholar] [CrossRef]
- Garrido-Bazán, V.; Téllez-Téllez, M.; Herrera-Estrella, A.; Díaz-Godínez, G.; Nava-Galicia, S.; Villalobos-López, M.Á.; Arroyo-Becerra, A.; Bibbins-Martínez, M. Effect of textile dyes on activity and differential regulation of laccase genes from Pleurotus ostreatus grown in submerged fermentation. AMB Express 2016, 6, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pezzella, C.; Lettera, V.; Piscitelli, A.; Giardina, P.; Sannia, G. Transcriptional analysis of Pleurotus ostreatus laccase genes. Appl. Microbiol. Biotechnol. 2013, 97, 705–717. [Google Scholar] [CrossRef]
- Daou, M.; Piumi, F.; Cullen, D.; Record, E.; Faulds, C.B. Heterologous production and characterization of two glyoxal oxidases from Pycnoporus cinnabarinus. Appl. Environ. Microbiol. 2016, 82, 4867–4875. [Google Scholar] [CrossRef] [Green Version]
- Fernandez Fueyo, E.; Ruiz-Duenas, F.J.; Lopez-Lucendo, M.F.; Perez-Boada, M.; Rencorat, J.; Gutierrez, A.; Pisabarro, A.G.; Ramirez, L.; Martinez, A.T. A secretomic view of woody and nonwoody lignocellulose degradation by Pleurotus ostreatus. Biotechnol. Biofuels 2016, 9, 49. [Google Scholar] [CrossRef] [Green Version]
- Pollegioni, L.; Tonin, F.; Rosini, E. The FEBS journal. FEBS J. 2015, 282, 1190–1213. [Google Scholar] [CrossRef]
- Sakai, K.; Matsuzaki, F.; Wise, L.; Sakai, Y.; Jindou, S.; Ichinose, H.; Takaya, N.; Kato, M.; Wariishi, H.; Shimizu, M. Biochemical characterization of CYP505D6, a self-sufficient cytochrome P450 from the white-rot fungus Phanerochaete chrysosporium. Appl. Environ. Microbiol. 2018, 84, e01091-18. [Google Scholar] [CrossRef] [Green Version]
- Hadibarata, T.; Kristanti, R.A. Fate and cometabolic degradation of benzo[a]pyrene by white-rot fungus Armillaria sp. F022. Bioresour. Technol. 2012, 107, 314–318. [Google Scholar] [CrossRef]
- Stoytchev, I.; Nerud, F. Ligninolytic enzyme complex of Armillaria spp. Folia Microbiol. (Praha) 2000, 45, 248–250. [Google Scholar] [CrossRef]
- Sośnicka, A.; Lubiński, O.; Turło, J. Erythromycin and penicillin toxic post-products management using Pleurotus ostreatus. Res. J. Biotechnol. 2019, 14, 29–36. [Google Scholar]
- Kózka, B.; Nałęcz-Jawecki, G.; Turło, J.; Giebułtowicz, J. Application of Pleurotus ostreatus to efficient removal of selected antidepressants and immunosuppressant. J. Environ. Manag. 2020, 273, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hariharan, S.; Nambisan, P. Optimization of Lignin Peroxidase, Manganese Peroxidase, and Lac Production from Ganoderma lucidum Under Solid State Fermentation of Pineapple Leaf. BioResources 2013, 8, 250–271. [Google Scholar] [CrossRef] [Green Version]
- Lung, M.Y.; Huang, P.C. Optimization of exopolysaccharide production from Armillaria mellea in submerged cultures. Lett. Appl. Microbiol. 2010, 50, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.H.; Liu, L.D.; Dong, H.X.; Qu, H.G.; Cai, D.H. Optimization of submerged culture condition for production of mycelial biomass by Armillaria mellea. Zhong Yao Cai 2007, 30, 509–512. [Google Scholar]
- Bhak, G.; Song, M.; Lee, S.; Hwang, S. Response Surface Analysis of Solid State Growth of Pleurotus ostreatus Mycelia utilizing Whey Permeate. Biotechnol. Lett. 2005, 27, 1537–1541. [Google Scholar] [CrossRef] [PubMed]
- Theodoros, A.; Argyro, B.; Stavros, P.; Athanasios, A.K.; Poonam, N. Upgrading of Mixed Food Industry Side-Streams by Solid-State Fermentation with P. ostreatus. Recycling 2018, 3, 12. [Google Scholar] [CrossRef] [Green Version]
- Kapich, A.N.; Prior, B.A.; Botha, A.; Galkin, S.; Lundell, T.; Hatakka, A. Effect of lignocellulose-containing substrates on production of ligninolytic peroxidases in submerged cultures of Phanerochaete chrysosporium ME-446. Enzym. Microb. Technol. 2004, 34, 187–195. [Google Scholar] [CrossRef]
- Kapich, A.N.; Prior, B.A.; Lundell, T.; Hatakka, A. A rapid method to quantify pro-oxidant activity in cultures of wood-decaying white-rot fungi. J. Microbiol. Methods 2005, 61, 261–271. [Google Scholar] [CrossRef]
- Pena, R.; Lú-Chau, T.; Lema, J. Use of White-Rot Fungi for Valorization of Stillage From Bioethanol Production. Waste Biomass Valori. 2012, 3, 295–303. [Google Scholar] [CrossRef]
- Zuo, S.; Niu, D.; Zheng, M.; Jiang, D.; Tian, P.; Li, R.; Xu, C. Effect of Irpex lacteus, Pleurotus ostreatus and Pleurotus cystidiosus pretreatment of corn stover on its improvement of the in vitro rumen fermentation. J. Sci. Food Agric. 2018, 98, 4287–4295. [Google Scholar] [CrossRef] [PubMed]
- Itelima, J. Cultivation of mushroom (Pleurotus ostreatus) using corn cobs and saw dust as the major substrates. Glob. J. Agric. Sci. 2012, 11, 51–56. [Google Scholar] [CrossRef]
- Thakkar, A.P.; Dhamankar, V.S.; Kapadnis, B.P. Biocatalytic decolourisation of molasses by Phanerochaete chrysosporium. Bioresour. Technol. 2006, 97, 1377–1381. [Google Scholar] [CrossRef] [PubMed]
- Vahabzadeh, F.; Mehranian, M.; Saatari, A.R. Color removal ability of Phanerochaete chrysosporium in relation to lignin peroxidase and manganese peroxidase produced in molasses wastewater. World J. Microbiol. Biotechnol. 2004, 20, 859–864. [Google Scholar] [CrossRef]
- Vahabzadeh, F.; Mogharei, A.; Mehranian, M. Decolorization of molasses waste water from an alcoholic fermentation process with Phanerochaete chrysosporium-Involvement of ligninase. Iran J. Chem. Chem. Eng. 2002, 21, 126–134. [Google Scholar] [CrossRef]
- Hoa, H.T.; Wang, C.-L. The Effects of Temperature and Nutritional Conditions on Mycelium Growth of Two Oyster Mushrooms (Pleurotus ostreatus and Pleurotus cystidiosus). Mycobiology 2015, 43, 14–23. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Reddy, K.; Oh, Y.; Jung, Y.; Yeo, J.; Choi, H. Rumen fermentation and digestibility of spent mushroom (Pleurotus ostreatus) substrate inoculated with Lactobacillus brevis for Hanwoo steers. Rev. Colomb. Cienc. Pecu. 2017, 30, 267–277. [Google Scholar] [CrossRef]
- Roldán-Carrillo, T.; Rodrıguez-Vázquez, R.; Dıaz-Cervantes, D.; Vázquez-Torres, H.; Manzur-Guzmán, A.; Torres-Domınguez, A. Starch-based plastic polymer degradation by the white rot fungus Phanerochaete chrysosporium grown on sugarcane bagasse pith: Enzyme production. Bioresour. Technol. 2003, 86, 1–5. [Google Scholar] [CrossRef]
- Wu, B.; Hu, G.-K.; Feng, H.; Wu, J.-M.; Zhang, Y.-Z. Cloning and Expression of an α-Amylase Gene from Phanerochaete chrysosporium. Curr. Microbiol. 2007, 55, 105–113. [Google Scholar] [CrossRef]
- Wirunpan, K.; Chinwang, S.; Chaikong, N.; Pukahuta, C. Increasing Nutritional Contents of Cassava Starch Wastes Using Pleurotus ostreatus (Jacq.) P. Kumm. and Lentinus squarrosulus Mont. J. Pure Appl. Microbiol. 2019, 13, 117–125. [Google Scholar] [CrossRef] [Green Version]
- Birch, P.R.J. Targeted differential display of abundantly expressed sequences from the basidiomycete Phanerochaete chrysosporium which contain regions coding for fungal cellulose-binding domains. Curr. Genet. 1998, 33, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Hou, Q.; Niu, T. Effect of cultivating Pleurotus ostreatus on substrates supplemented with herb residues on yield characteristics, substrates degradation, and fruiting bodies’ properties. J. Sci. Food Agric. 2020, 100, 4901–4910. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Ma, F.; Li, Y.; Yu, H.; Li, C.; Zhang, X. Differential proteomic profiles of Pleurotus ostreatus in response to lignocellulosic components provide insights into divergent adaptive mechanisms. Front. Microbiol. 2017, 8, 480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Düzkale Sözbir, G. Utilization of various lignocellulosic substrates for Pleurotus ostreatus mushroom cultivation in the manufacturing of polycaprolactone (PCL)-based biocomposite films. BioResources 2021, 16, 3783–3796. [Google Scholar] [CrossRef]
- Sachs, I.B.; Leatham, G.F.; Myers, G.C. Biomechanical Pulping of Aspen Chips by Phanerochaete-Chrysosporium-Fungal Growth-Pattern and Effects on Wood Cell-Walls. Wood Fiber Sci. 1989, 21, 331–342. [Google Scholar]
- Mata, G.; Perez-Torres, J.; Medel, R.; Perez-Merlo, R.; Salmones, D. Culture of Pleurotus ostreatus in pine shavings: Isolation of strains and evaluation of their productivity. Madera Bosques 2019, 25, e2521715. [Google Scholar] [CrossRef]
- Piškur, B.; Bajc, M.; Robek, R.; Humar, M.; Sinjur, I.; Kadunc, A.; Oven, P.; Rep, G.; Al Sayegh Petkovšek, S.; Kraigher, H.; et al. Influence of Pleurotus ostreatus inoculation on wood degradation and fungal colonization. Bioresour. Technol. 2011, 102, 10611–10617. [Google Scholar] [CrossRef]
- Ayata, U.; Akcay, C.; Esteves, B. Determination of decay resistance against Pleurotus ostreatus and Coniophora puteana fungus of heat-treated scotch pine, oak and beech wood species. Maderas. Cienc. Y Tecnol. 2017, 19, 309–316. [Google Scholar] [CrossRef] [Green Version]
- Reid, I.D. Effects of nitrogen supplements on degradation of aspen wood lignin and carbohydrate components by Phanerochaete chrysosporium. Appl. Environ. Microbiol. 1983, 45, 830–837. [Google Scholar] [CrossRef] [Green Version]
- Andrade, M.V.F.; da Silva, K.M.L.; Siqueira, J.P.D.; Wanderley, C.R.P.; Araujo, R.D.; Marinho, G.; Rodrigues, K. Azo Dye Degradation by Phanerochaete chrysosporium in the Medium Enriched with Nitrogen in the Presence of Primary Co-Substrate. Braz. Arch. Biol. Technol. 2013, 56, 867–874. [Google Scholar] [CrossRef]
- Haider, A.; Zulfiqar, M. Optimization of Cultural Conditions for the Treatment of Pulp and Paper Industrial Effluent by Pleurotus ostreatus (L.). Pak. J. Agri. Res. 2019, 32, 507–513. [Google Scholar] [CrossRef]
- Barraco-Vega, M.; Romero, H.; Richero, M.; Cerdeiras, M.P.; Cecchetto, G. Functional characterization of two novel purine transporters from the Basidiomycota Phanerochaete chrysosporium. Gene 2017, 601, 1. [Google Scholar] [CrossRef] [PubMed]
- Adebayo-Tayo, B.; Jonathan, S.G.; Popoola, O.; Egbomuche, R.C. Optimization of growth conditions for mycelial yield and exopolysaccharride production by Pleurotus ostreatus cultivated in Nigeria. Afr. J. Microbiol. Res. 2011, 5, 2130–2138. [Google Scholar] [CrossRef]
- El-Dein, M.N.; El-Fallal, A.A.; El-Shahat, A.; Hereher, F.E. Exopolysaccharides production by Pleurotus pulmonarius: Factors affecting formation and their structures. Pak. J. Biol. Sci. 2004, 7, 1078–1084. [Google Scholar] [CrossRef] [Green Version]
- Coelho-Moreira, J.D.S.; Bracht, A.; Souza, A.C.D.S.D.; Oliveira, R.F.; Sá-Nakanishi, A.B.D.; Souza, C.G.M.D.; Peralta, R.M. Degradation of diuron by Phanerochaete chrysosporium: Role of ligninolytic enzymes and cytochrome P450. BioMed Res. Int. 2013, 2013, 251354. [Google Scholar] [CrossRef] [Green Version]
- Li, X.-m.; Yang, Q.; Zhang, Y.; Zheng, W.; Yue, X.; Wang, D.-B.; Zeng, G.-M. Biodegradation of 2, 4-dichlorophenol in a fluidized bed reactor with immobilized Phanerochaete chrysosporium. Water Sci. Technol. 2010, 62, 947–955. [Google Scholar] [CrossRef]
- Shi, J.; Chinn, M.; Sharma-Shivappa, R. Interactions between fungal growth, substrate utilization, and enzyme production during solid substrate cultivation of Phanerochaete chrysosporium on cotton stalks. Enzyme Microb. Technol. 2014, 37, 2463–2473. [Google Scholar] [CrossRef]
- Zhang, Q.; Wei, Y.; Han, H.; Weng, C. Enhancing bioethanol production from water hyacinth by new combined pretreatment methods. Bioresour. Technol. 2018, 251, 358–363. [Google Scholar] [CrossRef]
- Tlecuitl-Beristain, S.; Sánchez, C.; Loera, O.; Robson, G.D.; Díaz-Godínez, G. Laccases of Pleurotus ostreatus observed at different phases of its growth in submerged fermentation: Production of a novel laccase isoform. Mycol. Res. 2008, 112, 1080–1084. [Google Scholar] [CrossRef]
- Arbanah, M.; Miradatul Najwa, M.; Ku Halim, K. Utilization of Pleurotus ostreatus in the removal of Cr (VI) from chemical laboratory waste. Int. Ref. J. Eng. Sci. 2013, 2, 29–39. [Google Scholar]
- Diaz, R.; Alonso, S.; Sanchez, C.; Tomasini, A.; Bibbins-Martinez, M.; Diaz-Godinez, G. Characterization of the Growth and Laccase Activity of Strains of Pleurotus ostreatus in Submerged Fermentation. BioResources 2011, 6, 282–290. [Google Scholar] [CrossRef]
- Tay, C.; Liew, H.; Yin, C.-Y.; Abdul-Talib, S.; Surif, S.; Suhaimi, A.; Yong, S. Biosorption of cadmium ions using Pleurotus ostreatus: Growth kinetics, isotherm study and biosorption mechanism. Korean J. Chem. Eng. 2011, 28, 825–830. [Google Scholar] [CrossRef] [Green Version]
- Téllez-Téllez, M.; Fernández, F.; Montiel-González, A.; Sánchez, C.; Díaz-Godínez, G. Growth and laccase production by Pleurotus ostreatus in submerged and solid-state fermentation. Appl. Microbiol. Biotechnol. 2008, 81, 675–679. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Chen, W.; Wang, L.; Sun, H.; Zhu, Z. Improvement of laccase production by Pleurotus ostreatus by means of agroindustrial waste and fermentation kinetics. Mycosphere 2017, 8, 147–161. [Google Scholar] [CrossRef]
- Vamanu, E. Biological activities of the polysaccharides produced in submerged culture of two edible Pleurotus ostreatus mushrooms. J. Biomed. Biotechnol. 2012, 2012, 565974. [Google Scholar] [CrossRef] [Green Version]
- Billal, F.; Thurston, C. Purification of laccase II from Armillaria mellea and comparison of its properties with those of laccase I. Mycol. Res. 1996, 100, 1099–1105. [Google Scholar] [CrossRef]
- Castanera, R.; Omarini, A.; Santoyo, F.; Pérez, G.; Pisabarro, A.; Ramírez, L. Non-Additive Transcriptional Profiles Underlie Dikaryotic Superiority in Pleurotus ostreatus Laccase Activity. PLoS ONE 2013, 8, e73282. [Google Scholar] [CrossRef]
- Barclay, C.D.; Legge, R.L.; Farquhar, G.F. Modeling the Growth-Kinetics of Phanerochaete-Chrysosporium in Submerged Static Culture. Appl. Environ. Microbiol. 1993, 59, 1887–1892. [Google Scholar] [CrossRef] [Green Version]
- Cruz-Morato, C.; Lucas, D.; Llorca, M.; Rodriguez-Mozaz, S.; Gorga, M.; Petrovic, M.; Barcelo, D.; Vicent, T.; Sarra, M.; Marco-Urrea, E. Hospital wastewater treatment by fungal bioreactor: Removal efficiency for pharmaceuticals and endocrine disruptor compounds. Sci. Total Environ. 2014, 493, 365–376. [Google Scholar] [CrossRef]
- Patel, M.; Kumar, R.; Kishor, K.; Mlsna, T.; Pittman, C.U., Jr.; Mohan, D. Pharmaceuticals of emerging concern in aquatic systems: Chemistry, occurrence, effects, and removal methods. Chem. Rev. 2019, 119, 3510–3673. [Google Scholar] [CrossRef] [Green Version]
- Legorreta-Castañeda, A.J.; Lucho-Constantino, C.A.; Beltrán-Hernández, R.I.; Coronel-Olivares, C.; Vázquez-Rodríguez, G.A. Biosorption of water pollutants by fungal pellets. Water (Basel) 2020, 12, 1155. [Google Scholar] [CrossRef] [Green Version]
- Xiong, J.-Q.; Kurade, M.B.; Jeon, B.-H. Can Microalgae Remove Pharmaceutical Contaminants from Water? Trends Biotechnol. 2018, 36, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Gornik, T.; Kovacic, A.; Heath, E.; Hollender, J.; Kosjek, T. Biotransformation study of antidepressant sertraline and its removal during biological wastewater treatment. Water Res. 2020, 181, 115864. [Google Scholar] [CrossRef] [PubMed]
- Osawa, R.A.; Carvalho, A.P.; Monteiro, O.C.; Oliveira, M.C.; Florêncio, M.H. Transformation products of citalopram: Identification, wastewater analysis and in silico toxicological assessment. Chemosphere 2019, 217, 858–868. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Arnold, W.A.; Hozalski, R.M. The relative roles of sorption and biodegradation in the removal of contaminants of emerging concern (CECs) in GAC-sand biofilters. Water Res. 2018, 146, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Drzewicz, P.; Drobniewska, A.; Sikorska, K.; Nałęcz-Jawecki, G. Analytical and ecotoxicological studies on degradation of fluoxetine and fluvoxamine by potassium ferrate. Environ. Technol. 2019, 40, 3265–3275. [Google Scholar] [CrossRef] [PubMed]
- Gornik, T.; Shinde, S.; Lamovsek, L.; Koblar, M.; Heath, E.; Sellergren, B.; Kosjek, T. Molecularly Imprinted Polymers for the Removal of Antide-Pressants from Contaminated Wastewater. Polymers 2020, 13, 120. [Google Scholar] [CrossRef]
- Dias, S.; Correia, B.; Fraga-Santiago, P.; Silva, C.; Baptista, P.C.; Gomes, C.R.; Almeida, C.M.R. Potential of an estuarine salt marsh plant (Phragmites australis (Cav.) Trin. Ex Steud10751) for phytoremediation of bezafibrate and paroxetine. Hydrobiologia 2021, 848, 3291–3304. [Google Scholar] [CrossRef]
- Gornik, T.; Vozic, A.; Heath, E.; Trontelj, J.; Roskar, R.; Zigon, D.; Vione, D.; Kosjek, T. Determination and photodegradation of sertraline residues in aqueous environment. Environ. Pollut. 2020, 256, 113431. [Google Scholar] [CrossRef]
- Nassar, R.; Trivella, A.; Mokh, S.; Al-Iskandarani, M.; Budzinski, H.; Mazellier, P. Photodegradation of sulfamethazine, sulfamethoxypiridazine, amitriptyline, and clomipramine drugs in aqueous media. J. Photochem. Photobiol. A Chem. 2017, 336, 176–182. [Google Scholar] [CrossRef]
- Wawryniuk, M.; Pietrzak, A.; Nalecz-Jawecki, G. Evaluation of direct and indirect photodegradation of mianserin with high-performance liquid chromatography and short-term bioassays. Ecotoxicol. Environ. Saf. 2015, 115, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Chang, B.; Sun, X.; Luo, H.; Wang, W.; Li, C. Chloride-mediated electrochemical degradation of the venlafaxine antidepressant. Environ. Technol. Innov. 2022, 25, 102189. [Google Scholar] [CrossRef]
- Akhtar, N.; Mannan, M.A.-u. Mycoremediation: Expunging environmental pollutants. Biotechnol. Rep. 2020, 26, e00452. [Google Scholar] [CrossRef] [PubMed]

| ID | Component 1 | Component 2 | Weight of Component 1 [g] | Weight of Component 2 [g] |
|---|---|---|---|---|
| 1 | Glucose | CSL | 1.387 | 1.602 |
| 2 | Molasses | 2.635 | 1.602 | |
| 3 | Sucrose | 1.317 | 1.602 | |
| 4 | Whey | 2.274 | 0.940 | |
| 5 | Soluble starch | 1.248 | 1.602 | |
| 6 | Insoluble starch | 1.248 | 1.602 | |
| 7 | Microcrystalline cellulose | 1.248 | 1.602 | |
| 8 | Cellulose | 1.248 | 1.602 | |
| 9 | Poplar | 1.109 | 1.602 | |
| 10 | Larch | 1.109 | 1.602 | |
| 11 | Pine | 1.109 | 1.602 | |
| 12 | Glucose | DDGS | 1.620 | 2.144 |
| 13 | Molasses | 3.079 | 2.144 | |
| 14 | Sucrose | 1.539 | 2.144 | |
| 15 | Whey | 2.484 | 1.176 | |
| 16 | Soluble starch | 1.458 | 2.144 | |
| 17 | Insoluble starch | 1.458 | 2.144 | |
| 18 | Microcrystalline cellulose | 1.458 | 2.144 | |
| 19 | Cellulose | 1.458 | 2.144 | |
| 20 | Poplar | 1.296 | 2.144 | |
| 21 | Larch | 1.296 | 2.144 | |
| 22 | Pine | 1.296 | 2.144 | |
| 23 | Glucose | NH4Cl | 2.900 | 0.397 |
| 24 | Molasses | 5.511 | 0.397 | |
| 25 | Sucrose | 2.755 | 0.397 | |
| 26 | Whey | 3.278 | 0.161 | |
| 27 | Soluble starch | 2.610 | 0.397 | |
| 28 | Insoluble starch | 2.610 | 0.397 | |
| 29 | Microcrystalline cellulose | 2.610 | 0.397 | |
| 30 | Cellulose | 2.610 | 0.397 | |
| 31 | Poplar | 2.320 | 0.397 | |
| 32 | Larch | 2.320 | 0.397 | |
| 33 | Pine | 2.320 | 0.397 | |
| 34 | Glucose | NH4NO3 | 2.900 | 0.297 |
| 35 | Molasses | 5.511 | 0.297 | |
| 36 | Sucrose | 2.755 | 0.297 | |
| 37 | Whey | 3.278 | 0.120 | |
| 38 | Soluble starch | 2.610 | 0.297 | |
| 39 | Insoluble starch | 2.610 | 0.297 | |
| 40 | Microcrystalline cellulose | 2.610 | 0.297 | |
| 41 | Cellulose | 2.610 | 0.297 | |
| 42 | Poplar | 2.320 | 0.297 | |
| 43 | Larch | 2.320 | 0.297 | |
| 44 | Pine | 2.320 | 0.297 | |
| 45 | Glucose | Soy peptone | 1.811 | 1.051 |
| 46 | Molasses | 3.442 | 1.051 | |
| 47 | Sucrose | 1.721 | 1.051 | |
| 48 | Whey | 2.637 | 0.547 | |
| 49 | Soluble starch | 1.630 | 1.051 | |
| 50 | Insoluble starch | 1.630 | 1.051 | |
| 51 | Microcrystalline cellulose | 1.630 | 1.051 | |
| 52 | Cellulose | 1.630 | 1.051 | |
| 53 | Poplar | 1.449 | 1.051 | |
| 54 | Larch | 1.449 | 1.051 | |
| 55 | Pine | 1.449 | 1.051 | |
| 56 | Glucose | Urea | 2.789 | 0.223 |
| 57 | Molasses | 5.299 | 0.223 | |
| 58 | Sucrose | 2.649 | 0.223 | |
| 59 | Whey | 3.226 | 0.092 | |
| 60 | Soluble starch | 2.510 | 0.223 | |
| 61 | Insoluble starch | 2.510 | 0.223 | |
| 62 | Microcrystalline cellulose | 2.510 | 0.223 | |
| 63 | Cellulose | 2.510 | 0.223 | |
| 64 | Poplar | 2.231 | 0.223 | |
| 65 | Pine | 2.231 | 0.223 | |
| 66 | Larch | 2.231 | 0.223 | |
| 67 | Glucose | Yeast extract | 1.990 | 0.929 |
| 68 | Molasses | 3.782 | 0.929 | |
| 69 | Sucrose | 1.891 | 0.929 | |
| 70 | Whey | 2.766 | 0.462 | |
| 71 | Soluble starch | 1.791 | 0.929 | |
| 72 | Insoluble starch | 1.791 | 0.929 | |
| 73 | Microcrystalline cellulose | 1.791 | 0.929 | |
| 74 | Cellulose | 1.791 | 0.929 | |
| 75 | Poplar | 1.592 | 0.929 | |
| 76 | Larch | 1.592 | 0.929 | |
| 77 | Pine | 1.592 | 0.929 | |
| RGM * | Glucose, yeast extract, casein peptone, KH2PO4 | 2 g of glucose, 0.4 g of yeast extract, 0.4 g of casein peptone, 0.04 g of KH2PO4 | ||
| H2O ** | Distilled water only | |||
| Components (C Source + N Source) | Comments | Percentage of C Source [% w/v] | Percentage of N Source [% w/v] |
|---|---|---|---|
| Molasses + CSL | - | 6.59 | 4.01 |
| Poplar + CSL | Sawdust as a biocarrier; deciduous tree preferred by WRF | 2.77 | 4.01 |
| Molasses + DDGS | - | 7.70 | 5.36 |
| Poplar + DDGS | Sawdust as a biocarrier; deciduous tree preferred by WRF | 3.24 | 5.36 |
| Nominal Concentration C*DDGS/CSL | 100% | 50% | 25% | 10% | 5% |
|---|---|---|---|---|---|
| mS fraction | mS | ⅟2 mS | ⅟4 mS | ⅟10 mS | ⅟20 mS |
| Actual concentration CCSL [% w/v] | 5.27 | 2.63 | 1.32 | 0.527 | 0.263 |
| Actual concentration CDDGS [% w/v] | 7.44 | 3.72 | 1.86 | 0.744 | 0.372 |
| Medium | Species | µ [d−1] | texp0 [d] | Δtexp [d] | mavS [g] | tA>0 [d] | Amax [U∙mL−1] | tAmax [d] |
|---|---|---|---|---|---|---|---|---|
| RGM (reference growing medium) | Am | 0.594 (74) | 8 | 6 | 0.0529 (80) | n.a.d. a | n.a.d. | n.a.d. |
| Pc | 0.65 (19) | 5 | 4 | 0.1656 (69) | n.a.d. | n.a.d. | n.a.d. | |
| Po | 0.00769 (46) | 11 | >3 | 0.0272 (27) | n.a.d. | n.a.d. | n.a.d. | |
| Molasses + CSL | Am | 1.46 (61) | 11 | 2 | 0.942 (63) | 13 | 0.0078 (34) | 13 |
| Pc | 0.839 (70) | 5 | 2 | 0.780 (33) | n.a.d. | n.a.d. | n.a.d. | |
| Po | 0.97 (18) | 5 | 5 | 0.826 (11) | 7 | 0.050 (11) | 14 | |
| Molasses + DDGS | Am | 0.69 (18) | 7 | 5 | 0.671 (27) | <5 | 0.051 (50) | 11 |
| Pc | 0.318 (35) | <7 | 3 | 0.3872 (37) | n.a.d. | n.a.d. | n.a.d. | |
| Po | 0.77 (11) | 6 | 4 | 0.533 (22) | <5 | 0.00764 (86) | 14 | |
| Poplar + CSL | Am | 0.87 (14) | 8 | 4 | 0.530 (77) | 11 | 0.167 (15) | 13 |
| Pc | 0.90 (27) | 5 | 6 | 0.751 (19) | n.a.d. | n.a.d. | n.a.d. | |
| Po | 1.47 (36) | 6 | 8 | 0.770 (25) | 7 | 1.41 (34) | 14 | |
| Poplar + DDGS | Am | 0.747 (74) | 8 | 4 | 0.579 (16) | 9 | 0.0163 (22) | 14 |
| Pc | 0.54 (18) | 5 | 5 | 0.3831 (80) | n.a.d. | n.a.d. | n.a.d. | |
| Po | 1.47 (70) | 5 | >9 | 0.854 (18) | 8 | 0.92 (24) | 14 | |
| CSL 100% b | Am | 0.454 (34) | 11 | 3 | 0.0656 (31) | n.a.d. | n.a.d. | n.a.d. |
| Pc | 1.24 (50) | 5 | 2 | 0.4606 (87) | n.a.d. | n.a.d. | n.a.d. | |
| Po | 1.37 (26) | 5 | 2 | 0.4381 (62) | 7 | 0.491 (54) | 12 | |
| CSL 50% d | Am | 0.503 (93) | 10 | >4 | 0.2106 (35) | 10 | 0.156 (13) | 14 |
| Pc | 0.65 (19) | <5 | 6 | 0.2911 (87) | n.a.d. | n.a.d. | n.a.d. | |
| Po | 1.504 (38) | 5 | 2 | 0.2191 (17) | 6 | 0.873 (33) | 10 | |
| CSL 25% | Am | 0.65 (11) | 9 | >5 | 0.0795 (27) | 12 | 0.0135 (68) | 14 |
| Pc | 0.136 (19) | <5 | 8 | 0.1545 (77) | n.a.d. | n.a.d. | n.a.d. | |
| Po | 1.30 (16) | 5 | 3 | 0.1502 (12) | <5 | 0.076 (21) | 12 | |
| CSL 10% | Am | 0.364 (25) | 6 | >8 | 0.0312 (17) | 10 | 0.0057 (19) | 14 |
| Pc | 0.101 (20) | 6 | 6 | 0.0512 (25) | n.a.d. | n.a.d. | n.a.d. | |
| Po | 0.362 (54) | 6 | 2 | 0.0678 (56) | <5 | 0.0213 (42) | 12 | |
| CSL 5% | Am | 0.521 (96) | 5 | 6 | 0.01772 (70) | <5 | 0.0043 (12) | 14 |
| Pc | ND | 11 | >3 | 0.0168 (11) | n.a.d. | n.a.d. | n.a.d. | |
| Po | 0.5265 (15) | 5 | 4 | 0.02208 (71) | <5 | 0.0084 (21) | 8 | |
| DDGS 100% c | Am | 0.8268 (30) | 12 | 2 | 0.1199 (41) | 10 | 0.0531 (58) | 14 |
| Pc | 0.334 (36) | <5 | 3 | 0.770 (27) | n.a.d. | n.a.d. | n.a.d. | |
| Po | 1.14 (12) | 6 | 6 | 0.893 (25) | 11 | 0.263 (37) | 12 | |
| DDGS 50% d | Am | 1.19 (28) | 8 | 6 | 0.336 (12) | 14 | 0.040 (14) | 14 |
| Pc | 0.3953 (31) | <5 | 3 | 0.2920 (22) | n.a.d. | n.a.d. | n.a.d. | |
| Po | 1.75 (26) | 5 | 3 | 0.2500 (77) | 7 | 0.33 (14) | 14 | |
| DDGS 25% | Am | 0.730 (57) | 10 | 4 | 0.0415 (39) | 11 | 0.00186 (84) | 14 |
| Pc | 0.83 (35) | 5 | 2 | 0.08093 (67) | n.a.d. | n.a.d. | n.a.d. | |
| Po | 0.81 (13) | 5 | 3 | 0.2071 (33) | 6 | 0.403 (55) | 10 | |
| DDGS 10% | Am | 0.490 (67) | 12 | 2 | 0.00413 (47) | n.a.d. | n.a.d. | n.a.d. |
| Pc | 0.424 (67) | 6 | 2 | 0.0407 (18) | n.a.d. | n.a.d. | n.a.d. | |
| Po | 0.484 (37) | 8 | 2 | 0.0154 (12) | <5 | 0.051 (10) | 11 | |
| DDGS 5% | Am | 0.521 (96) | 8 | 5 | 0.0245 (11) | 12 | 0.0068 (12) | 13 |
| Pc | ND | ND | ND | 0.01500 (36) | n.a.d. | n.a.d. | n.a.d. | |
| Po | 0.402 (55) | 5 | 3 | 0.0277 (19) | <5 | 0.0088 (15) | 11 |
| Mathematical Model | CSL | DDGS | ||||||
|---|---|---|---|---|---|---|---|---|
| (Non)inhibitory | Name | Equation | A. mellea | P. chrysosporium | P. ostreatus | A. mellea | P. chrysosporium | P. ostreatus |
| Non-inhibitory models | Monod | µ = µm · CS/(KS + CS) | 0.690 | 0.928 * | 0.845 | 0.887 | 0.355 | 0.678 |
| Tessier | µ = µm · [1 − exp(−CS/KS)] | 0.895 | 0.963 | 0.948 | 0.941 | 0.722 | 0.853 | |
| Moser | µ = µm · (CS)m/[KS + (CS)m] m > 0 | 0.866 | 0.964 | 0.960 | 0.957 | 0.817 | 0.838 | |
| Inhibitory models | Haldane- Andrews | µ = µm·CS/[KS + CS + (CS)2/KI] | 0.904 | 0.932 | 0.917 | 0.888 | 0.841 | 0.767 |
| Aiba | µ = µm · CS · exp(−CS/KI)/(KS + CS) | 0.861 | 0.925 | 0.938 | 0.890 | 0.838 | 0.831 | |
| Edwards | µ = µm · [exp(−CS/KI) − exp(−CS/KS)] | 0.801 | 0.926 | 0.899 | 0.833 | 0.531 | 0.728 | |
| Luong | µ = µm · CS · (1 − CS/K2)m/(K1 + CS) | 0.902 | 0.999 | 0.946 | 0.876 | 0.804 | 0.833 | |
| Yamane | µ = µm · CS/[KS + CS + (CS/KI)m] | 0.909 | 0.928 | 0.991 | 0.889 | 0.968 | 0.982 | |
| Han- Levenspiel | µ = µm · CS· · (1 − CS/K2)m/[CS + K1 · (1 − CS/K2)n] | 0.893 | 0.931 | 0.963 | 0.974 | 0.994 | 0.965 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sośnicka, A.; Kózka, B.; Makarova, K.; Giebułtowicz, J.; Klimaszewska, M.; Turło, J. Optimization of White-Rot Fungi Mycelial Culture Components for Bioremediation of Pharmaceutical-Derived Pollutants. Water 2022, 14, 1374. https://doi.org/10.3390/w14091374
Sośnicka A, Kózka B, Makarova K, Giebułtowicz J, Klimaszewska M, Turło J. Optimization of White-Rot Fungi Mycelial Culture Components for Bioremediation of Pharmaceutical-Derived Pollutants. Water. 2022; 14(9):1374. https://doi.org/10.3390/w14091374
Chicago/Turabian StyleSośnicka, Agata, Bartosz Kózka, Katerina Makarova, Joanna Giebułtowicz, Marzenna Klimaszewska, and Jadwiga Turło. 2022. "Optimization of White-Rot Fungi Mycelial Culture Components for Bioremediation of Pharmaceutical-Derived Pollutants" Water 14, no. 9: 1374. https://doi.org/10.3390/w14091374
APA StyleSośnicka, A., Kózka, B., Makarova, K., Giebułtowicz, J., Klimaszewska, M., & Turło, J. (2022). Optimization of White-Rot Fungi Mycelial Culture Components for Bioremediation of Pharmaceutical-Derived Pollutants. Water, 14(9), 1374. https://doi.org/10.3390/w14091374






