Assessment of Bilge Water Degradation by Isolated Citrobacter sp. and Two Indigenous Strains and Identification of Organic Content by GC-MS
Abstract
1. Introduction
2. Material and Methods
2.1. Collection of Real Bilge Water
2.2. Isolation of Pure Strains from Environmental Samples and Identification of Indigenous Bacterial Strains
2.3. Strain Characterization by 16S rRNA Sequence Analysis
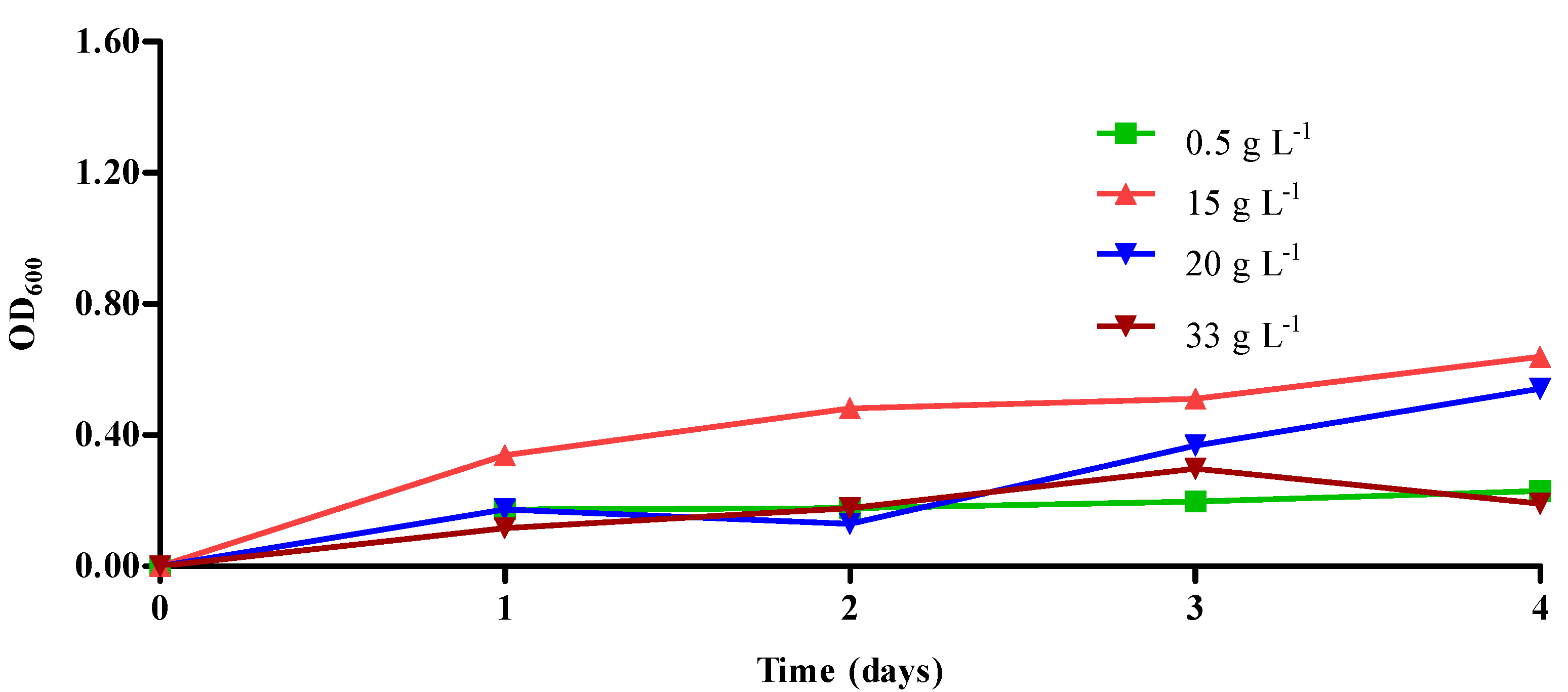
2.4. Cultivation of Bacterial Strain under Different Environmental Conditions
2.5. Biodegradation of Real Bilge Water by Different Isolated Bacterial Strains
2.6. HPLC-UV/VIS Analysis
2.7. GC-MS Analysis
3. Results and Discussion
3.1. Isolation and Characterization of Pure Bacterial Strains
3.2. Biodegradation of Phenanthrene by Citrobacter sp. D2
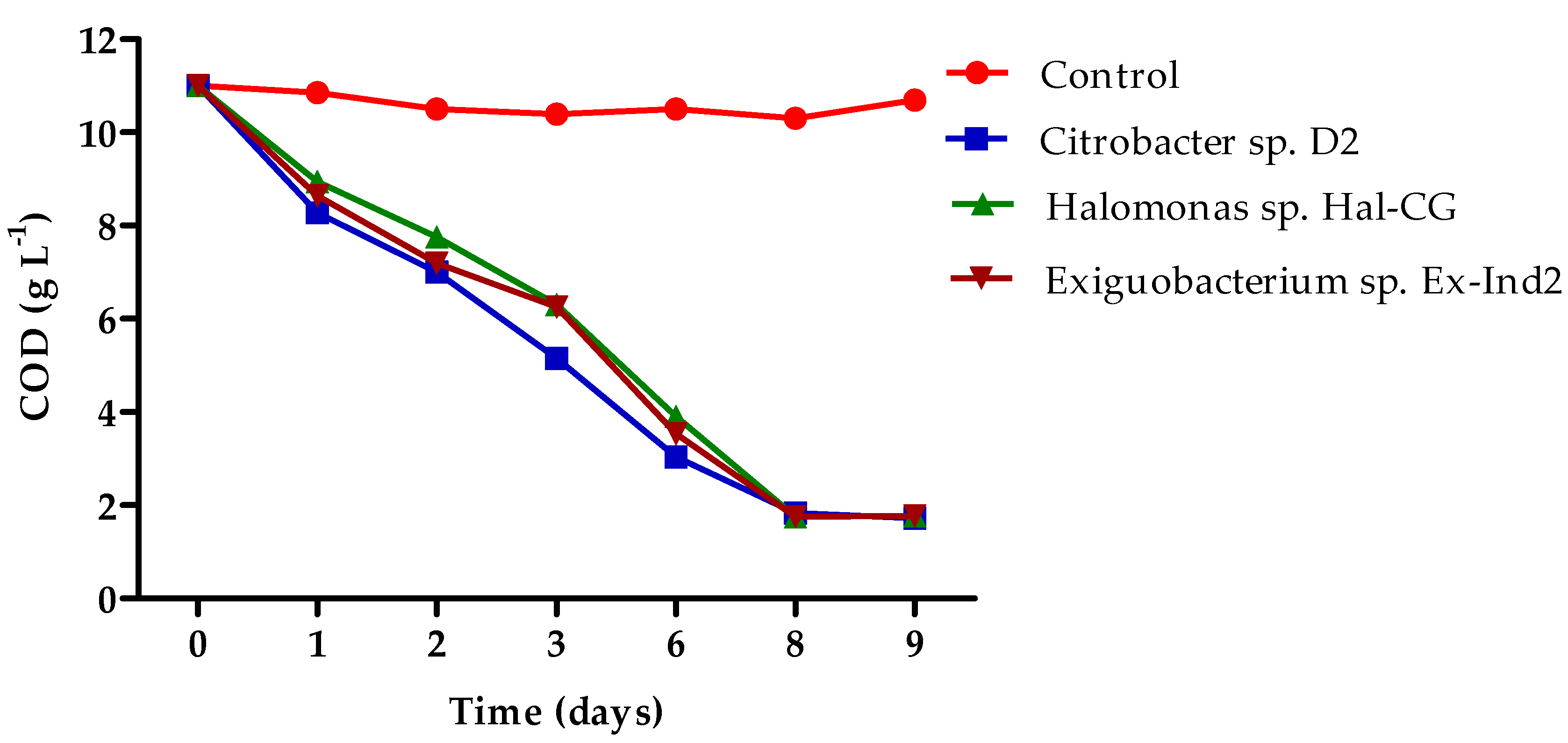
3.3. Biodegradation of Bilge Water by Isolated Strains
3.4. GC-MS Analysis of Real Bilge Water
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McLaughlin, C.; Falatko, D.; Danesi, R.; Albert, R. Characterizing shipboard bilgewater effluent before and after treatment. Environ. Sci. Pollut. Res. 2014, 21, 5637–5652. [Google Scholar] [CrossRef] [PubMed]
- IMO. International Convention for the Prevention of Pollution from Ships (MARPOL). 1973. Available online: https://www.imo.org/en/About/Conventions/Pages/International-Convention-for-the-Prevention-of-Pollution-from-Ships-(MARPOL).aspx (accessed on 15 March 2022).
- Gryta, M. Separation of saline oily wastewater by membrane distillation. Chem. Pap. 2020, 74, 2277–2286. [Google Scholar] [CrossRef]
- Han, M.; Zhang, J.; Chu, W.; Chen, J.; Zhou, G. Research progress and prospects of marine oily wastewater treatment: A review. Water 2019, 11, 2517. [Google Scholar] [CrossRef]
- Church, J.; Lundin, J.G.; Diaz, D.; Mercado, D.; Willner, M.R.; Lee, W.H.; Paynter, D.M. Identification and characterization of bilgewater emulsions. Sci. Total Environ. 2019, 691, 981–995. [Google Scholar] [CrossRef] [PubMed]
- Tummons, E.N.; Tarabara, V.V.; Chew, J.W.; Fane, A.G. Behavior of oil droplets at the membrane surface during crossflow microfiltration of oil-water emulsions. J. Membr. Sci. 2016, 500, 211–224. [Google Scholar] [CrossRef]
- Rincón, G.J.; La Motta, E.J. Simultaneous removal of oil and grease, and heavy metals from artificial bilge water using electro-coagulation/flotation. J. Environ. Manag. 2014, 144, 42–50. [Google Scholar] [CrossRef]
- Corti-Monzón, G.; Nisenbaum, M.; Villegas-Plazas, M.; Junca, H.; Murialdo, S. Enrichment and characterization of a bilge microbial consortium with oil in water-emulsions breaking ability for oily wastewater treatment. Biodegradation 2020, 31, 57–72. [Google Scholar] [CrossRef]
- Nisenbaum, M.; Corti-Monzón, G.; Villegas-Plazas, M.; Junca, H.; Mangani, A.; Patat, M.L.; Gonzá, J.F.; Murialdo, S.E. Enrichment and key features of a robust and consistent indigenous marine-cognate microbial consortium growing on oily bilge wastewaters. Biodegradation 2020, 31, 91–108. [Google Scholar] [CrossRef]
- Fowler, S.J.; Toth, C.R.A.; Gieg, L.M. Community structure in methanogenic enrichments provides insight into syntrophic interactions in hydrocarbon-impacted environments. Front. Microbiol. 2016, 7, 1–13. [Google Scholar] [CrossRef]
- Mazioti, A.A.; Notarides, G.; Symeou, G.; Vyrides, I. Improving biological treatment of real bilge wastewater with zero valent iron and activated charcoal addition. Front. Bioeng. Biotechnol. 2020, 8, 1462. [Google Scholar] [CrossRef]
- Gouveia, V.; Almeida, C.M.R.; Almeida, T.; Teixeira, C.; Mucha, A.P. Indigenous microbial communities along the NW Portuguese Coast: Potential for hydrocarbons degradation and relation with sediment contamination. Mar. Pollut. Bull. 2018, 131, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.W.; Lee, H.; Kwon, B.-O.; Khim, J.S.; Yim, U.H.; Kim, B.S.; Kim, J.J. Biosurfactant-assisted bioremediation of crude oil by indigenous bacteria isolated from Taean beach sediment. Environ. Pollut. 2018, 241, 254–264. [Google Scholar] [CrossRef] [PubMed]
- Mazioti, A.A.; Vasquez, M.I.; Vyrides, I. Comparison of different cultures and culturing conditions for the biological deterioration of organic load from real saline bilge wastewater: Microbial diversity insights and ecotoxicity assessment. Environ. Sci. Pollut. Res. 2021, 28, 36506–36522. [Google Scholar] [CrossRef] [PubMed]
- Uma, V.; Gandhimathi, R. Organic removal and synthesis of biopolymer from synthetic oily bilge water using the novel mixed bacterial consortium. Bioresour. Technol. 2019, 273, 169–176. [Google Scholar] [CrossRef]
- Feknous, N.; Branes, Z.; Rouabhia, K.; Batisson, I.; Amblard, C. Isolation characterization and growth of locally isolated hydrocarbonoclastic marine bacteria (Eastern Algerian Coast). Environ. Monit. Assess. 2017, 189, 49. [Google Scholar] [CrossRef]
- Primeia, S.; Inoue, C.; Chien, M.F. Potential of biosurfactants’ production on degrading heavy oil by bacterial consortia obtained from tsunami-induced oil-spilled beach areas in Miyagi, Japan. J. Mar. Sci. Eng. 2020, 8, 577. [Google Scholar] [CrossRef]
- Rizzo, C.; Rappazzo, A.C.; Michaud, L.; De Domenico, E.; Rochera, C.; Camacho, A.; Lo Giudice, A. Efficiency in hydrocarbon degradation and biosurfactant production by Joostella sp. A8 when grown in pure culture and consortia. J. Environ. Sci. 2018, 67, 115–126. [Google Scholar] [CrossRef]
- Boraha, D.; Yadav, R.N.S. Bioremediation of petroleum based contaminants with biosurfactant produced by a newly isolated petroleum oil degrading bacterial strain. Egypt. J. Pet. 2017, 26, 181–188. [Google Scholar] [CrossRef]
- Liu, H.; Yang, G.; Jia, H.; Sun, B. Crude Oil Degradation by a Novel Strain Pseudomonas aeruginosa AQNU-1 Isolated from an Oil-Contaminated Lake Wetland. Processes 2022, 10, 307. [Google Scholar] [CrossRef]
- Lenchi, N.; Kebbouche-Gana, S.; Servais, P.; Gana, M.L.; Lliros, M. Diesel biodegradation capacities and biosurfactant production in saline-alkaline conditions by Delftia sp. NL1, Isolated from an Algerian Oilfield. Geomicrobiol. J. 2020, 37, 454–466. [Google Scholar] [CrossRef]
- Olivera, N.L.; Commendatore, M.G.; Delgado, O.; Esteves, J.L. Microbial characterization and hydrocarbon biodegradation potential of natural bilge waste microflora. J. Ind. Microbiol. Biotechnol. 2003, 30, 542–548. [Google Scholar] [CrossRef] [PubMed]
- Santisi, S.; Gentile, G.; Volta, A.; Bonsignore, M.; Mancini, G.; Quatrini, P.; Cappello, S. Isolation and Characterization of Oil-Degrading Bacteria from Bilge Water. Int. J. Microbiol. Appl. 2015, 2, 45–49. [Google Scholar]
- Nievas, M.L.; Commendatore, M.G.; Olivera, N.L.; Esteves, J.L.; Bucala, V. Biodegradation of bilge waste from Patagonia with an indigenous microbial Community. Bioresour. Technol. 2006, 97, 2280–2290. [Google Scholar] [CrossRef] [PubMed]
- Sivaraman, C.; Ganguly, A.; Nikolausz, M.; Mutnuri, S. Isolation of hydrocarbonoclastic bacteria from bilge oil contaminated water. Int. J. Environ. Sci. Technol. 2011, 8, 461–470. [Google Scholar] [CrossRef][Green Version]
- Cappello, S.; Santisi, S.; Calogero, R.; Hassanshahian, M.; Yakimov, M.M. Characterisation of Oil-Degrading Bacteria Isolated from Bilge Water. Water Air Soil Pollut. 2012, 223, 3219–322628. [Google Scholar] [CrossRef]
- Corti-Monzón, G.; Nisenbaum, M.; Peressutti, S.; Junca, H.; García-Bonilla, E.; Murialdo, S.E. Oily Bilge Wastes Harbor a Set of Persistent Hydrocarbonoclastic Bacteria Accompanied by a Variable alkB Gene Composition in Marine Vessel Samples from Southwestern Atlantic Port of Mar del Plata, Argentina. Water Air Soil Pollut. 2021, 232, 1–19. [Google Scholar] [CrossRef]
- Tiselius, P.; Magnusson, K. Toxicity of treated bilge water: The need for revised regulatory control. Mar. Pollut. Bull. 2017, 114, 860–866. [Google Scholar] [CrossRef]
- Drakou, E.; Koutinas, M.; Pantelides, I.; Tsolakidou, M.; Vyrides, I. Insights into the metabolic basis of the halotolerant Pseudomonas aeruginosa strain LVD-10 during toluene biodegradation. Int. Biodeter. Biodegr. 2015, 99, 85–94. [Google Scholar] [CrossRef]
- Koutinas, M.; Kyriakou, M.; Andreou, K.; Hadjicharalambous, M.; Kaliviotis, E.; Pasias, D.; Kazamias, G.; Varavvas, C.; Vyrides, I. Enhanced biodegradation and valorization of drilling wastewater via simultaneous production of bio-surfactants and polyhydroxyalkanoates by Pseudomonas citronellolis SJTE-3. Bioresour. Technol. 2021, 340, 340125679. [Google Scholar] [CrossRef]
- APHA. Standard Methods for the Examination of Water and Wastewater, 21st ed.; American Public Health Association: Washington, DC, USA, 2005. [Google Scholar]
- Vyrides, I.; Drakou, E.-M.; Ioannou, S.; Michael, F.; Gatidou, G.; Stasinakis, A.S. Biodegradation of bilge water: Batch test under anaerobic and aerobic conditions and performance of three pilot aerobic Moving Bed Biofilm Reactors (MBBRs) at different filling fractions. J. Environ. Manag. 2018, 217, 356. [Google Scholar] [CrossRef]
- Deng, T.; Wang, H.; Yan, K. Phenol biodegradation by isolated Citrobacter strain under hypersaline conditions. Water Sci. Technol. 2018, 77, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Adebajo, S.O.; Akintokun, A.K.; Ojo, A.E.; Dami, M.; Egbagbe, D.M.; Akintokun, P.O.; Adebajo, L.O. Biosurfactant Production by Rhizospheric Bacteria Isolated from Biochar Amended Soil Using Different Extraction Solvents. Appl. Environ. Res. 2019, 41, 72–82. [Google Scholar] [CrossRef]
- Anaukwu, C.G.; Ezemba, C.C.; Anakwenze, V.N.; Agu, K.C.; Okeke, B.C.; Awah, N.S.; Ekwealor, I.A. Effect of biosurfactant produced by Citrobacter murliniaeAF025369 and a synthetic surfactant on degradation of crude oil. Edorium. J. Microbiol. 2016, 1, 1–6. [Google Scholar]
- Kaczorek, E.; Sałek, K.; Guzi, U.; Dudzinska-Bajorek, B. Cell surface properties and fatty acids composition of Stenotropho-monas maltophilia under the influence of hydrophobic compounds and surfactants. New Biotechnol. 2013, 30, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; He, J.; Liu, T.; Xin, X.; Hu, H. Removal of heavy metal from sludge by the combined application of a biodegradable biosurfactant and complexing agent in enhanced electrokinetic treatment. Chemosphere 2017, 189, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Cazoir, D.; Fine, L.; Ferronato, C.; Chovelon, J.M. Hydrocarbon removal from bilgewater by a combination of air-stripping and photocatalysis. J. Hazard. Mater. 2012, 235–236, 159–168. [Google Scholar] [CrossRef]
- Shen, Y.; Yao, J.; Son, J.; Zhu, Z.; Yu, X.Y. Liquid ToF-SIMS revealing the oil, water, and surfactant interface evolution. Phys. Chem. Chem. Phys 2020, 22, 11771–11782. [Google Scholar] [CrossRef]
- Woiski, C.; Dobslaw, D.; Engesser, K.-H. Isolation and characterization of 2-butoxyethanol degrading bacterial strains. Biodegradation 2020, 31, 153–169. [Google Scholar] [CrossRef]
- O’Reilly, K.T.; Mohler, R.E.; Zemo, D.A.; Ahn, S.; Tiwary, A.K.; Magaw, R.I.; Devine, C.E.; Synowiec, K.A. Identification of ester metabolites from petroleum hydrocarbon biodegradation in groundwater using GC x GC-TOFMS. Environ. Toxicol. Chem. 2015, 34, 1959–1961. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]



| Name | Molecular Formula | RT | Probability (%) |
|---|---|---|---|
| Heptane | C7H16 | 7.05 | 81.8 |
| Methylyclohexane (toluene) | C7H14 | 7.68 | 55.6 |
| 2,3,4-trimethyl-pentane | C8H18 | 8.43 | 55.4 |
| 3-methyl-heptane (octane) | C8H18 | 9.16 | 59 |
| Octane | C8H18 | 10.14 | 65.3 |
| 1-butoxy-2-propanol | C7H16O2 | 15.26 | 93.7 |
| 2-ethyl-1-hexanol | C8H18O | 18.36 | 71.6 |
| Methyl isobutyl ketone | C6H12O | 18.92 | 92.9 |
| 2-methyl-phenol | C7H8O | 19.31 | 59.0 |
| Acetophenone | C8H8O | 19.71 | 52.3 |
| Pentadecane | C15H32 | 31.98 | 52.5 |
| Hexadecane | C16H34 | 34.42 | 56.3 |
| Hexadecanoic acid, methyl ester (palmitic acid) | C17H34O2 | 41.40 | 83.9 |
| Docosane | C22H46 | 46.55 | 58.5 |
| Tricosane | C23H48 | 48.26 | 56.0 |
| Compound | Untreated BW | Citrobacter sp. D2. | Exiguobacterium sp. Ex-Ind2 | Halomonas sp. Hal-CG |
|---|---|---|---|---|
| Heptane | 32008407 (81.8%) | n.d. | 14717758 (80,7%) | 6195579 (83.7%) |
| Hexane | n.d. | 5182909 | n.d. | n.d. |
| N-hexadecanoic acid | 2092353 (83.9%) | n.d. | n.d. | n.d. |
| N-hexadecanoic, methyl ester | n.d. | 37200692 (80%) | 5698795 (81.0) | n.d. |
| N-hexadecanoic, ethyl ester | n.d. | 2945055 (86.4%) | 14717758 (83.2%) | 4921636 (87.4%) |
| Methyl isobutyl Ketone | 8198875 (92.9%) | n.d. | n.d. | n.d. |
| 1-butoxy-2-propanol | 728060 (93.7%) | n.d. | n.d. | n.d. |
| Octadecanoic acid methyl ester | n.d. | 9251735 (82.7%) | 4955474 (84.1%) | 2722022 (80.5%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gatidou, G.; Drakou, E.-M.; Vyrides, I. Assessment of Bilge Water Degradation by Isolated Citrobacter sp. and Two Indigenous Strains and Identification of Organic Content by GC-MS. Water 2022, 14, 1350. https://doi.org/10.3390/w14091350
Gatidou G, Drakou E-M, Vyrides I. Assessment of Bilge Water Degradation by Isolated Citrobacter sp. and Two Indigenous Strains and Identification of Organic Content by GC-MS. Water. 2022; 14(9):1350. https://doi.org/10.3390/w14091350
Chicago/Turabian StyleGatidou, Georgia, Efi-Maria Drakou, and Ioannis Vyrides. 2022. "Assessment of Bilge Water Degradation by Isolated Citrobacter sp. and Two Indigenous Strains and Identification of Organic Content by GC-MS" Water 14, no. 9: 1350. https://doi.org/10.3390/w14091350
APA StyleGatidou, G., Drakou, E.-M., & Vyrides, I. (2022). Assessment of Bilge Water Degradation by Isolated Citrobacter sp. and Two Indigenous Strains and Identification of Organic Content by GC-MS. Water, 14(9), 1350. https://doi.org/10.3390/w14091350








