The Ability of a Bacterial Strain to Remove a Phenolic Structure as an Approach to Pulp and Paper Mill Wastewater Treatment: Optimization by Experimental Design
Abstract
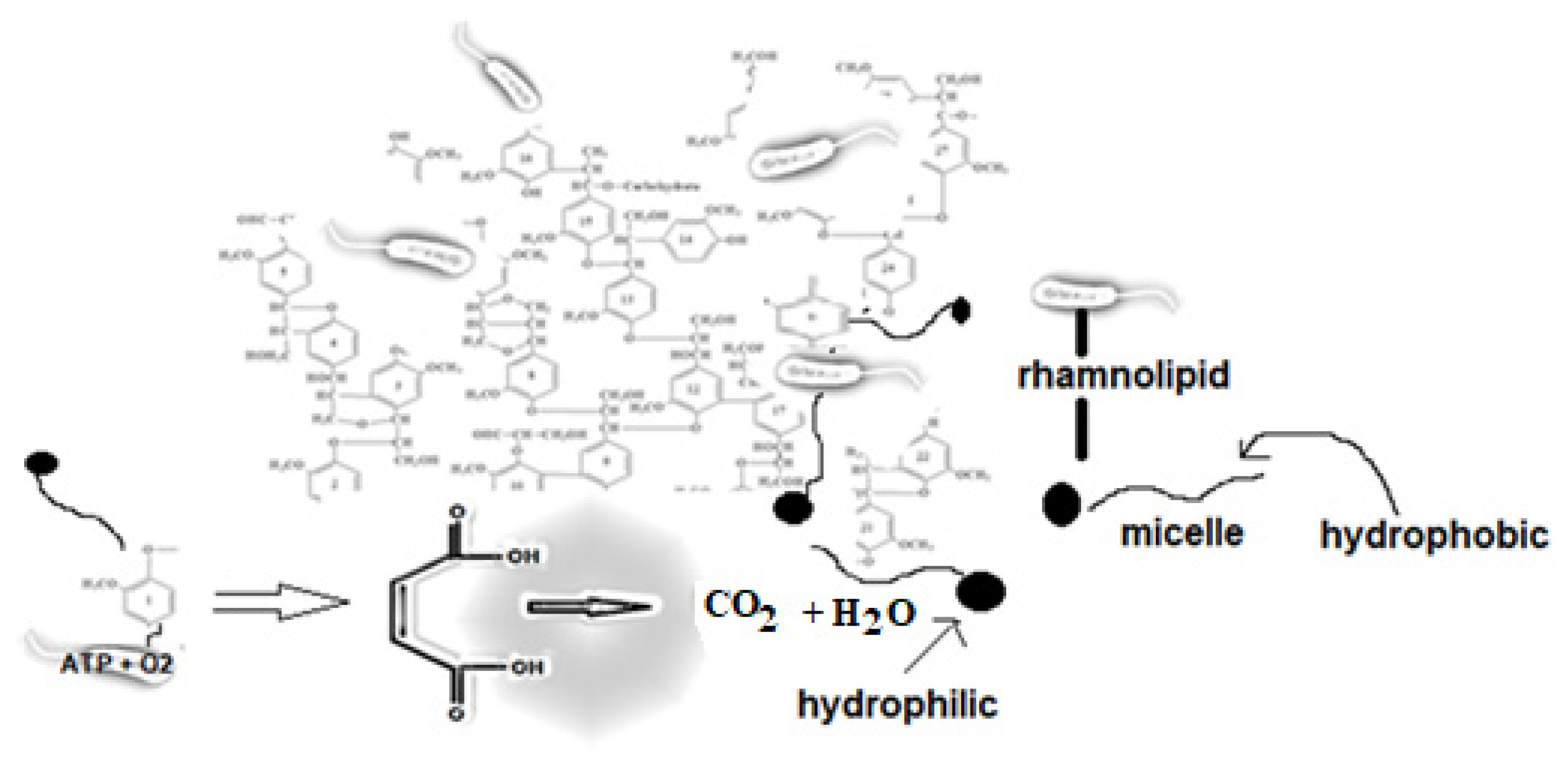
1. Introduction
2. Materials and Methods
2.1. Chemical Oxygen Demand (COD)
2.2. Total Organic Carbon (TOC)
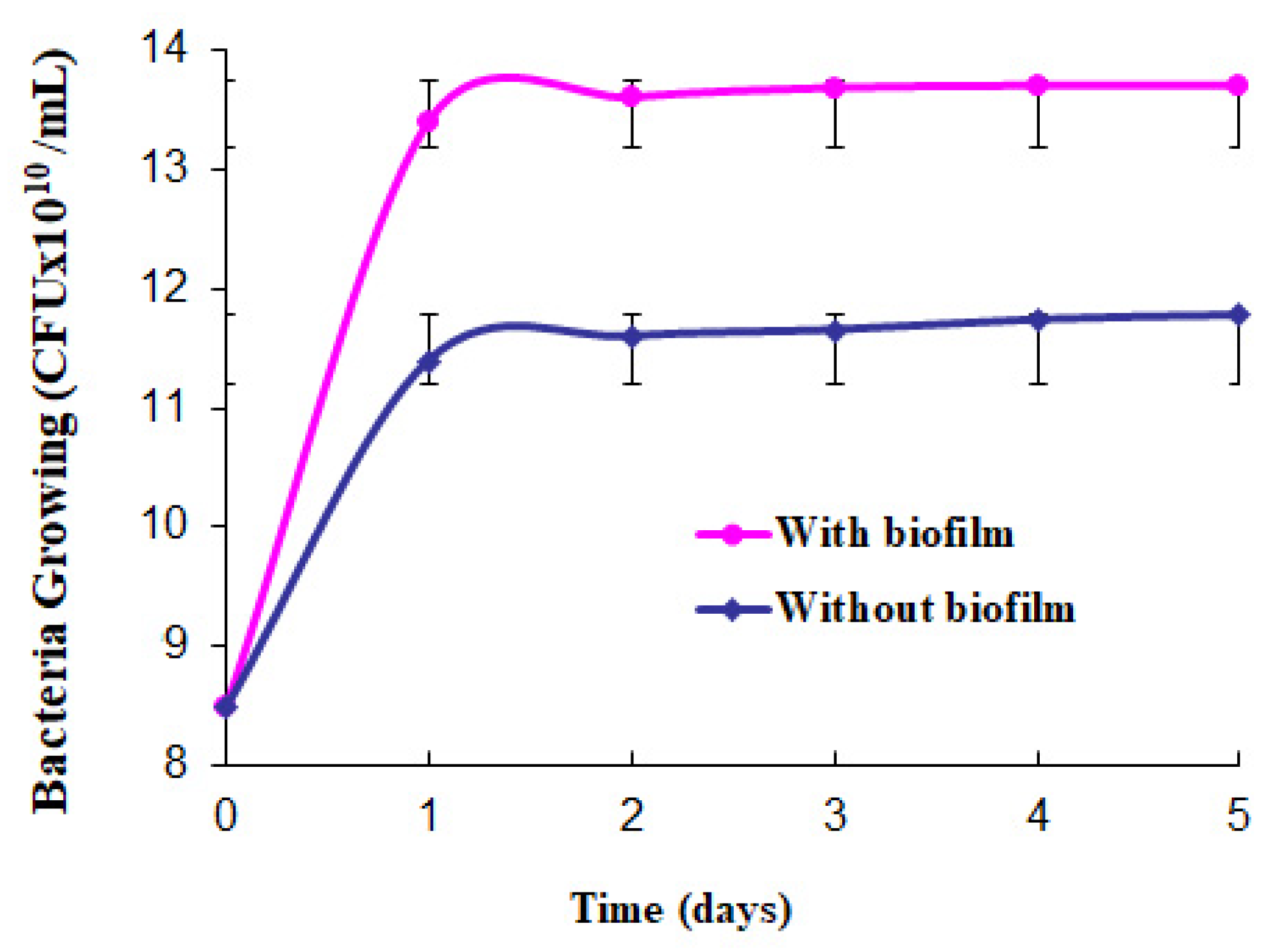
2.3. Bacterial Growth
2.4. Multivariate Analysis
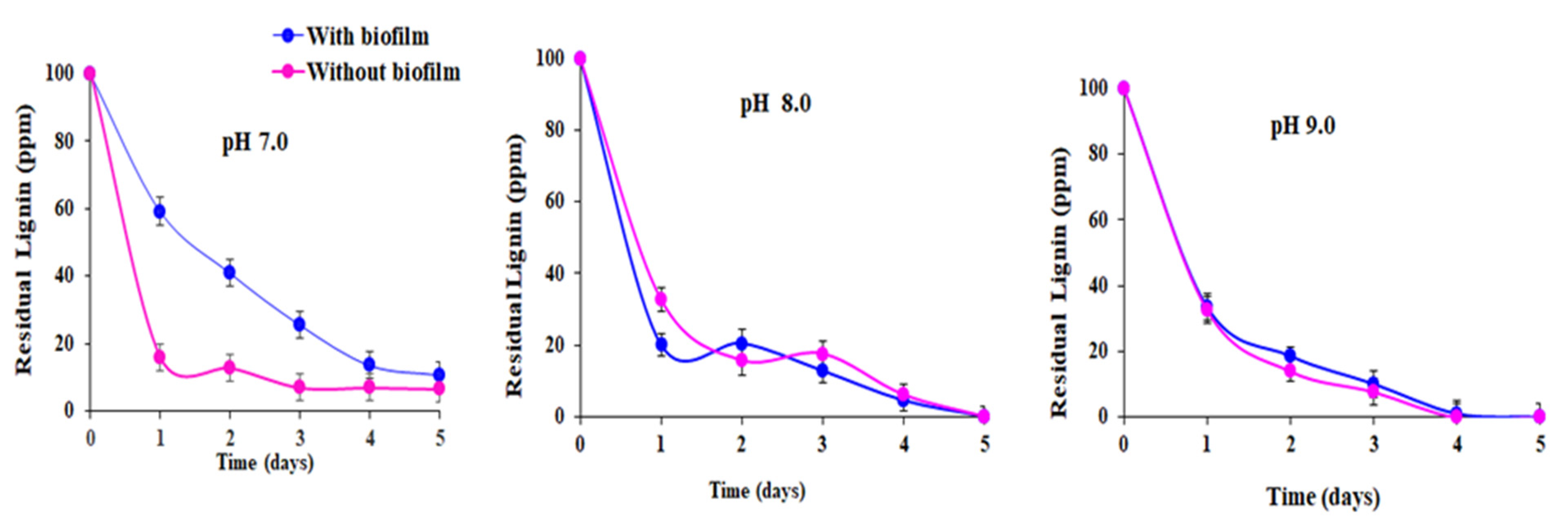
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ashrafi, O.; Yerushalmi, L.; Haghighat, F. Wastewater treatment in the pulp-and-paper industry: A review of treatment processes and the associated greenhouse gas emission. J. Environ. Manag. 2015, 158, 146–157. [Google Scholar] [CrossRef]
- Macdonald, G.Z.; Hogan, N.S.; Van Den Heuvel, M.R. Effects of habitat and pulp and paper mill contamination on a population of brook stickleback (Culaea inconstans). Water Qual. Res. J. Can. 2020, 55, 52–66. [Google Scholar] [CrossRef]
- Singh, A.K.; Chandra, R. Pollutants released from the pulp paper industry: Aquatic toxicity and their health hazards. Aquat. Toxicol. 2019, 211, 202–216. [Google Scholar] [CrossRef]
- Merayo, N.; Hermosilla, D.; Blanco, L.; Cortijo, L.; Blanco, A. Assessing the application of advanced oxidation processes, and their combination with biological treatment, to effluents from pulp and paper industry. J. Hazard. Mater. 2013, 262, 420–427. [Google Scholar] [CrossRef]
- Krishna, K.; Sarkar, O.; Mohan, S. Bioelectrochemical treatment of paper and pulp wastewater in comparison with anaerobic process: Integrating chemical coagulation with simultaneous power production. Bioresour. Technol. 2014, 174, 142–151. [Google Scholar] [CrossRef]
- Rybczynska-Tkaczyk, K.; Korniłłowicz-Kowalska, T. Biotransformation and ecotoxicity evaluation of alkali lignin in optimized cultures of microscopic fungi. Int. Biodeterior. Biodegrad. 2017, 117, 131–140. [Google Scholar] [CrossRef]
- Abedinzadeh, N.; Shariat, M.; Monavari, S.; Pendashte, A. Evaluation of color and COD removal by Fenton from biologically (SBR) pre-treated pulp and paper wastewater. Process Saf. Environ. Prot. 2018, 116, 82–91. [Google Scholar] [CrossRef]
- Goswami, L.; Kumar, R.; Pakshirajan, K.; Pugazhenthi, G. A novel integrated biodegradation—Microfiltration system for sustainable wastewater treatment and energy recovery. J. Hazard. Mater. 2019, 365, 707–715. [Google Scholar] [CrossRef]
- Huang, H.; Peng, C.H.; Peng, P.; Lin, Y.; Zhang, X.; Ren, H. Towards the biofilm characterization and regulation in biological wastewater treatment. Appl. Microbiol. Biotechnol. 2019, 103, 1115–1129. [Google Scholar] [CrossRef]
- Valderrama, O.; Zedda, K.; Velizarov, S. Membrane Filtration Opportunities for the Treatment of Black Liquor in the Paper and Pulp Industry. Water 2021, 13, 2270. [Google Scholar] [CrossRef]
- Lopes, G.; Pereira, M.d.S.; Borges, A. Dairy Wastewater Treatment with Organic Coagulants: A Comparison of Factorial Designs. Water 2021, 13, 2240–2307. [Google Scholar]
- Liu, J.Z.; Wang, Q.; Yan, J.B.; Qin, X.R.; Li, L.; Xu, W.; Subramaniam, R.; Bajpai, R. Isolation and characterization of a novel phenol degrading bacterial strain WUST-C1. Ind. Eng. Chem. Res. 2013, 52, 258–265. [Google Scholar] [CrossRef]
- Chakraborty, J.; DAS, S. Characterization of the metabolic pathway and catabolic gene expression in biphenyl degrading marine bacterium pseudomonas aeruginosa JP-11. Chemosphere 2016, 144, 1706–1714. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; Chandra, R. Syntrophic co-culture of Bacillus subtilis and Klebsiella pneumonia for degradation of kraft lignin discharged from rayon grade pulp industry. J. Environ. Sci. 2015, 33, 229–238. [Google Scholar] [CrossRef]
- Fang, X.; Li, Q.; Lin, Y.; Lin, X.; Dai, Y.; Guo, Z.; Pan, D. Screening of a microbial consortium for selective degradation of lignin from tree trimmings. Bioresour. Technol. 2018, 254, 247–255. [Google Scholar] [CrossRef]
- Zhao, J.; Li, Y.; Li, Y.; Yu, Z.; Chen, X. Effects of 4-chlorophenol wastewater treatment on sludge acute toxicity, microbial diversity and functional genes expression in an activated sludge process. Bioresour. Technol. 2018, 265, 39–44. [Google Scholar] [CrossRef]
- Chao, Y.; Guo, F.; Fang, H.; Zhang, T. Hydrophobicity of diverse bacterial populations in activated sludge and biofilm revealed by microbial adhesion to hydrocarbons assay and high-throughput sequencing. Colloids Surf. B Biointerfaces 2014, 114, 379–385. [Google Scholar] [CrossRef]
- Moussavi, G.; Ghodrati, S.; Mohseni-Bandpei, A. The biodegradation and COD removal of 2-chlorophenol in a granularanoxic baffled reactor. J. Biotechnol. 2014, 184, 111–117. [Google Scholar]
- Wu, H.; Shen, J.; Jiang, X.; Liu, X.; Sun, X.; Li, J.; Han, W.; Mu, Y.; Wang, L. Bioaugmentation potential of a newly isolated strain Sphingomonas sp. NJUST37 for the treatment of wastewater containing highly toxic and recalcitrant tricyclazole. Bioresour. Technol. 2018, 264, 98–105. [Google Scholar] [CrossRef]
- Wu, H.; Shen, J.; Jiang, X.; Liu, X.; Sun, X.; Li, J.; Han, W.; Wang, L. Bioaugmentation strategy for the treatment of fungicide wastewater by two triazole-degrading strains. Chem. Eng. J. 2018, 349, 17–24. [Google Scholar] [CrossRef]
- Buntic, A.V.; Pavlovic, M.D.; Antonovic, D.G.; Siler-Marinkovic, S.S.; Dimitrijevic-Brankovic, S.I. A treatment of wastewater containing basic dyes by the use of new strain Streptomyces microflavus CKS6. J. Clean. Prod. 2017, 148, 347–354. [Google Scholar]
- Haritash, A.K.; Kaushik, C.P. Biodegradation aspects of Polycyclic Aromatic Hydrocarbons (PAHs) A review. J. Hazard. Mater. 2009, 169, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Horváthová, H.; Lászlová, K.; Dercová, K. Bioremediation of PCB-contaminated shallow river sediments: The efficacy of biodegradation using individual bacterial strains and their consortia. Chemosphere 2018, 193, 270–277. [Google Scholar] [CrossRef]
- Papazi, A.; Pappas, I.; Kotzabasis, K. Combinational system for biodegradation of olive oil mill wastewater phenolics and high yield of bio-hydrogen production. J. Biotechnol. 2019, 306, 47–53. [Google Scholar] [CrossRef]
- Policastro, G.; Giugliano, M.; Luongo, V.; Napolitano, R.; Fabbricino, M. Enhancing photo fermentative hydrogen production using ethanol rich dark fermentation effluents. Int. J. Hydrog. Energy 2022, 47, 117–126. [Google Scholar] [CrossRef]
- Yeber, M.C.; Soto, C. Bioremediation Method to Degrade Chemical Contaminants from Industrial Effluents. (PCT/IB2016/050279). WIPO. United States Patent US10,883,150. Europe Patent EP3,406,739, 2021. [Google Scholar]
- Takur, I.S. Screening and identification of microbial strains for removal of colour and adsorbable organic halogens in pulp and paper mill effluent. Process Biochem. 2004, 39, 1693–1699. [Google Scholar] [CrossRef]
- Chandra, R.; Raj, A.; Purohit, H.J.; Kapley, A. Characterization and optimization of three potential aerobic bacterial strains kraft lignin degradation from pulp paper waste. Chemosphere 2007, 67, 839–846. [Google Scholar] [CrossRef]
- Garcha, S.; Verma, N.; Brar, S.K. Isolation, characterization and identification of microorganisms from unorganized dairy sector wastewater and sludge samples and evaluation of their biodegradability. Water Resour. Ind. 2016, 16, 19–28. [Google Scholar] [CrossRef]
- Singh, P.; Thakur, I.S. Colour removal of anaerobically treated pulp and paper mill effluent by microorganisms in two steps bioreactor. Bioresour. Technol. 2006, 97, 218–223. [Google Scholar] [CrossRef]
- Raj, A.; Krishna, R.; Reddy, M.; Chandra, R.; Purohit, h.; Kapley, A. Biodegradation of kraft-lignin by Bacillus sp. isolated from sludge of pulp and paper mill. Biodegradation 2007, 18, 783–792. [Google Scholar] [CrossRef]
- Tiku, D.K.; Kumar, A.; Chaturvedi, R.; Makhijani, S.D.; Manoharan, A.; Kumar, R. Holistic Bioremediation of Pulp Mill Effluents Using Autochthonous Bacteria. Int. Biodeterior. Biodegrad. 2010, 64, 173–183. [Google Scholar] [CrossRef]
- Kumar, A.; Dhall, P.; Kumar, R. Redefining BOD: COD ratio of pulp mill industrial wastewaters in BOD analysis by formulating a specific microbial seed. Int. Biodeterior. Biodegrad. 2010, 64, 197–202. [Google Scholar] [CrossRef]
- Majumdar, S.; Priyadarshine, R.; Kumar, A.; Mandal, T.; Mandal, D. Exploring Planococcus sp. TRC1, a bacterial isolate, for carotenoid pigment production and detoxification of paper mill effluent in immobilized fluidized bed reactor. J. Clean. Prod. 2019, 211, 1389–1402. [Google Scholar] [CrossRef]
- Yang, C.; Cao, G.; Li, Y.; Zhang, X.; Ren, H.; Wang, X.; Feng, J.; Zhao, L.; XU, P. A Constructed Alkaline Consortium and Its Dynamics in Treating Alkaline Black Liquor with Very High Pollution Load. PLoS ONE 2008, 3, e3777. [Google Scholar] [CrossRef]
- Neoh, C.; Lam, C.; Ghani, S.; Ware, I.; Sarip, S.; Ibrahim, Z. Bioremediation of high-strength agricultural wastewater using Ochrobactrum sp strain SZ1. 3 Biotech. 2016, 6, 2–9. [Google Scholar] [CrossRef]
- Haq, I.; Kumar, S.; Kumari, V.; Singh, S.; Raj, A. Evaluation of bioremediation potentiality of ligninolytic Serratia liquefaciens for detoxification of pulp and paper mill effluent. J. Hazard. Mater. 2016, 305, 190–199. [Google Scholar] [CrossRef]
- Budiman, P.M.; WU, T.Y.; Ramanan, R.N.; Hay, J. Treatment and Reuse of Effluents from Palm Oil, Pulp and Paper Mills as a Combined Substrate by Using Purple Nonsulfur Bacteria. Ind. Eng. Chem. Res. 2014, 53, 14921–14931. [Google Scholar] [CrossRef]
- Gaur, N.; Narasimhulu, K.; Pydisetty, Y. Recent advances in the bio-remediation of persistent organic pollutants and its effect on environment. J. Clean. Prod. 2018, 198, 1602–1631. [Google Scholar] [CrossRef]
- Balabanič, D.; Filipič, M.; Klemenčič, A.; Žegura, B. Raw and biologically treated paper mill wastewater effluents and the recipient surface waters: Cytotoxic and genotoxic activity and the presence of endocrine disrupting compounds. Sci. Total Environ. 2017, 574, 78–89. [Google Scholar] [CrossRef]
- Cai, F.; Lei, L.; LI, Y. Different bioreactors for treating secondary effluent from the recycled paper mill. Sci. Total Environ. 2019, 667, 49–56. [Google Scholar] [CrossRef]
- Câmara, J.; Sousa, M.; Barros, E. Modeling of rhamnolipid biosurfactant production: Estimation of kinetic parameters by genetic algorithm. J. Surfactants Deterg. 2020, 23, 705–714. [Google Scholar] [CrossRef]
- Hussain, A.; Dubey, S.K.; Kumar, V. Kinetic study for aerobic treatment of phenolic wastewater. Water Resour. Ind. 2015, 11, 81–90. [Google Scholar] [CrossRef]
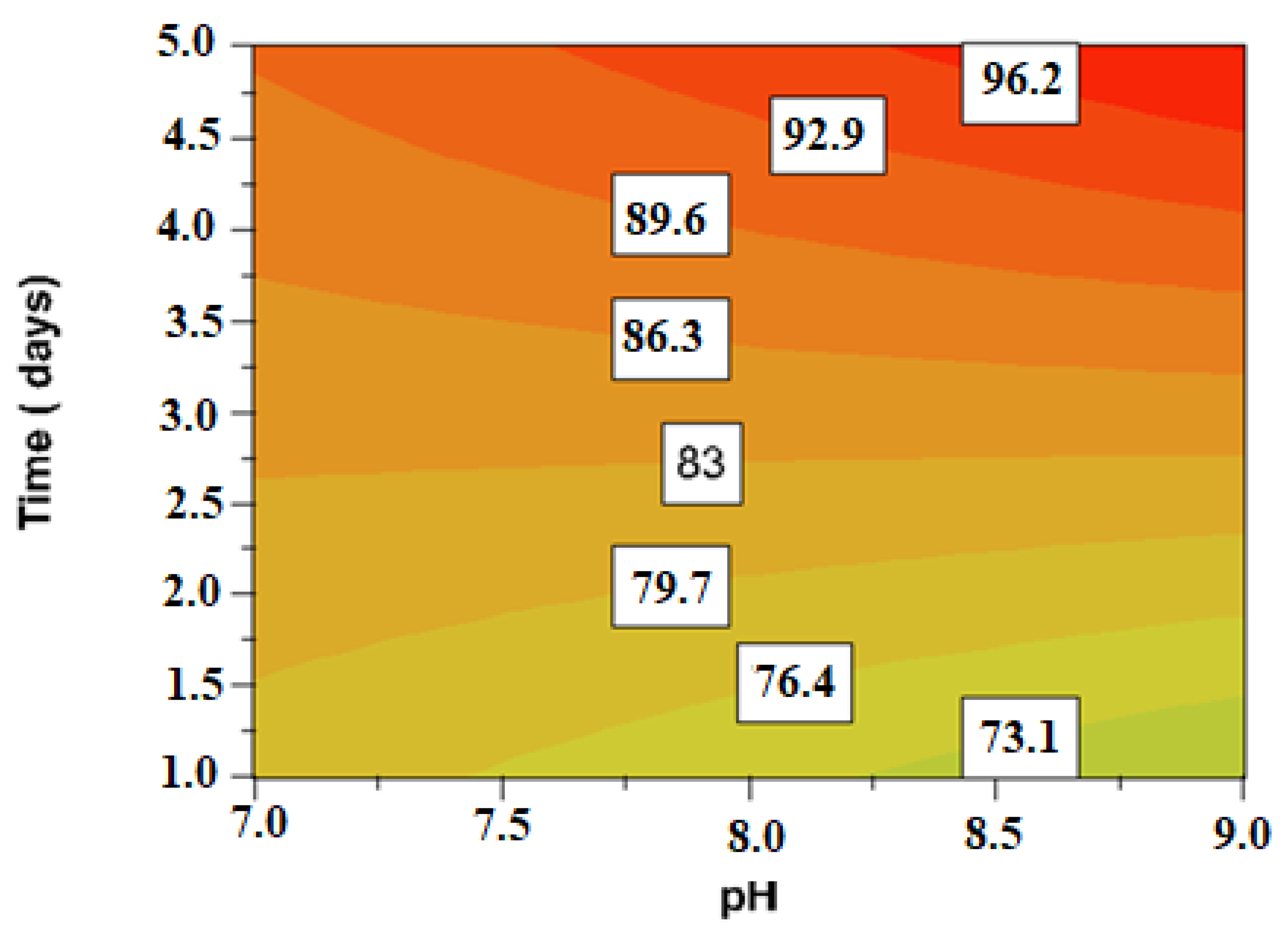


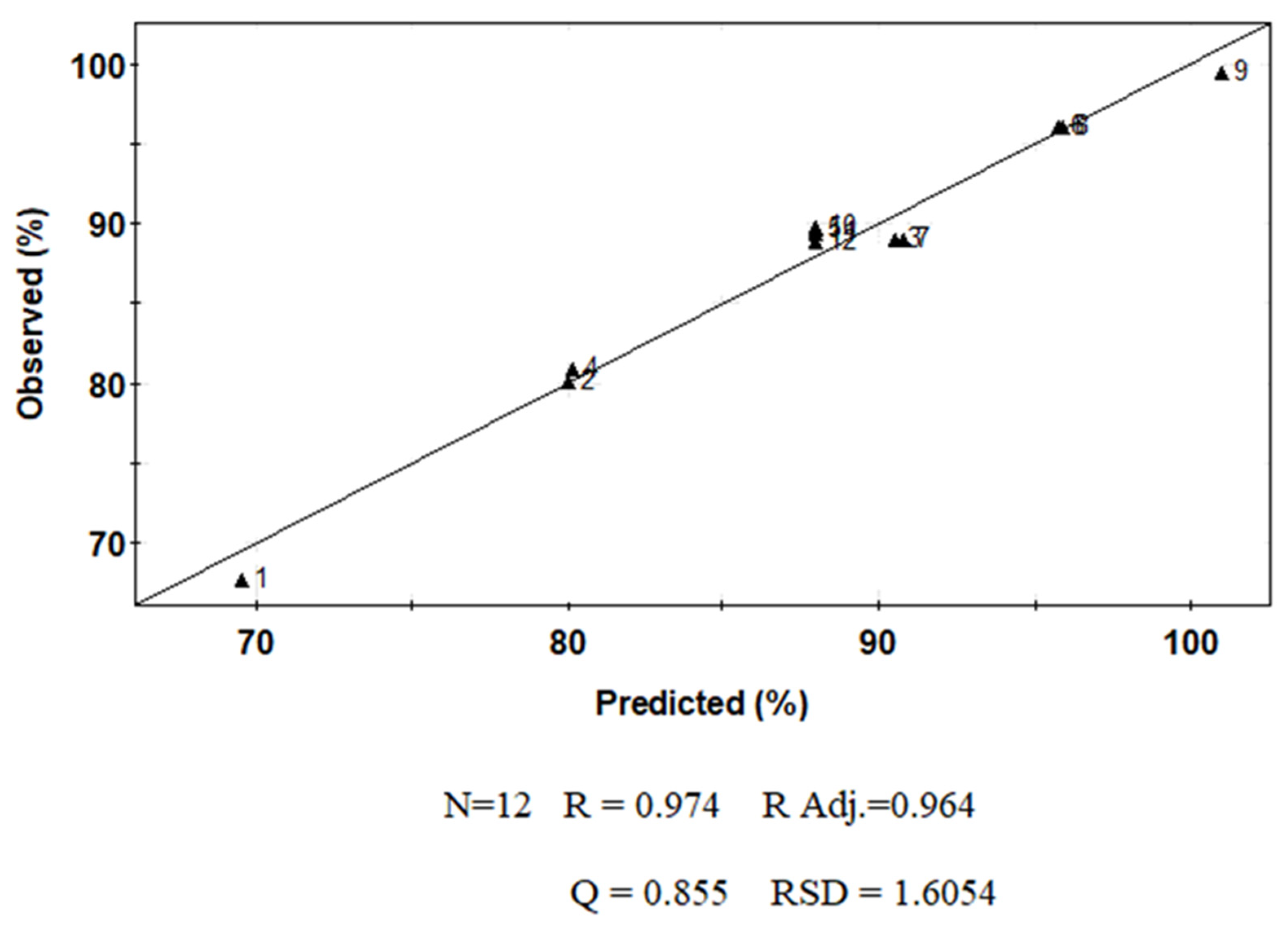


| Experimental Name | Run Order | pH Coded (Uncoded Value) | Time (Days) Coded (Uncoded Value) |
|---|---|---|---|
| N1 | 6 | 7 (−1) | 1 (−1) |
| N2 | 10 | 9 (+1) | 1 (−1) |
| N3 | 8 | 7 (−1) | 5 (+1) |
| N4 | 1 | 9 (+1) | 5 (+1) |
| N5 | 2 | 8 (0) | 3 (0) |
| N6 | 4 | 8 (0) | 3 (0) |
| N7 | 9 | 8 (0) | 3 (0) |
| N8 | 3 | 8 (0) | 3 (0) |
| N9 | 12 | 8 (0) | 1 (−1) |
| N10 | 5 | 7 (−1) | 3 (0) |
| N11 | 11 | 9 (+1) | 3 (0) |
| N12 | 7 | 8 (0) | 5 (+1) |
| Lignin Removed (%) | pH | With Biofilm | Without Biofilm | ||
|---|---|---|---|---|---|
| Kv/day | R2 | Kv/day | R2 | ||
| 90.0 | 7.0 | 11.0 | 0.60 | 17.0 | 0.99 |
| 100.0 | 8.0 | 12.4 | 0.90 | 12.4 | 0.86 |
| 100.0 | 9.0 | 32.4 | 0.94 | 32.4 | 0.92 |
| Color Removed (%) | |||||
| 90.0 | 7.0 | 9.60 | 0.89 | 9.60 | 0.67 |
| 100.0 | 8.0 | 12.0 | 0.60 | 14.9 | 0.74 |
| 100.0 | 9.0 | 12.0 | 0.60 | 14.9 | 0.74 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yeber, M.C.; Silva, T. The Ability of a Bacterial Strain to Remove a Phenolic Structure as an Approach to Pulp and Paper Mill Wastewater Treatment: Optimization by Experimental Design. Water 2022, 14, 3296. https://doi.org/10.3390/w14203296
Yeber MC, Silva T. The Ability of a Bacterial Strain to Remove a Phenolic Structure as an Approach to Pulp and Paper Mill Wastewater Treatment: Optimization by Experimental Design. Water. 2022; 14(20):3296. https://doi.org/10.3390/w14203296
Chicago/Turabian StyleYeber, María Cristina, and Tatiana Silva. 2022. "The Ability of a Bacterial Strain to Remove a Phenolic Structure as an Approach to Pulp and Paper Mill Wastewater Treatment: Optimization by Experimental Design" Water 14, no. 20: 3296. https://doi.org/10.3390/w14203296
APA StyleYeber, M. C., & Silva, T. (2022). The Ability of a Bacterial Strain to Remove a Phenolic Structure as an Approach to Pulp and Paper Mill Wastewater Treatment: Optimization by Experimental Design. Water, 14(20), 3296. https://doi.org/10.3390/w14203296






