Eukaryotic Diversity Based on High-Throughput 18S rRNA Sequencing and Its Relationship with Environmental Factors in a Salt Lake in Tibet, China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Physicochemical Factor Determination
2.2. DNA Extraction and Sequencing of Benthic Eukaryotes
2.3. Data Analysis and Processing
3. Results
3.1. Analysis of Environmental Factors
3.2. Analysis of Eukaryotic Community Structure in Kyêbxang Co
3.2.1. Sequencing Statistics, Eukaryotic Species Composition
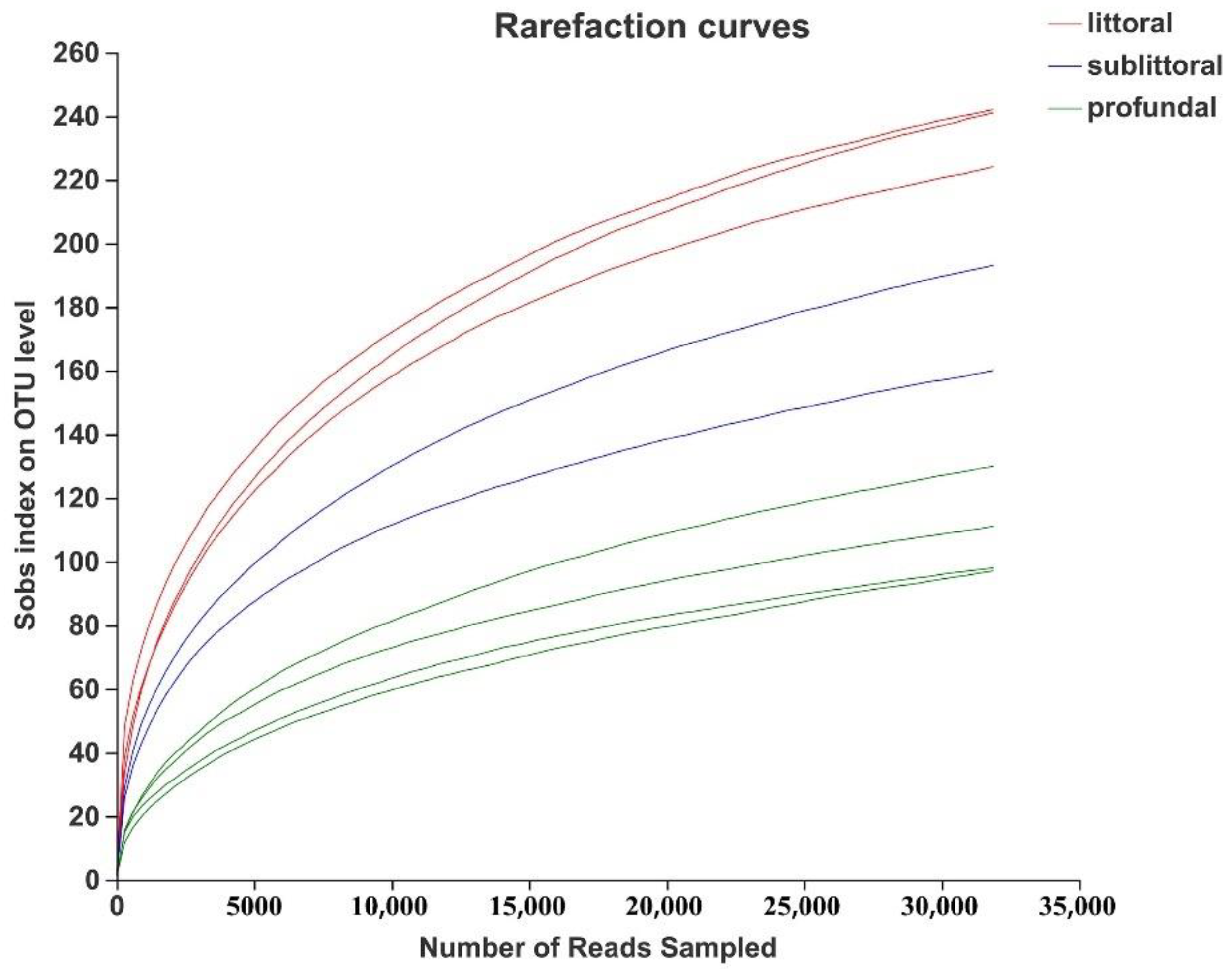
3.2.2. Analysis of the Alpha Diversity of Eukaryotes
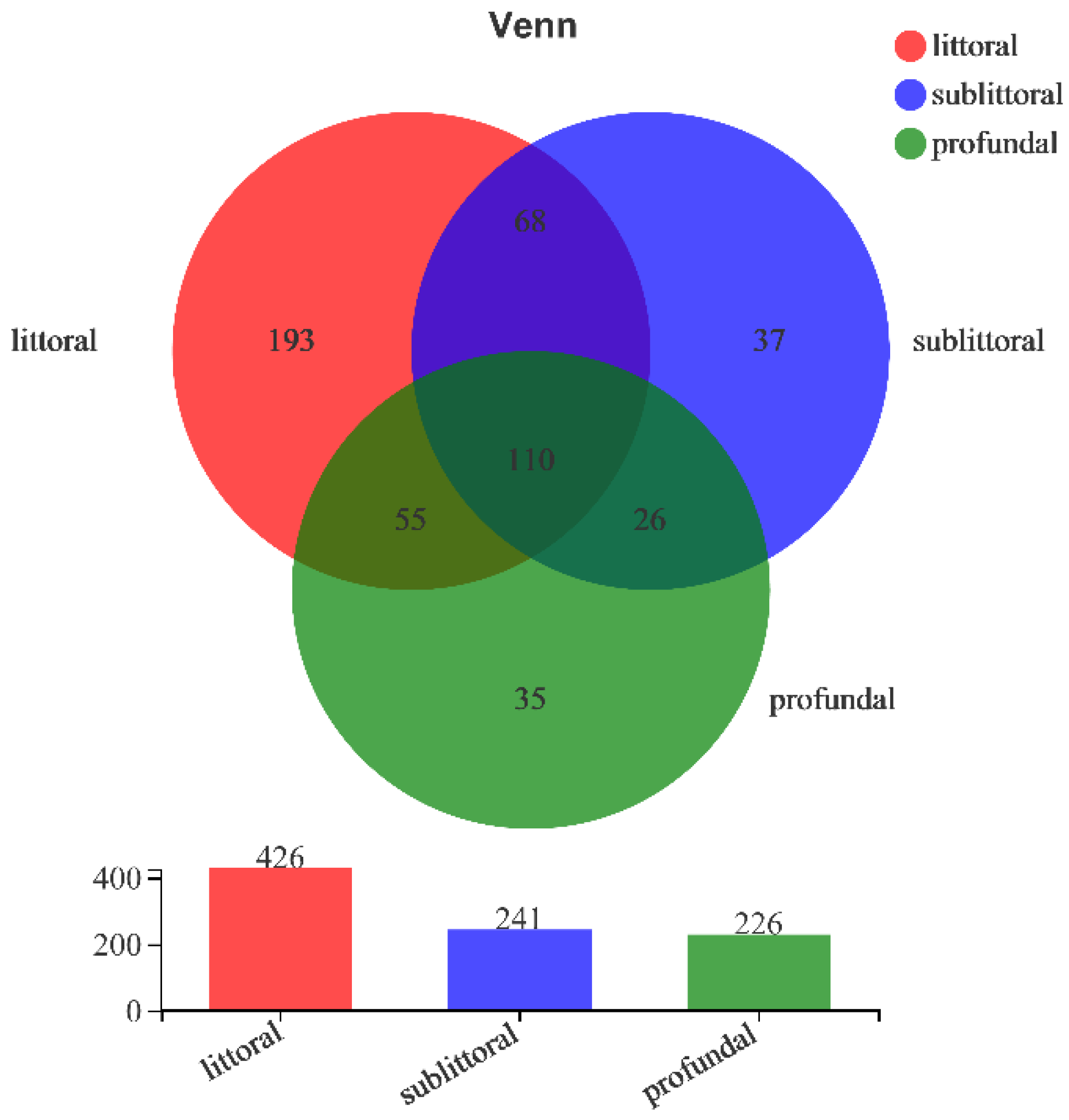
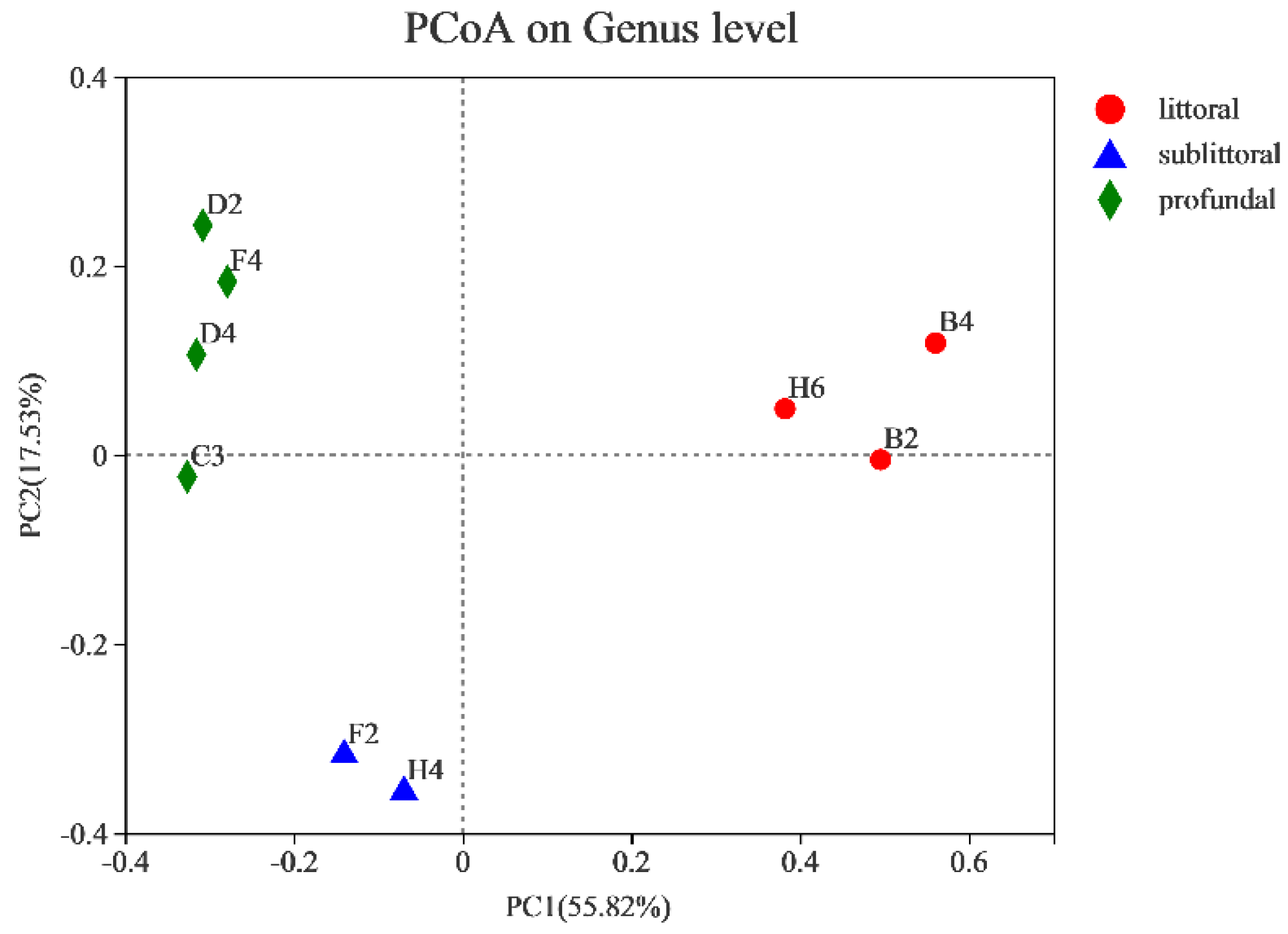
3.2.3. Analysis of the Beta Diversity of Eukaryotes
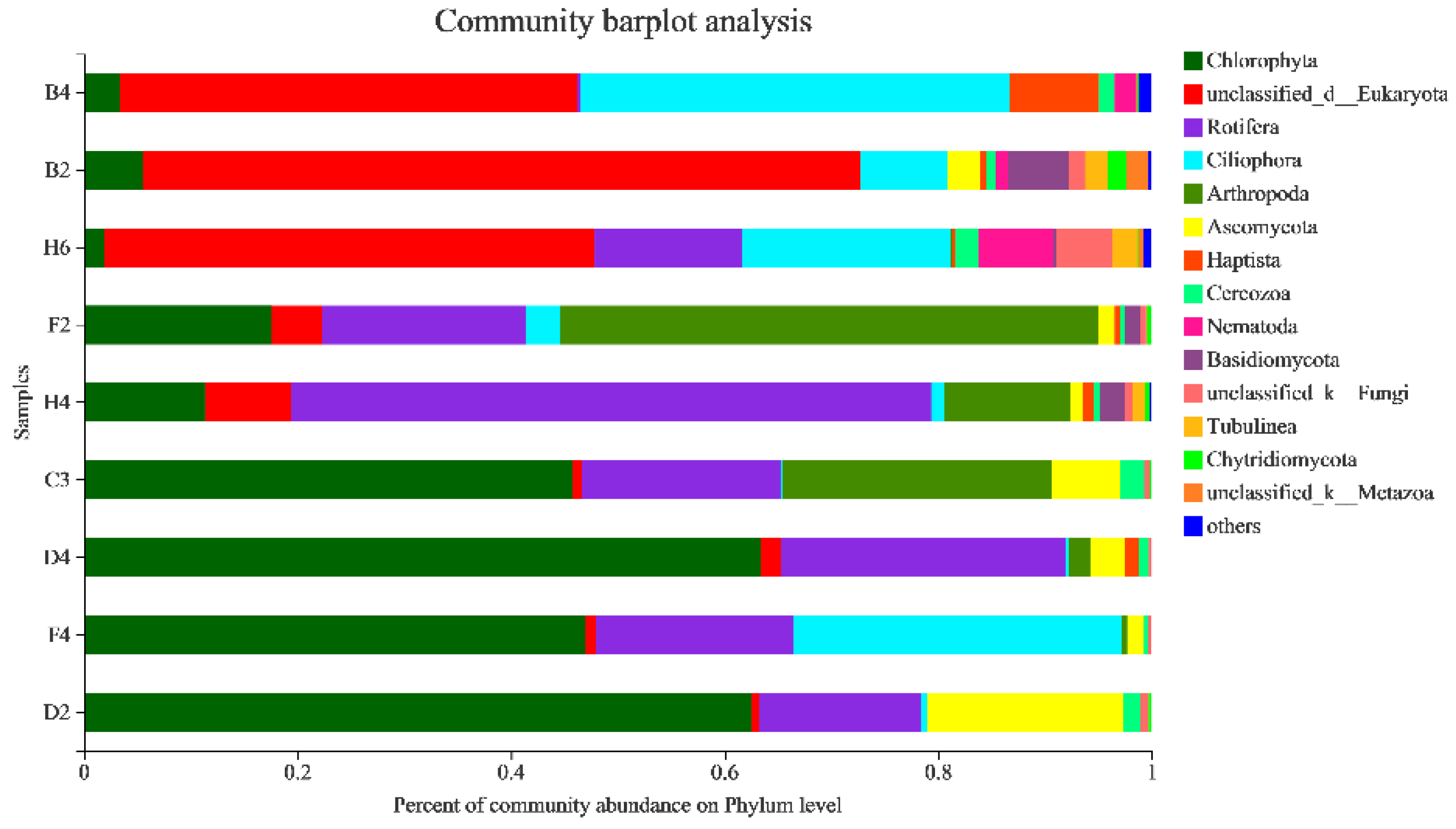
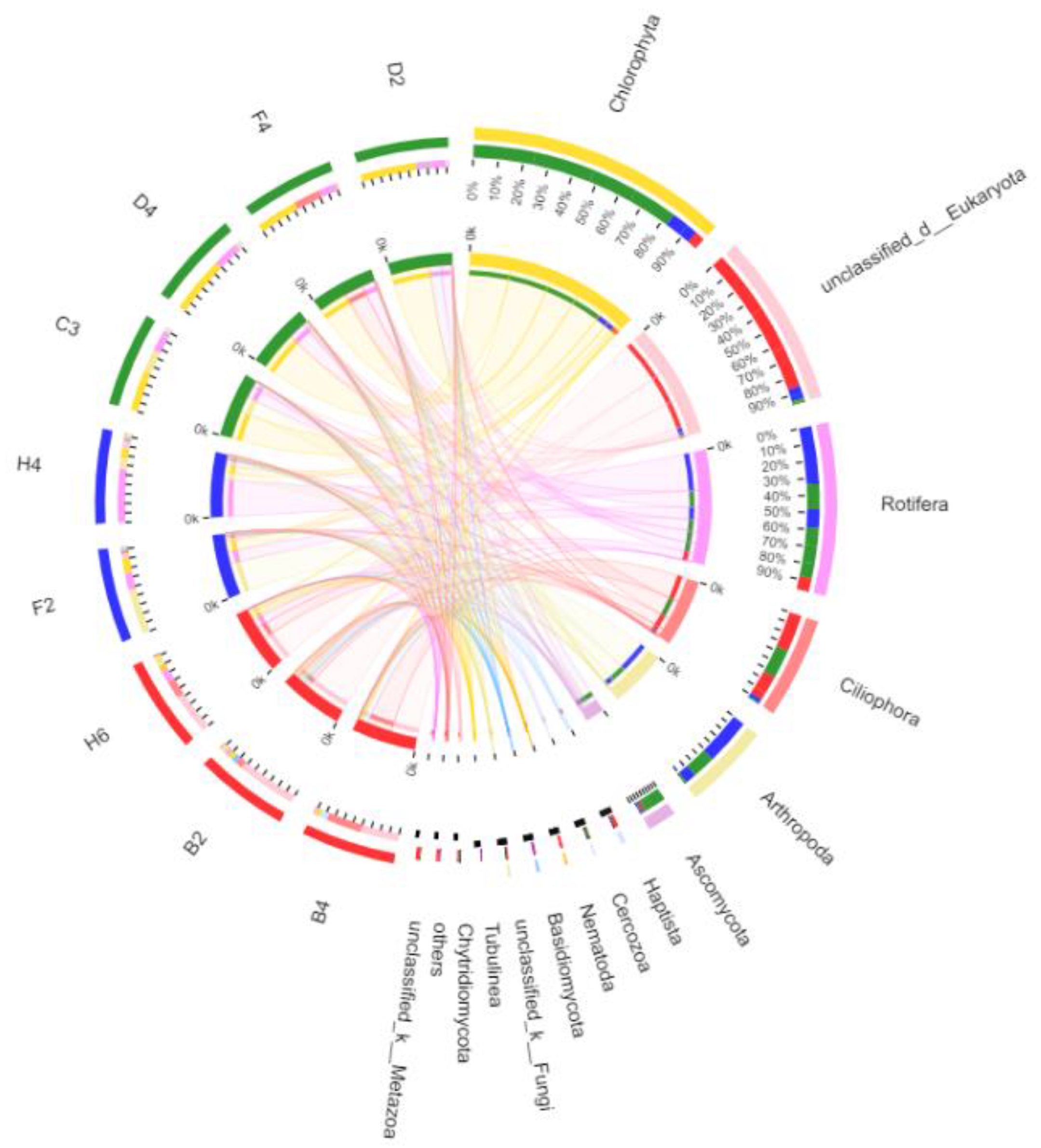
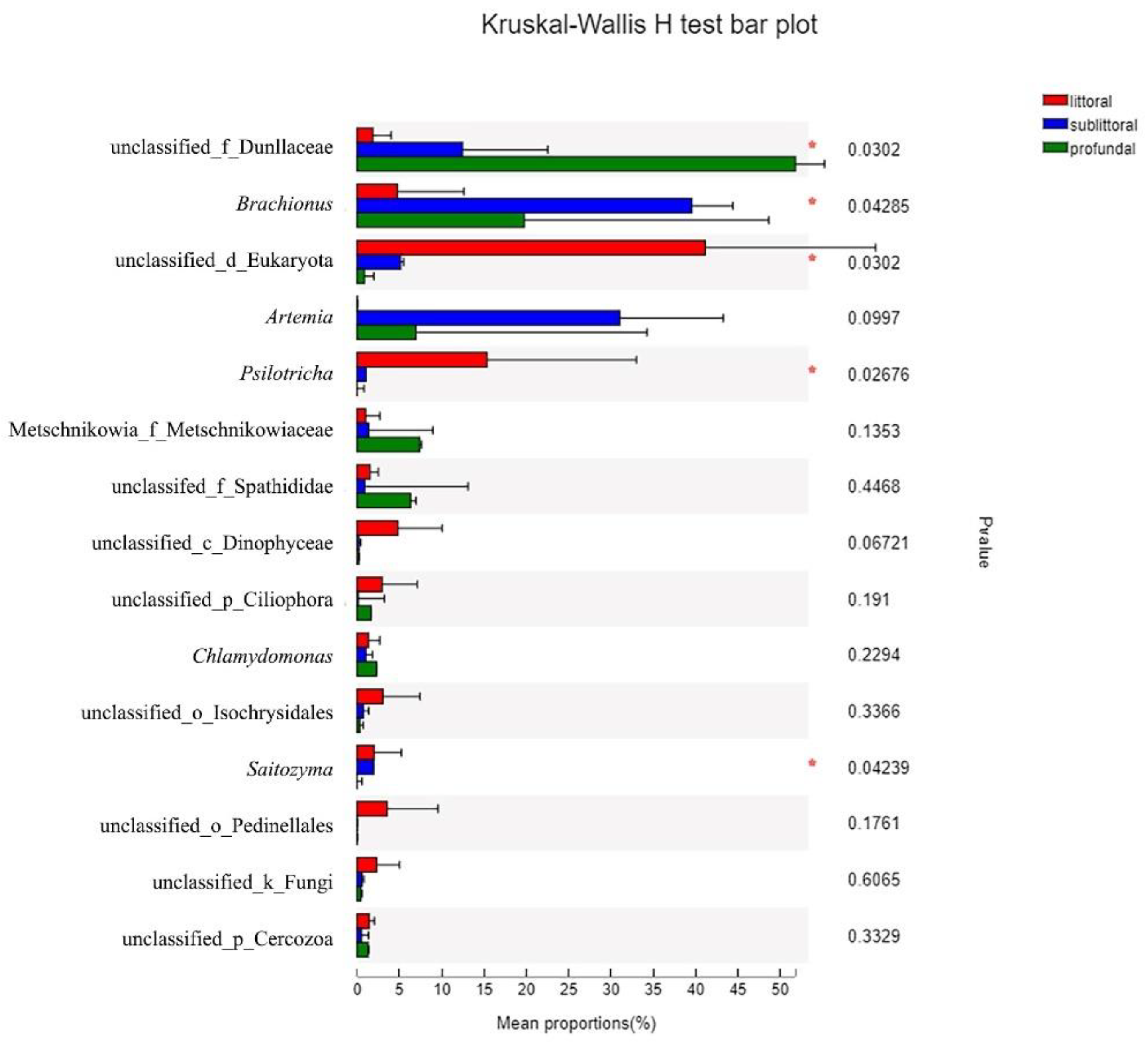
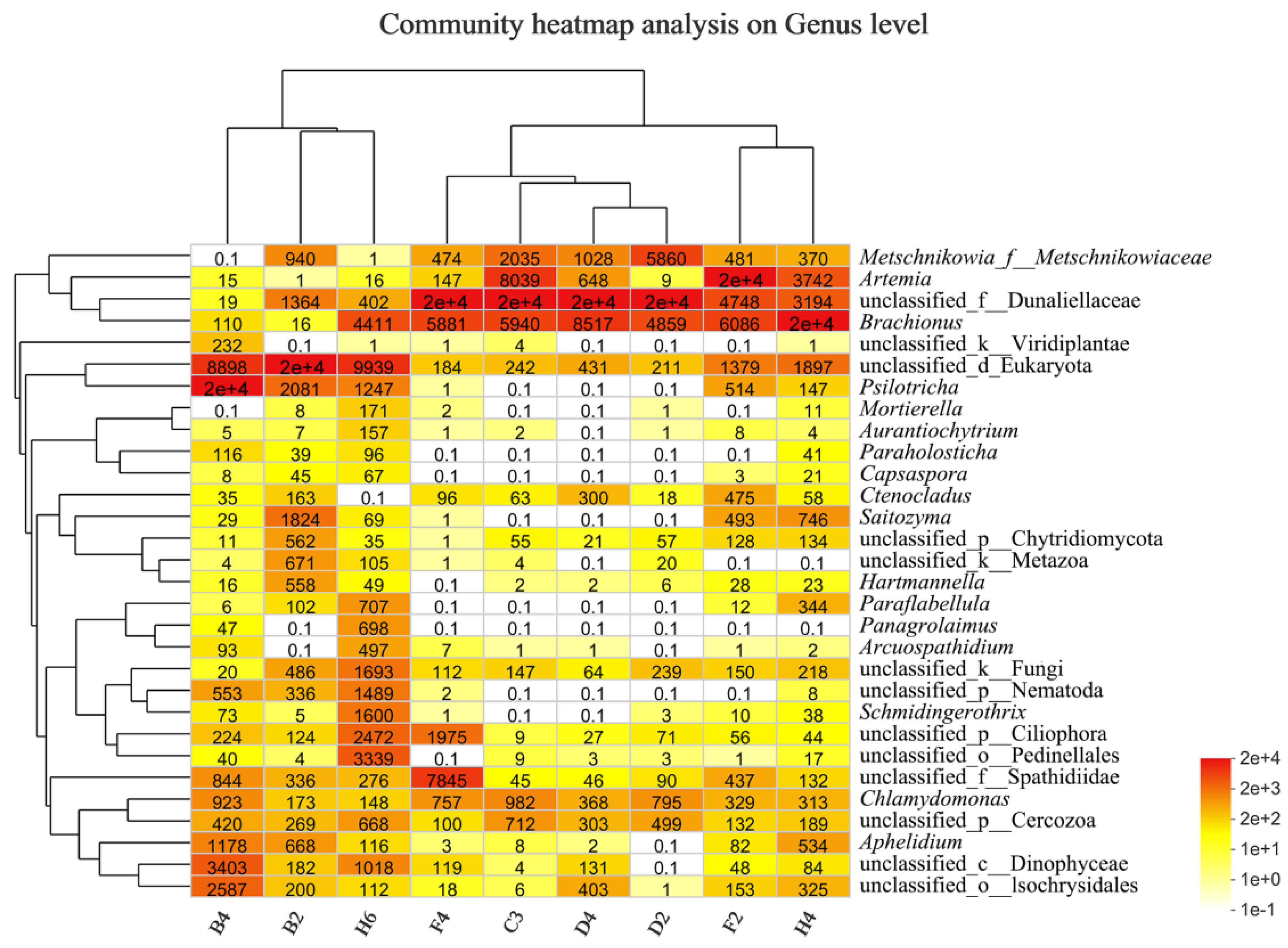
3.2.4. Taxonomic Composition and Difference Analysis of Eukaryotes
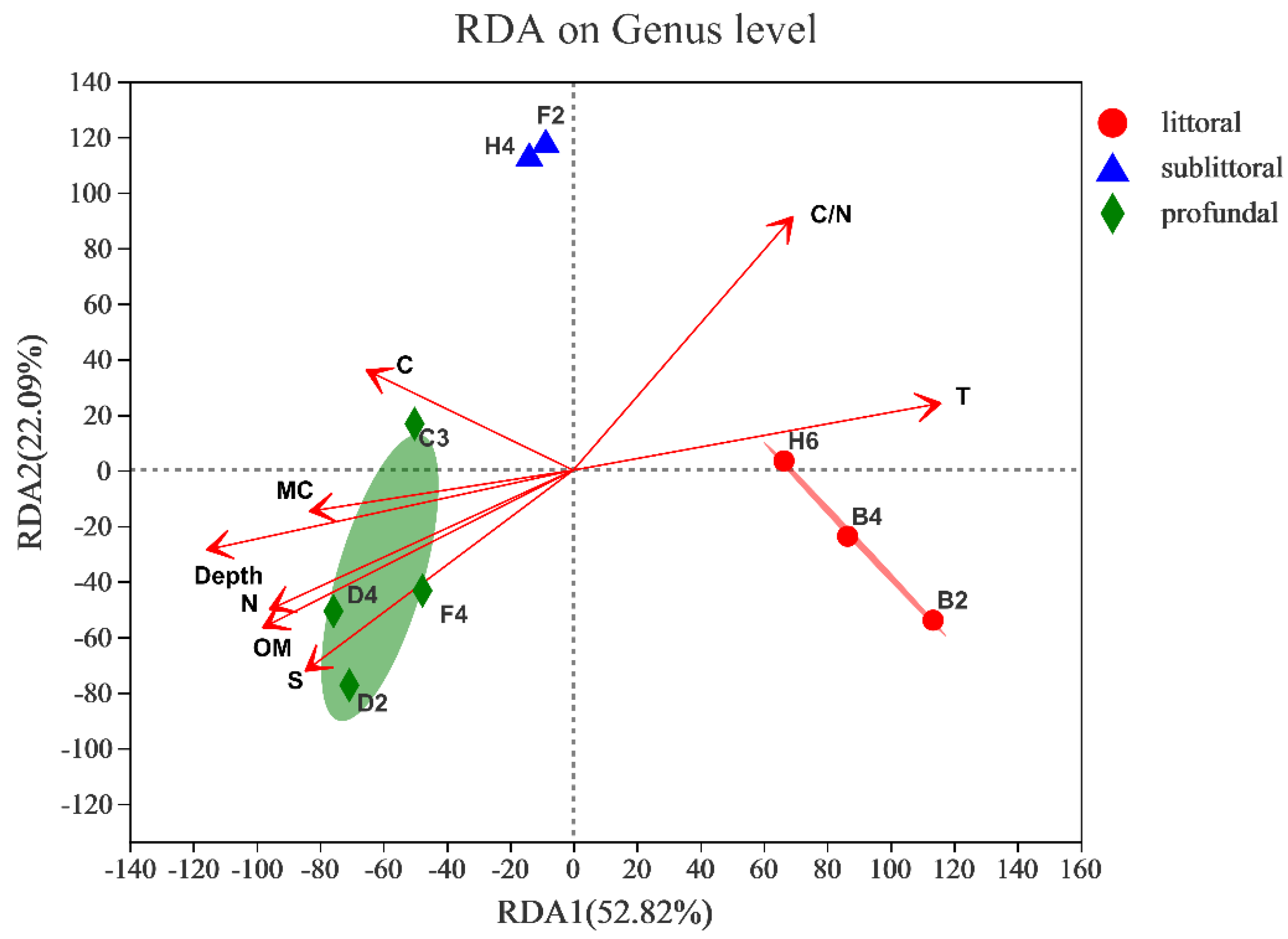
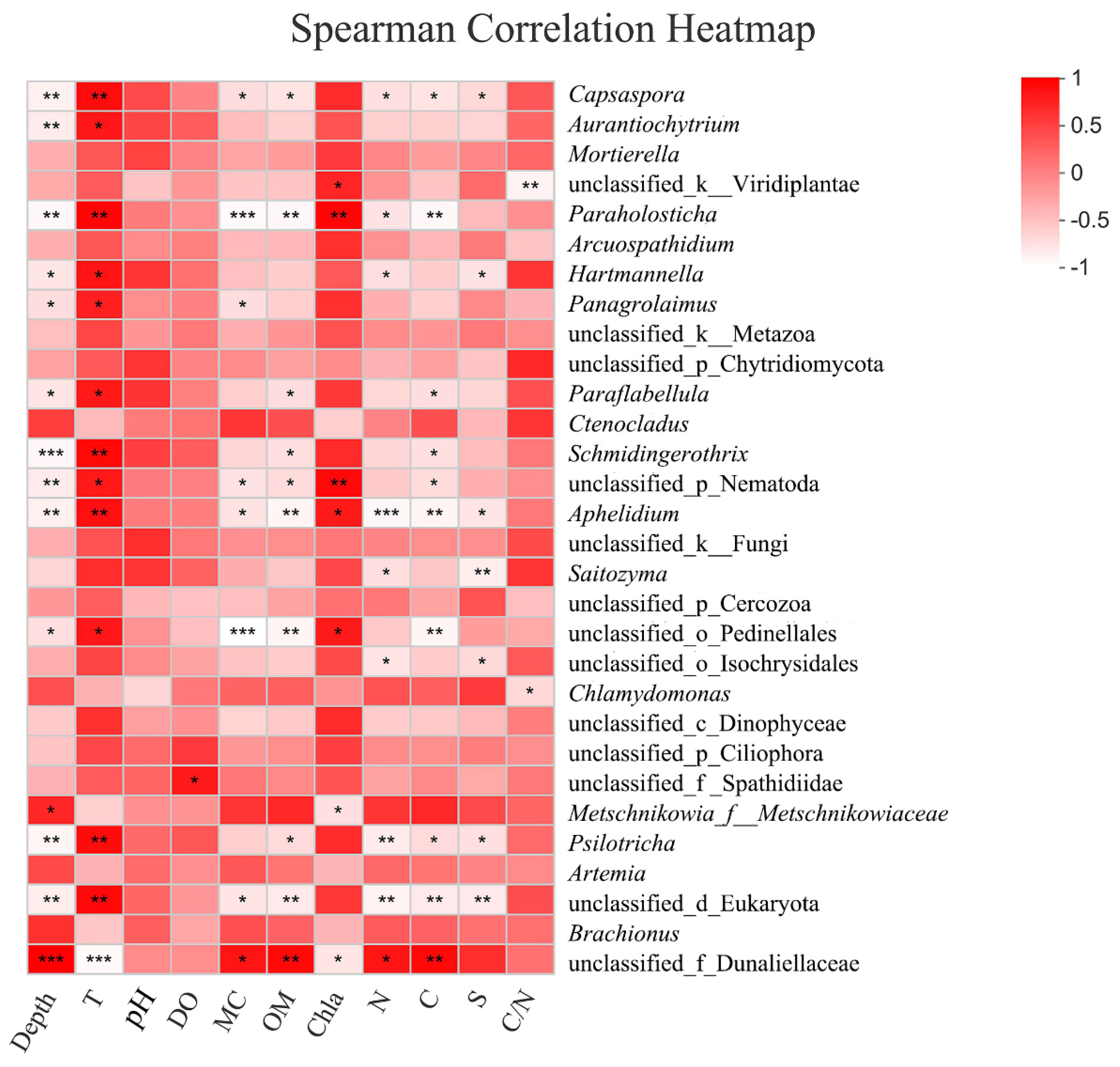
3.2.5. Response to Environmental Factors
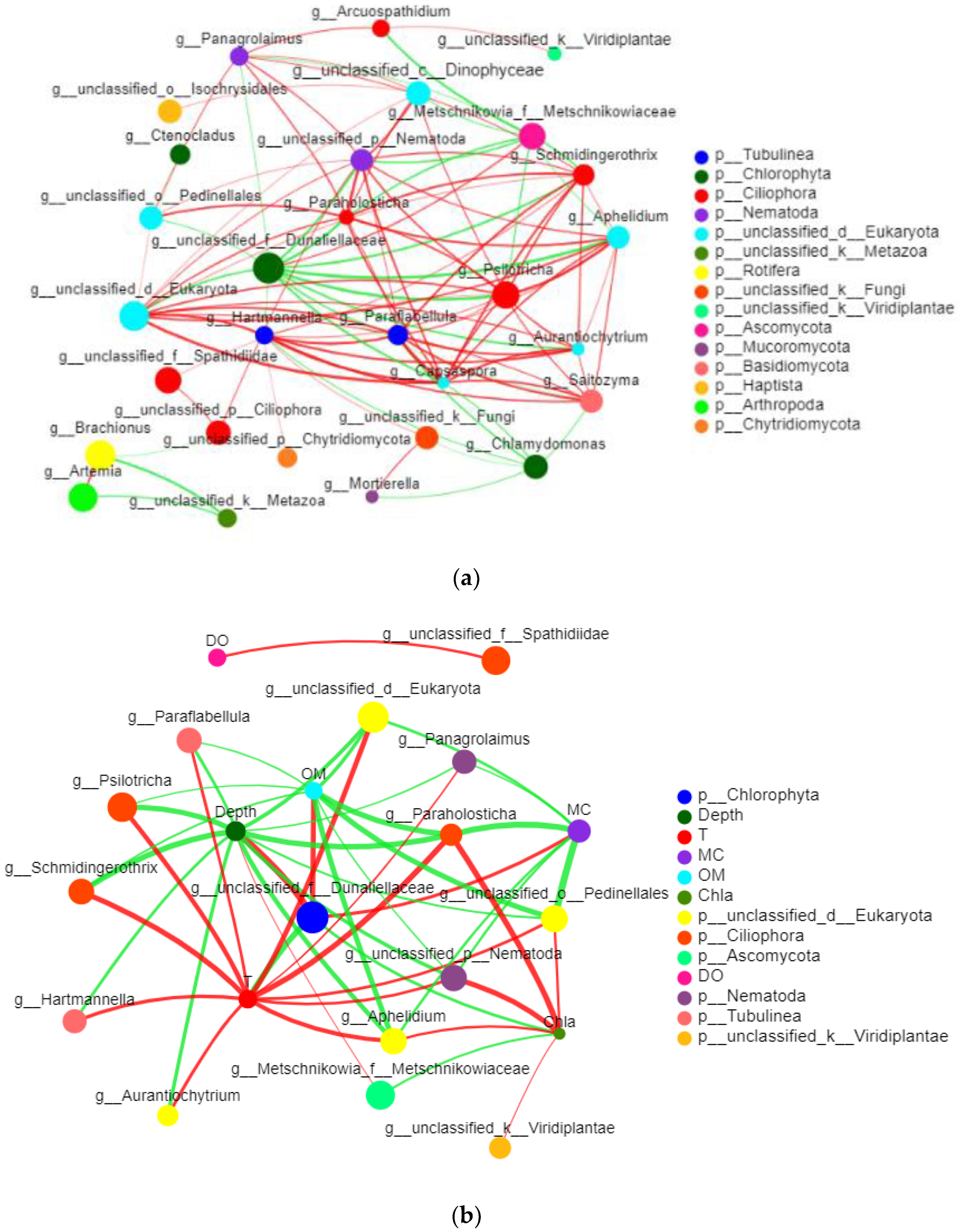
3.2.6. Network Analysis of Eukaryotes
4. Discussion
4.1. Community Composition of Eukaryotes in Kyêbxang Co Salt Lake
4.2. Relationship between Eukaryotes and Environmental Factors
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zheng, M.; Liu, X. Hydrochemistry of Salt Lakes of the Qinghai-Tibet Plateau, China. Aquat. Geochem. 2009, 15, 293–320. [Google Scholar] [CrossRef]
- Chang, Q.; Lai, Z.; An, F.; Wang, H.; Lei, Y.; Han, F. Chronology for terraces of the Nalinggele River in the north Qinghai-Tibet Plateau and implications for salt lake resource formation in the Qaidam Basin. Quatern. Int. 2017, 430, 12–20. [Google Scholar] [CrossRef]
- Jiang, T.; Ji, L.; Cheng, H.; Li, B.; Li, G.; Ma, H.; Zhang, X.; Li, C.; Ma, X.; Zhang, P. Algae mineralization experiment and genetic analysis of hydromagnesite in Bangor Lake, Xizang (Tibet). Geol. Rev. 2021, 67, 1709–1726. [Google Scholar]
- Li, H.; Cheng, F.; Wei, Y.; Lydy, M.J.; You, J. Global occurrence of pyrethroid insecticides in sediment and the associated toxicological effects on benthic invertebrates: An overview. J. Hazard. Mater. 2017, 324, 258–271. [Google Scholar] [CrossRef]
- Chariton, A.A.; Stephenson, S.; Morgan, M.J.; Steven, A.D.L.; Colloff, M.J.; Court, L.N.; Hardy, C.M. Metabarcoding of benthic eukaryote communities predicts the ecological condition of estuaries. Environ. Pollut. 2015, 203, 165–174. [Google Scholar] [CrossRef]
- Wilden, B.; Traunspurger, W.; Geisen, S. Inventory of the benthic eukaryotic diversity in the oldest European lake. Ecol. Evol. 2021, 11, 11207–11215. [Google Scholar] [CrossRef]
- Sbrocca, C.; De Troch, M.; Losi, V.; Grassi, E.; Balsamo, M.; Semprucci, F. Habitat-Diversity Relations between Sessile Macrobenthos and Benthic Copepods in the Rocky Shores of a Marine Protected Area. Water 2021, 13, 1020. [Google Scholar] [CrossRef]
- Tsikopoulou, I.; Lampa, M.; Tsiola, A.; Pitta, P.; Tsapakis, M.; Karakassis, I. Functional adaptations of benthic communities to organic matter enrichment at the edge of an allowable zone of effect (AZE). Estuar. Coast. Shelf Sci. 2021, 262, 10756. [Google Scholar] [CrossRef]
- Berezina, A.; Yakushev, E.; Savchuk, O.; Vogelsang, C.; Staalstrom, A. Modelling the Influence from Biota and Organic Matter on the Transport Dynamics of Microplastics in the Water Column and Bottom Sediments in the Oslo Fjord. Water 2021, 13, 2690. [Google Scholar] [CrossRef]
- De Troyer, N.; Mereta, S.T.; Goethals, P.; Boets, P. Water Quality Assessment of Streams and Wetlands in a Fast Growing East African City. Water 2016, 8, 123. [Google Scholar] [CrossRef]
- Zhu, P.; Wang, Y.; Shi, T.; Huang, G.; Gong, J. Genetic Diversity of Benthic Microbial Eukaryotes in Response to Spatial Heterogeneity of Sediment Geochemistry in a Mangrove Ecosystem. Estuar. Coast. 2018, 41, 751–764. [Google Scholar] [CrossRef]
- Vignatti, A.M.; Paggi, J.C.; Cabrera, G.C.; Echaniz, S.A. Zooplankton diversity and its relationship with environmental changes after the filling of a temporary saline lake in the semi-arid region of La Pampa, Argentina. Lat. Am. J. Aquat. Res. 2012, 40, 1005–1016. [Google Scholar] [CrossRef]
- Moscatello, S.; Belmonte, G. Active and resting stages of zooplankton and its seasonal evolution in a hypersaline temporary pond of the Mediterranean coast (the Vecchia Salina, SE Italy). Sci. Mar. 2004, 68, 491–500. [Google Scholar] [CrossRef]
- Bik, H.M.; Halanych, K.M.; Sharma, J.; Thomas, W.K. Dramatic shifts in benthic microbial eukaryote communities following the Deepwater Horizon oil spill. PLoS ONE 2012, 7, e38550. [Google Scholar] [CrossRef]
- Pareek, C.S.; Smoczynski, R.; Tretyn, A. Sequencing technologies and genome sequencing. J. Appl. Genet. 2011, 52, 413–435. [Google Scholar] [CrossRef]
- Metzker, M.L. Sequencing technologies—The next generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef]
- Baird, D.J.; Hajibabaei, M. Biomonitoring 2.0: A new paradigm in ecosystem assessment made possible by next-generation DNA sequencing. Mol. Ecol. 2012, 21, 2039–2044. [Google Scholar]
- Shendure, J.; Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 2008, 26, 1135–1145. [Google Scholar] [CrossRef]
- Chen, M.; Chen, F.; Yu, Y.; Ji, J.; Kong, F. Genetic Diversity of Eukaryotic Microorganisms in Lake Taihu, a Large Shallow Subtropical Lake in China. Microb. Ecol. 2008, 56, 572–583. [Google Scholar] [CrossRef]
- Mianping, Z.; Yongsheng, Z.; Xifang, L.; Zhen, N.; Fanjing, K.; Wen, Q.I.; Qingxian, J.; Linzhong, P.U.; Xianhua, H.; Hailei, W.; et al. Progress and Prospects of Salt Lake Research in China. Acta Geol. Sin. 2016, 90, 1195–1235. [Google Scholar]
- Golubkov, S.M.; Shadrin, N.V.; Golubkov, M.S.; Balushkina, E.V.; Litvinchuk, L.F. Food Chains and Their Dynamics in Ecosystems of Shallow Lakes with Different Water Salinities. Russ. J. Ecol.+ 2018, 49, 442–448. [Google Scholar] [CrossRef]
- Meng, K.; Shi, X.; Wang, E.; Liu, F. High-altitude salt lake elevation changes and glacial ablation in Central Tibet, 2000–2010. Chin. Sci. Bull. 2012, 57, 525–534. [Google Scholar] [CrossRef]
- Wang, R.L.; Brassell, S.C.; Scarpitta, S.C.; Zheng, M.P.; Zhang, S.C.; Hayde, P.R.; Muench, L.M. Steroids in sediments from Zabuye Salt Lake, western Tibet: Diagenetic, ecological or climatic signals? Org. Geochem. 2004, 35, 157–168. [Google Scholar] [CrossRef]
- Shadrin, N.; Zheng, M.; Oren, A. Past, present and future of saline lakes: Research for global sustainable development. Chin. J. Oceanol. Limn. 2015, 33, 1349–1353. [Google Scholar] [CrossRef]
- Jia, Q.; Liu, X.; Wang, H.; Liu, S.; Luo, Y.; Chen, L.; Zheng, M. Bio-ecological resources of saline lakes in Tibet and their economic prospect. Sci. Technol. Rev. 2017, 35, 19–26. [Google Scholar]
- Lin, Q.; Xu, L.; Hou, J.; Liu, Z.; Jeppesen, E.; Han, B.P. Responses of trophic structure and zooplankton community to salinity and temperature in Tibetan lakes: Implication for the effect of climate warming. Water Res. 2017, 124, 618–629. [Google Scholar] [CrossRef]
- Zhao, Z.; He, J.; Geisen, S.; Han, L.; Wang, J.; Shen, J.; Wei, W.; Fang, Y.; Li, P.; Zhang, L. Protist communities are more sensitive to nitrogen fertilization than other microorganisms in diverse agricultural soils. Microbiome 2019, 7, 33. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Sahl, J.W.; Stres, B.; Thallinger, G.G.; VAN Horn, D.J.; Weber, C.F.; Ryabin, T.; Hall, J.R.; Hartmann, M.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microb. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Lejzerowicz, F.; Esling, P.; Pillet, L.; Wilding, T.A.; Black, K.D.; Pawlowski, J. High-throughput sequencing and morphology perform equally well for benthic monitoring of marine ecosystems. Sci. Rep.-UK 2015, 5, 13932. [Google Scholar] [CrossRef]
- Liao, W.; Tong, D.; Li, Z.; Nie, X.; Liu, Y.; Ran, F.; Liao, S. Characteristics of microbial community composition and its relationship with carbon, nitrogen and sulfur in sediments. Sci. Total Environ. 2021, 795, 148848. [Google Scholar] [CrossRef]
- Hondeveld, B.J.M.; Bak, R.P.M.; Van Raaphorst, W.; Van Duyl, F.C. Impact of grazing by benthic eukaryotic organisms on the nitrogen sediment–water exchange in the North Sea. J. Sea Res. 1999, 41, 255–268. [Google Scholar]
- Liu, K.; Ding, X.; Wang, H.; Zhang, X.; Hozzein, W.N.; Wadaan, M.A.M.; Lan, A.; Zhang, B.; Li, W. Eukaryotic microbial communities in hypersaline soils and sediments from the alkaline hypersaline Huama Lake as revealed by 454 pyrosequencing. Antonie Van Leeuwenhoek 2014, 105, 871–880. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Ding, X.; Tang, X.; Wang, J.; Li, W.; Yan, Q.; Liu, Z. Macro and Microelements Drive Diversity and Composition of Prokaryotic and Fungal Communities in Hypersaline Sediments and Saline-Alkaline Soils. Front. Microbiol. 2018, 9, 352. [Google Scholar] [CrossRef] [PubMed]
- Hugoni, M.; Escalas, A.; Bernard, C.; Nicolas, S.; Jezequel, D.; Vazzoler, F.; Sarazin, G.; Leboulanger, C.; Bouvy, M.; Got, P.; et al. Spatiotemporal variations in microbial diversity across the three domains of life in a tropical thalassohaline lake (Dziani Dzaha, Mayotte Island). Mol. Ecol. 2018, 27, 4775–4786. [Google Scholar] [CrossRef] [PubMed]
- Clark, D.E.; Pilditch, C.A.; Pearman, J.K.; Ellis, J.I.; Zaiko, A. Environmental DNA metabarcoding reveals estuarine benthic community response to nutrient enrichment—Evidence from an in-situ experiment. Environ. Pollut. 2020, 267, 115472. [Google Scholar] [CrossRef] [PubMed]
- Oren, A. A hundred years of Dunaliella research: 1905–2005. Saline Syst. 2005, 1, 2. [Google Scholar] [CrossRef]
- Pawlowski, J.; Christen, R.; Lecroq, B.; Bachar, D.; Shahbazkia, H.R.; Amaral-Zettler, L.; Guillou, L. Eukaryotic Richness in the Abyss: Insights from Pyrotag Sequencing. PLoS ONE 2011, 6, e18169. [Google Scholar] [CrossRef]
- Piredda, R.; Sarno, D.; Lange, C.B.; Tomasino, M.P.; Zingone, A.; Montresor, M. Diatom resting stages in surface sediments: A pilot study comparing Next Generation Sequencing and Serial Dilution Cultures. Cryptogam. Algol. 2017, 38, 31–46. [Google Scholar] [CrossRef]
- Burlakova, L.E.; Barbiero, R.P.; Karatayev, A.Y.; Daniel, S.E.; Hinchey, E.K.; Warren, G.J. The benthic community of the Laurentian Great Lakes: Analysis of spatial gradients and temporal trends from 1998 to 2014. J. Great Lakes Res. 2018, 44, 600–617. [Google Scholar] [CrossRef]
- Geisen, S.; Tveit, A.T.; Clark, I.M.; Richter, A.; Svenning, M.M.; Bonkowski, M.; Urich, T. Metatranscriptomic census of active protists in soils. ISME J. 2015, 9, 2178–2190. [Google Scholar] [CrossRef]
- Harder, C.B.; Ronn, R.; Brejnrod, A.; Bass, D.; Abu Al-Soud, W.; Ekelund, F. Local diversity of heathland Cercozoa explored by in-depth sequencing. ISME J. 2016, 10, 2488–2497. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Huang, B. Protist diversity and community assembly in surface sediments of the South China Sea. MicrobiologyOpen 2019, 8, e891. [Google Scholar] [CrossRef] [PubMed]
- Karpov, S.A.; Mamkaeva, M.A.; Benzerara, K.; Moreira, D.; López-García, P. Molecular Phylogeny and Ultrastructure of Aphelidium aff. melosirae (Aphelida, Opisthosporidia). Protist 2014, 165, 512–526. [Google Scholar] [CrossRef]
- Wu, P.; Shi, X.; Lv, X.; Gao, Y.; Li, H.; Zhang, J. Distribution of benthic eukaryote communities in intertidal zone of mangrove in the Beilun Estuary by high-throughput sequencing analysis. J. Appl. Oceanogr. 2019, 38, 232–238. [Google Scholar]
- Ouyang, J.; Chen, S.; Wei, N.; Zhang, S.; Yan, Q.; Ling, M.; Ma, Y. Microeukaryotic community structure and response to nereid bioturbation in mudflat sediments of Xiamen Bay. J. Fish. China 2017, 41, 1237–1245. [Google Scholar]
- Zhang, L.; Qi, Y.; Jiao, J.; Li, C. Microbial community diversity of reed rhizosphere soil in different sandy land habitats of Hexi Corridor, Gansu, China. J. Desert Res. 2021, 41, 1–9. [Google Scholar]
- Schenk, J.; Kleinbölting, N.; Traunspurger, W. Comparison of morphological, DNA barcoding, and metabarcoding characterizations of freshwater nematode communities. Ecol. Evol. 2020, 10, 2885–2899. [Google Scholar] [CrossRef]
- Shu, L.; Lin, J.; Xu, Y.; Cao, T.; Feng, J.; Peng, Z. Investigating the fish diversity in Erhai lake based on environmental DNA metabarcoding. Acta Hydrobiol. Sin. 2020, 44, 1080–1086. [Google Scholar]
- Barnes, M.A.; Turner, C.R. The ecology of environmental DNA and implications for conservation genetics. Conserv. Genet. 2016, 17, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Nascimento, F.; Lallias, D.; Bik, H.M.; Creer, S. Sample size effects on the assessment of eukaryotic diversity and community structure in aquatic sediments using high-throughput sequencing. Sci. Rep. 2018, 8, 11737. [Google Scholar] [CrossRef]
- Keeley, N.; Wood, S.A.; Pochon, X. Development and preliminary validation of a multi-trophic metabarcoding biotic index for monitoring benthic organic enrichment. Ecol. Indic. 2018, 85, 1044–1057. [Google Scholar] [CrossRef]
- Wang, Z.; Kang, W. Distribution of dinocysts and germination of phytoplankton resting spores in surface sediments from Zhelin Bay, the South China Sea. Acta Sci. Circumstantiae 2014, 34, 2043–2050. [Google Scholar]
- Obrezkova, M.S.; Pospelova, V.Y. Distribution of Diatoms and Dinocysts in Surface Sediments from the East Siberian and Chukchi Seas. Paleontol. J. 2019, 53, 790–794. [Google Scholar] [CrossRef]
- Rubino, F.; Belmonte, M.; Galil, B.S. Plankton resting stages in recent sediments of Haifa port, Israel (Eastern Mediterranean)—Distribution, viability and potential environmental consequences. Mar. Pollut. Bull. 2017, 116, 258–269. [Google Scholar] [CrossRef]
- Morard, R.L.; Lejzerowicz, F.; Darling, K.F.; Lecroq-Bennet, B.; Winther Pedersen, M.; Orlando, L.; Pawlowski, J.; Mulitza, S.; de Vargas, C.; Kucera, M. Planktonic foraminifera-derived environmental DNA extracted from abyssal sediments preserves patterns of plankton macroecology. Biogeosciences 2017, 14, 2741–2754. [Google Scholar] [CrossRef]
- Vehmaa, A.; Katajisto, T.; Candolin, U. Long-term changes in a zooplankton community revealed by the sediment archive. Limnol. Oceanogr. 2018, 63, 2126–2139. [Google Scholar] [CrossRef]
- Holm, M.W.; Kiørboe, T.; Brun, P.; Licandro, P.; Almeda, R.; Hansen, B.W. Resting eggs in free living marine and estuarine copepods. J. Plankton Res. 2018, 40, 2–15. [Google Scholar] [CrossRef]
- Belmonte, G.; Rubino, F. Resting cysts from coastal marine plankton. Oceanogr. Mar. Biol. Annu. Rev. 2019, 57, 1–88. [Google Scholar]
- Mcmahon, K.W.; Ambrose, W.G.; Johnson, B.J.; Sun, M.Y.; Lopez, G.R.; Clough, L.M.; Carroll, M.L. Benthic community response to ice algae and phytoplankton in Ny Alesund, Svalbard. Mar. Ecol. Prog. Ser. 2006, 310, 1–14. [Google Scholar] [CrossRef]
- Moscatello, S.; Rubino, F.; Saracino, O.D.; Fanelli, G.; Belmonte, G.; Boero, F. Plankton biodiversity around the Salento Peninsula (South East Italy): An integrated water/sediment approach. Sci. Mar. 2004, 68, 85–102. [Google Scholar] [CrossRef]
- Medeiros, J.P.; Chaves, M.L.; Silva, G.; Azeda, C.; Costa, J.L.; Marques, J.C.; Costa, M.J.; Chainho, P. Benthic condition in low salinity areas of the Mira estuary (Portugal): Lessons learnt from freshwater and marine assessment tools. Ecol. Indic. 2012, 19, 79–88. [Google Scholar] [CrossRef]
- Reich, M.; Wichels, A.; Panzer, K.; Krause, E.; Gimenez, L.; Gerdts, G. Impacts of a reduction in seawater pH mimicking ocean acidification on the structure and diversity of mycoplankton communities. Aquat. Microb. Ecol. 2017, 79, 221–233. [Google Scholar] [CrossRef]
- Cudowski, A.; Pietryczuk, A.; Hauschild, T. Aquatic fungi in relation to the physical and chemical parameters of water quality in the Augustów Canal. Fungal Ecol. 2015, 13, 193–204. [Google Scholar] [CrossRef]
- Bik, H.M.; Sung, W.; De Ley, P.; Baldwin, J.G.; Sharma, J.; Rocha-Olivares, A.; Thomas, W.K. Metagenetic community analysis of microbial eukaryotes illuminates biogeographic patterns in deep-sea and shallow water sediments. Mol. Ecol. 2012, 21, 1048–1059. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Shi, F.; Ma, B.; Dong, J.; Pachiadaki, M.; Zhang, X.; Edgcomb, V.P. Depth shapes alpha- and beta-diversities of microbial eukaryotes in surficial sediments of coastal ecosystems. Environ. Microbiol. 2015, 17, 3722–3737. [Google Scholar] [CrossRef]
- Gu, R.; Sun, P.; Wang, Y.; Yu, F.L.; Jiao, N.Z.; Xu, D.P. Genetic Diversity, Community Assembly, and Shaping Factors of Benthic Microbial Eukaryotes in Dongshan Bay, Southeast China. Front. Microbiol. 2020, 11, 592489. [Google Scholar] [CrossRef]
- Cariou, M.; Francois, C.M.; Voisin, J.; Pigneret, M.; Hervant, F.; Volatier, L.; Mermillod-Blondin, F. Effects of bioturbation by tubificid worms on biogeochemical processes, bacterial community structure and diversity in heterotrophic wetland sediments. Sci. Total Environ. 2021, 795, 148842. [Google Scholar] [CrossRef]
- Gillespie, D. Phytoplankton production in the Great Salt Lake, Utah, and a laboratory study of algal response to enrichment1. Limnol. Oceanogr. 1976, 21, 74–87. [Google Scholar]
- Prazukin, A.; Shadrin, N.; Balycheva, D.; Firsov, Y.; Lee, R.; Anufriieva, E. Cladophora spp. (Chlorophyta) modulate environment and create a habitat for microalgae in hypersaline waters. Eur. J. Phycol. 2021, 56, 231–243. [Google Scholar] [CrossRef]
- Dai, W.; Yang, S.; Que, Z.; Xiong, J. Responses of Plankton Microeukaryotic Community to Increasing Temperatures Created by Power Plant Thermal Discharges. Huanjing Kexue 2016, 37, 2696–2704. [Google Scholar] [CrossRef]
- Karpinsky, M.G. Review: The Caspian Sea benthos: Unique fauna and community formed under strong grazing pressure. Mar. Pollut. Bull. 2010, 61, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.; Martens, K.; Goddeeris, B. Oligochaeta from the abyssal zone of Lake Baikal (Siberia, Russia). Hydrobiologia 1999, 406, 165–174. [Google Scholar] [CrossRef]
- Sierszen, M.E.; Peterson, G.S.; Scharold, J.V. Depth-specific patterns in benthic-planktonic food web relationships in Lake Superior. Can. J. Fish. Aquat. Sci. 2006, 63, 1496–1503. [Google Scholar] [CrossRef]
- Parmar, T.K.; Rawtani, D.; Agrawal, Y.K. Bioindicators: The natural indicator of environmental pollution. Front. Life Sci. 2016, 9, 110–118. [Google Scholar]
- Gaedke, U.; Seifried, A.; Adrian, R. Biomass Size Spectra and Plankton Diversity in a Shallow Eutrophic Lake. Int. Rev. Hydrobiol. 2004, 89, 1–20. [Google Scholar] [CrossRef]
- Zheng, B.; Chen, Z.; Li, Y.; Fohrer, N.; Zhang, Y.; Wu, D.; Yan, X.; Li, B. Structural Characteristics and Driving Factors of the Planktonic Eukaryotic Community in the Danjiangkou Reservoir, China. Water 2020, 12, 3499. [Google Scholar] [CrossRef]
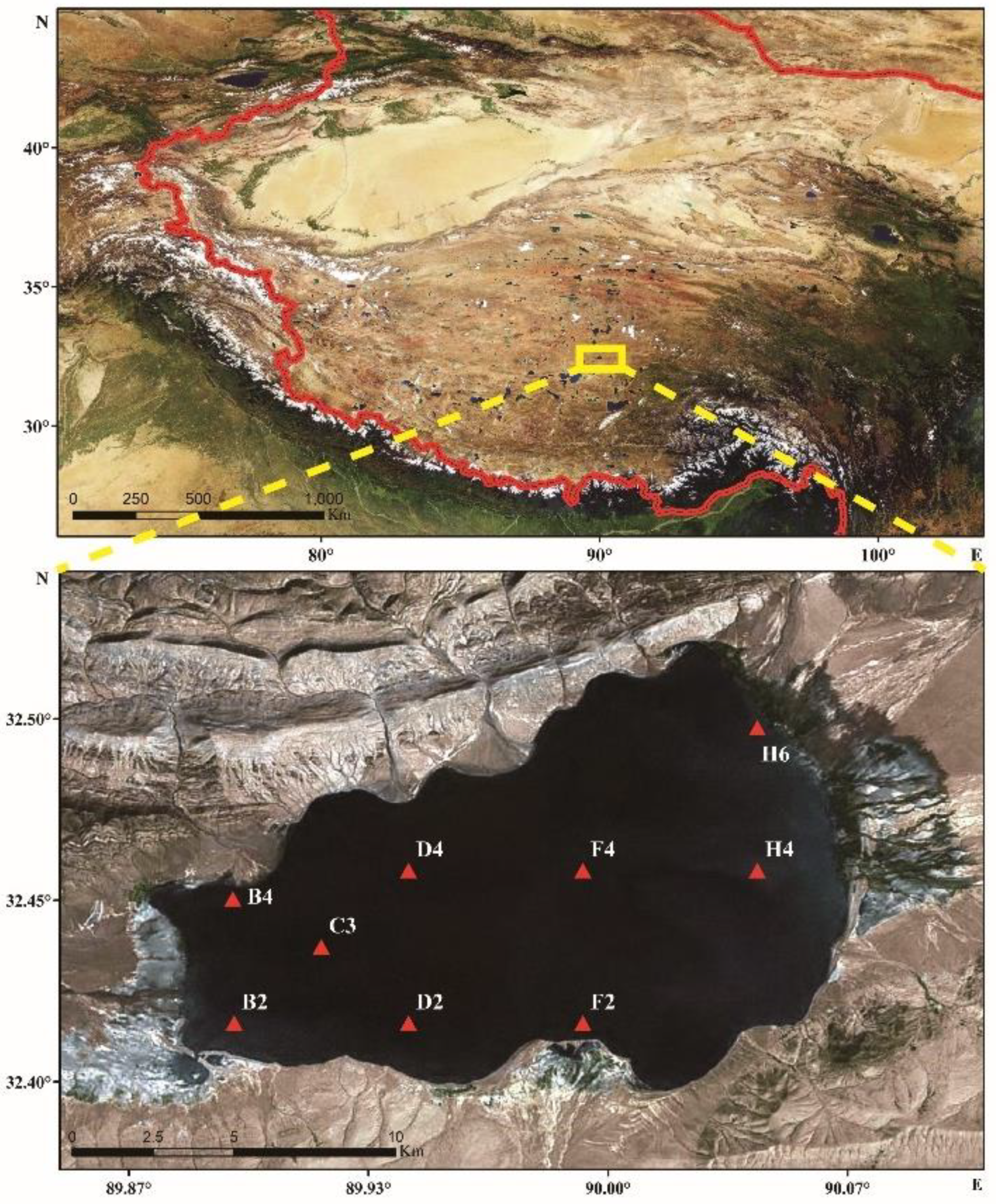

| B2 | B4 | C3 | D2 | D4 | F2 | F4 | H4 | H6 | |
|---|---|---|---|---|---|---|---|---|---|
| WD (m) | 7 | 1.5 | 16.3 | 16.2 | 23 | 12.8 | 21.5 | 9.5 | 5.4 |
| T (°C) | 13.76 | 15.29 | 1.07 | 1.25 | 0.95 | 3.85 | 0.73 | 13.04 | 13.81 |
| pH | 8.95 | 8.84 | 8.81 | 9 | 8.88 | 9.07 | 8.92 | 9.04 | 9 |
| DO (mg/L) | 4.84 | 6.06 | 1.19 | 6.08 | 0.08 | 11.7 | 9.71 | 2.06 | 4.67 |
| MC (%) | 46.32 | 36.87 | 46.42 | 51.48 | 49.64 | 52.99 | 60.64 | 44.11 | 40.72 |
| OM (%) | 2.86 | 0.43 | 3.58 | 7.8 | 7.47 | 4.51 | 8.6 | 1.93 | 2.83 |
| Chla (ug/L) | 0.61 | 1.8 | 0.56 | 0.47 | 0.4 | 0.34 | 0.6 | 0.89 | 0.71 |
| N (%) | 0.09 | 0.08 | 0.26 | 0.27 | 0.2 | 0.13 | 0.37 | 0.11 | 0.14 |
| C (%) | 4.84 | 1.89 | 5.73 | 8.89 | 8.67 | 6.29 | 10.04 | 3.84 | 3.98 |
| S (%) | 0.35 | 0.38 | 0.87 | 1.29 | 0.73 | 0.36 | 1.24 | 0.38 | 0.73 |
| C/N | 53.78 | 23.63 | 22.04 | 32.93 | 43.35 | 48.38 | 27.14 | 34.91 | 28.43 |
| Sediment type | Sand | Silty sand | Silty sand | Silty sand | Silty sand | Silty sand | Silty sand | Silty sand | Silty sand |
| Sample | Sobs | Shannon | Simpson | Ace | Chao | Coverage |
|---|---|---|---|---|---|---|
| C3 | 130 | 1.6340 | 0.2785 | 220.3709 | 181 | 0.9985 |
| F4 | 98 | 1.5741 | 0.2907 | 135.2014 | 133 | 0.9989 |
| D2 | 97 | 1.2520 | 0.4156 | 193.8154 | 179 | 0.9987 |
| F2 | 160 | 1.8418 | 0.2993 | 256.5098 | 231 | 0.9984 |
| B2 | 224 | 2.2485 | 0.3082 | 267.1607 | 266 | 0.9983 |
| H4 | 193 | 1.7892 | 0.3772 | 257.9494 | 259 | 0.9981 |
| D4 | 111 | 1.2460 | 0.4458 | 196.7759 | 160 | 0.9988 |
| H6 | 242 | 3.3565 | 0.0587 | 292.8188 | 281 | 0.9982 |
| B4 | 241 | 2.2952 | 0.2007 | 309.3496 | 304 | 0.9979 |
| Zone | Sobs | Shannon | Simpson | Ace | Chao | Coverage |
|---|---|---|---|---|---|---|
| littoral zone | 235.6667 ± 8.2597 | 2.6334 ± 0.5117 | 0.1892 ± 0.1022 | 289.7764 ± 17.3574 | 283.5118 ± 15.8694 | 0.9981 ± 0.0002 |
| sublittoral zone | 176.5000 ± 16.5000 | 1.8155 ± 0.0263 | 0.3383 ± 0.0390 | 257.2296 ± 0.7198 | 244.6945 ± 13.8611 | 0.9983 ± 0.0001 |
| profundal zone | 109.0000 ± 13.3229 | 1.4265 ± 0.1788 | 0.3577 ± 0.0740 | 186.5409 ± 31.3763 | 163.4338 ± 19.3142 | 0.9987 ± 0.0002 |
| RDA1 | RDA2 | r2 | p_Values | |
|---|---|---|---|---|
| Depth | −0.9706 | −0.2406 | 0.8259 | 0.008 |
| T | 0.979 | 0.2037 | 0.8172 | 0.008 |
| MC | −0.9845 | −0.1753 | 0.408 | 0.214 |
| OM | −0.8654 | −0.5012 | 0.7438 | 0.022 |
| N | −0.8858 | −0.464 | 0.6788 | 0.033 |
| C | −0.8754 | 0.4833 | 0.3149 | 0.334 |
| S | −0.7598 | −0.6502 | 0.7164 | 0.037 |
| C/N | 0.6046 | 0.7965 | 0.7605 | 0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, L.; Wang, Q.; Wang, Z.; Wang, F.; Sun, S.; Liu, X. Eukaryotic Diversity Based on High-Throughput 18S rRNA Sequencing and Its Relationship with Environmental Factors in a Salt Lake in Tibet, China. Water 2022, 14, 2724. https://doi.org/10.3390/w14172724
He L, Wang Q, Wang Z, Wang F, Sun S, Liu X. Eukaryotic Diversity Based on High-Throughput 18S rRNA Sequencing and Its Relationship with Environmental Factors in a Salt Lake in Tibet, China. Water. 2022; 14(17):2724. https://doi.org/10.3390/w14172724
Chicago/Turabian StyleHe, Lele, Qi Wang, Zhe Wang, Fang Wang, Shichun Sun, and Xiaoshou Liu. 2022. "Eukaryotic Diversity Based on High-Throughput 18S rRNA Sequencing and Its Relationship with Environmental Factors in a Salt Lake in Tibet, China" Water 14, no. 17: 2724. https://doi.org/10.3390/w14172724
APA StyleHe, L., Wang, Q., Wang, Z., Wang, F., Sun, S., & Liu, X. (2022). Eukaryotic Diversity Based on High-Throughput 18S rRNA Sequencing and Its Relationship with Environmental Factors in a Salt Lake in Tibet, China. Water, 14(17), 2724. https://doi.org/10.3390/w14172724








