Effects of Selenium in Different Valences on the Community Structure and Microbial Functions of Biofilms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biofilms Cultivation
2.2. Sodium Selenate and Sodium Selenite Exposure
2.3. DNA Extraction and High-Throughput Sequencing
2.4. Analysis Method of Functional Prediction
2.5. Statistical Methods
3. Results and Discussion
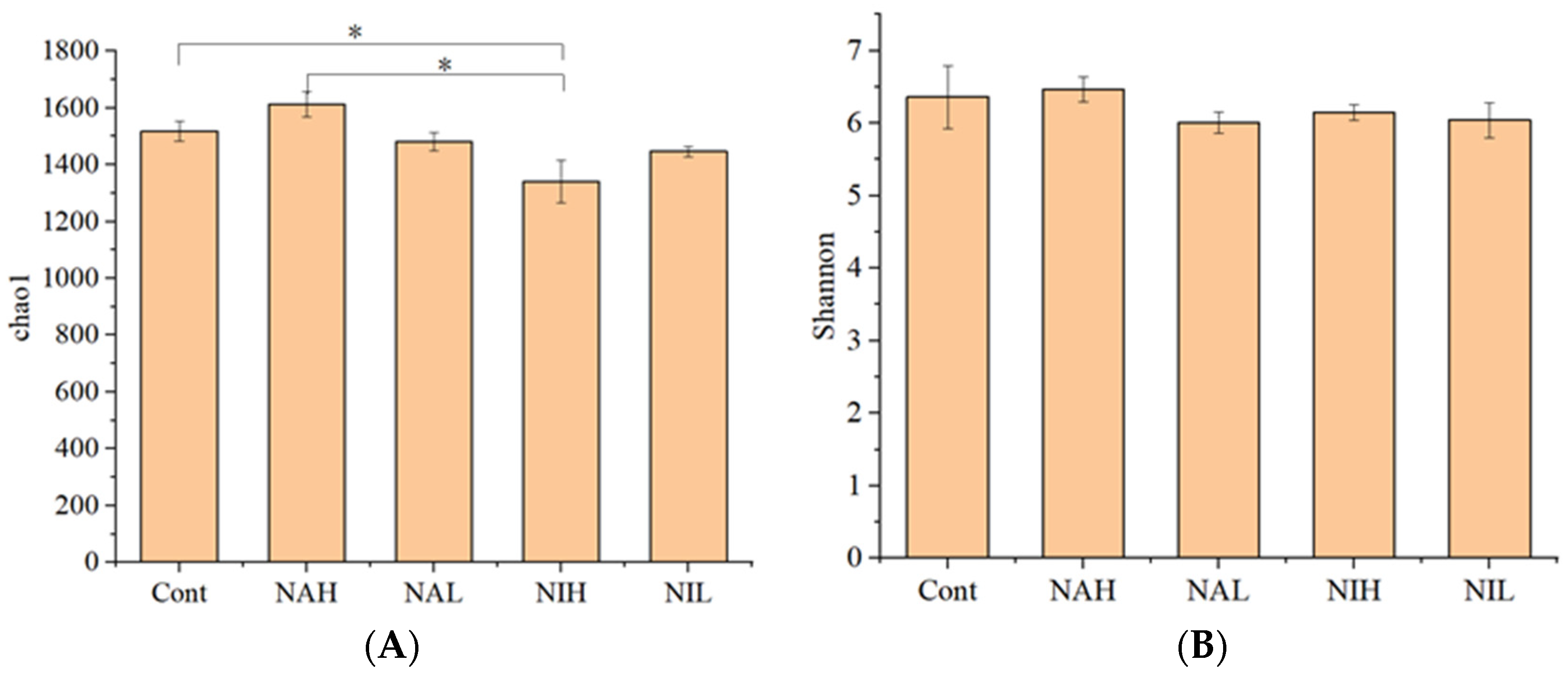
3.1. Taxonomic Annotation and Alpha Diversity
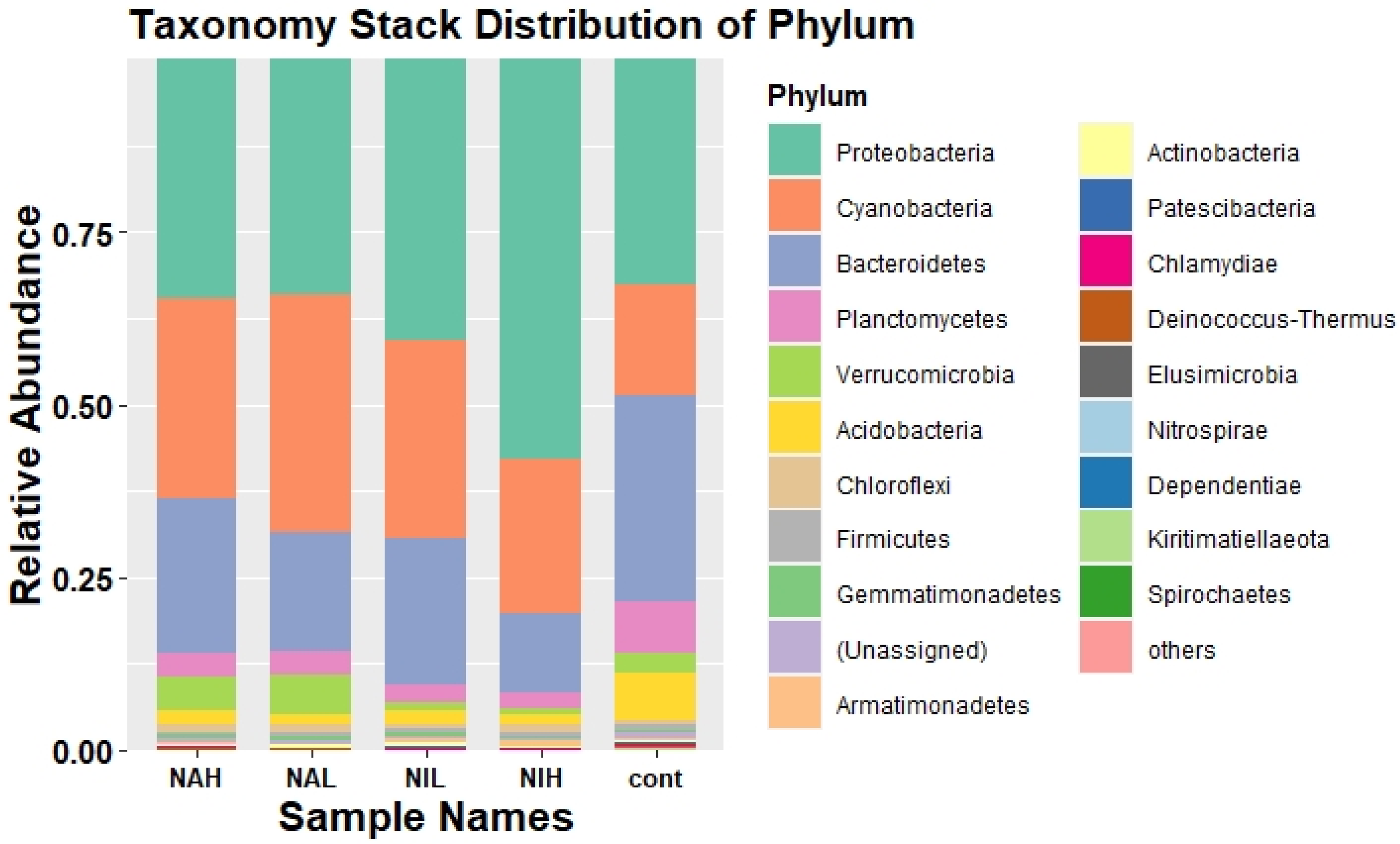
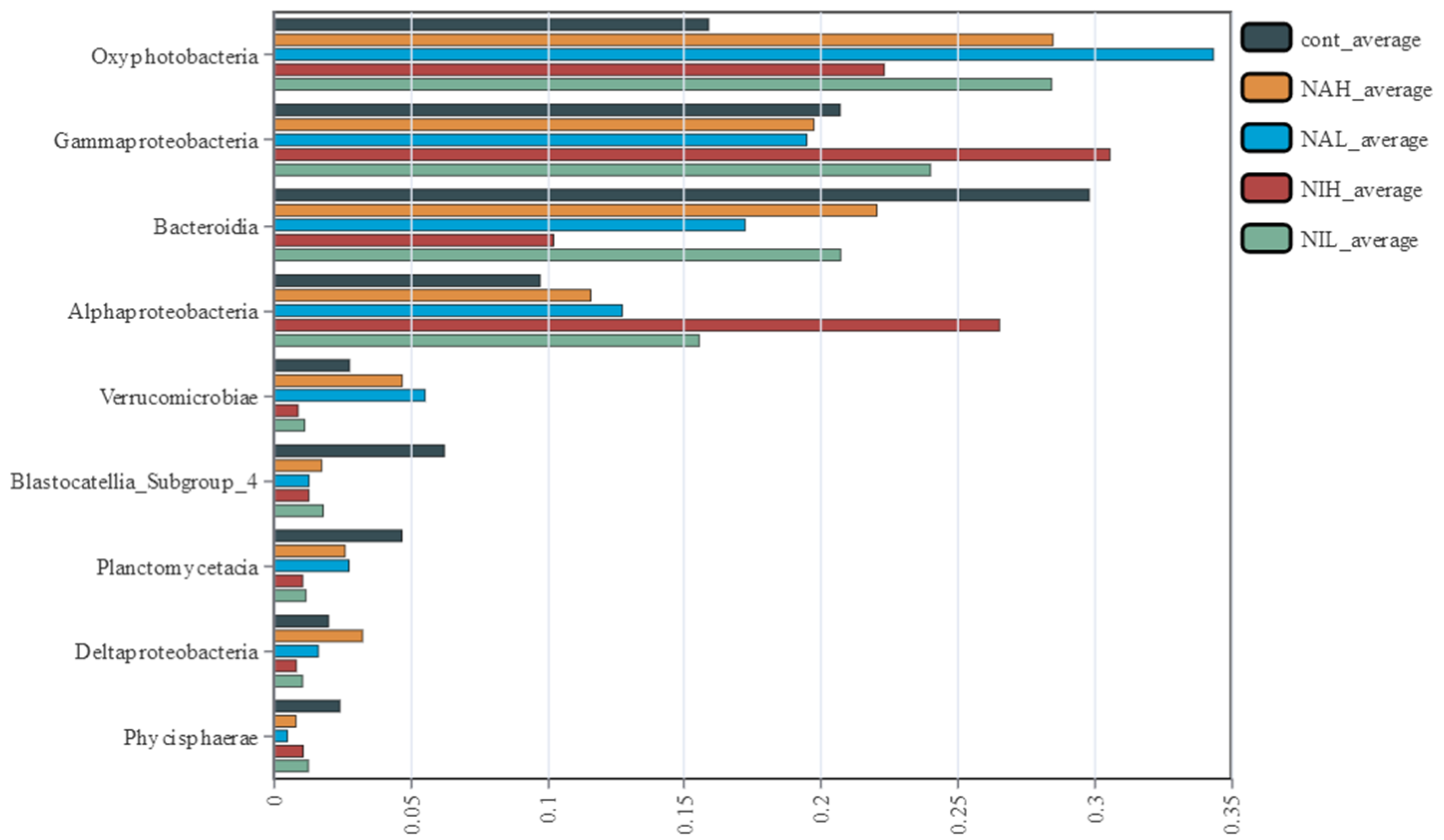
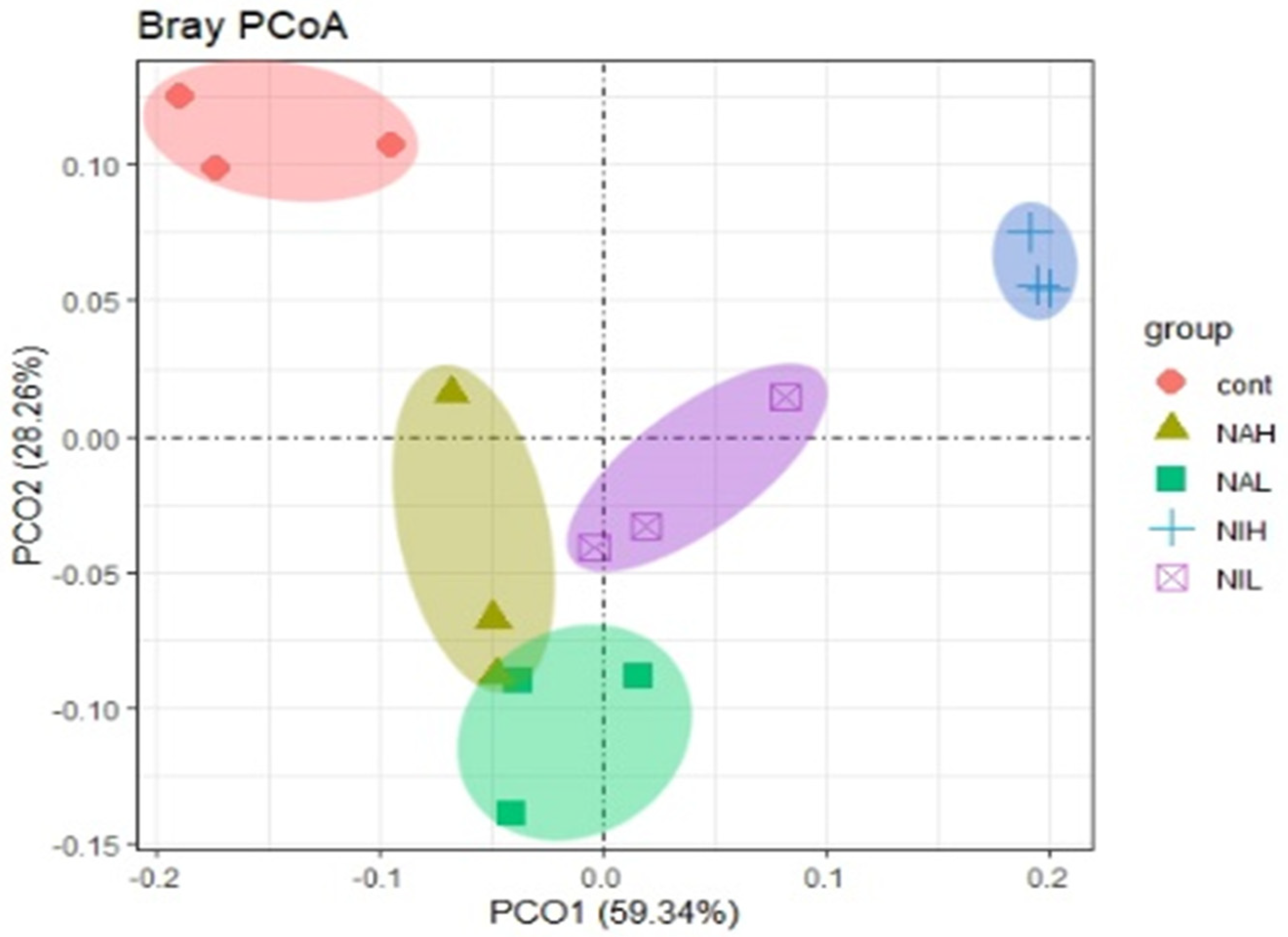
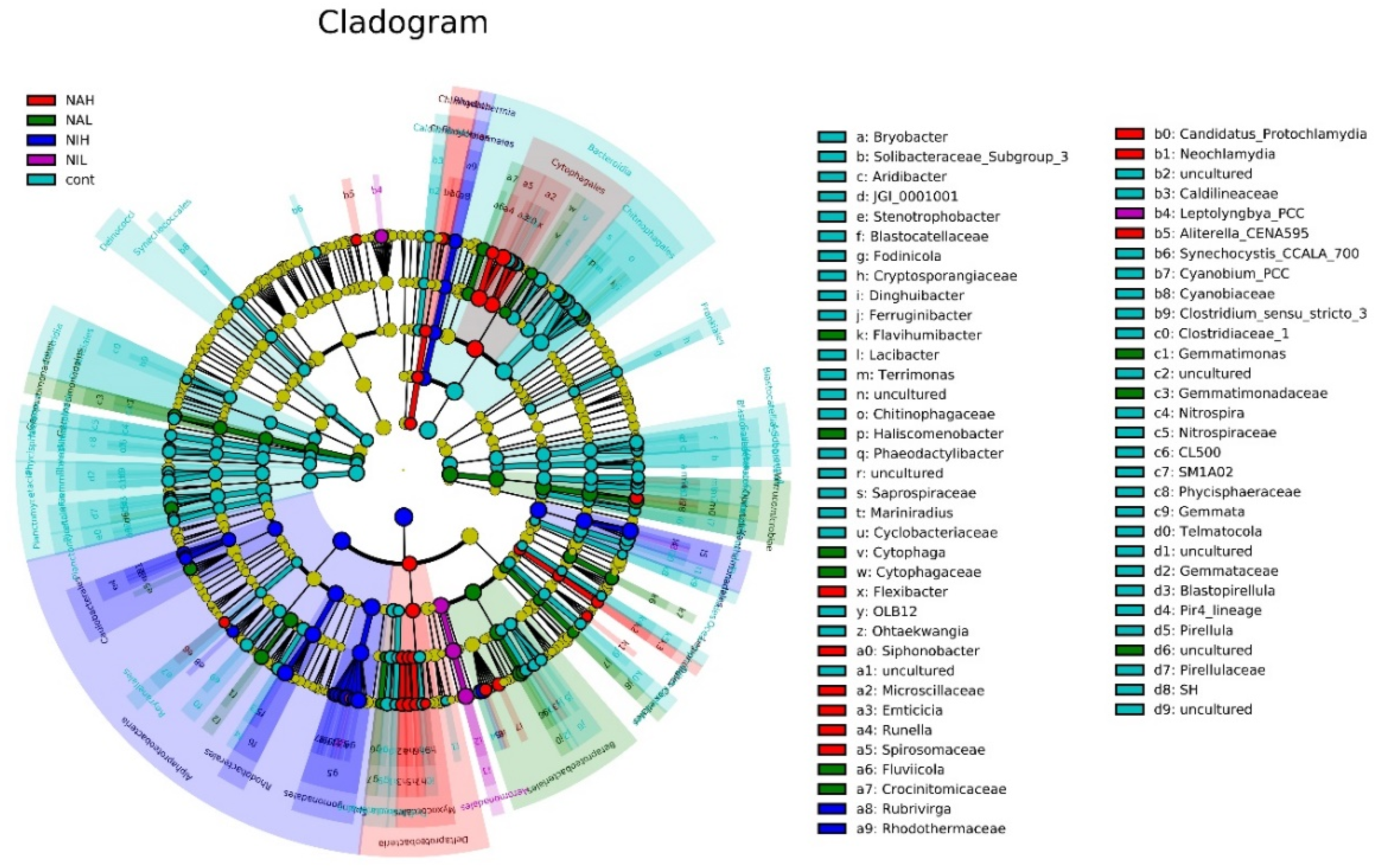
3.2. Bacterial Community Composition and Structure in Biofilms
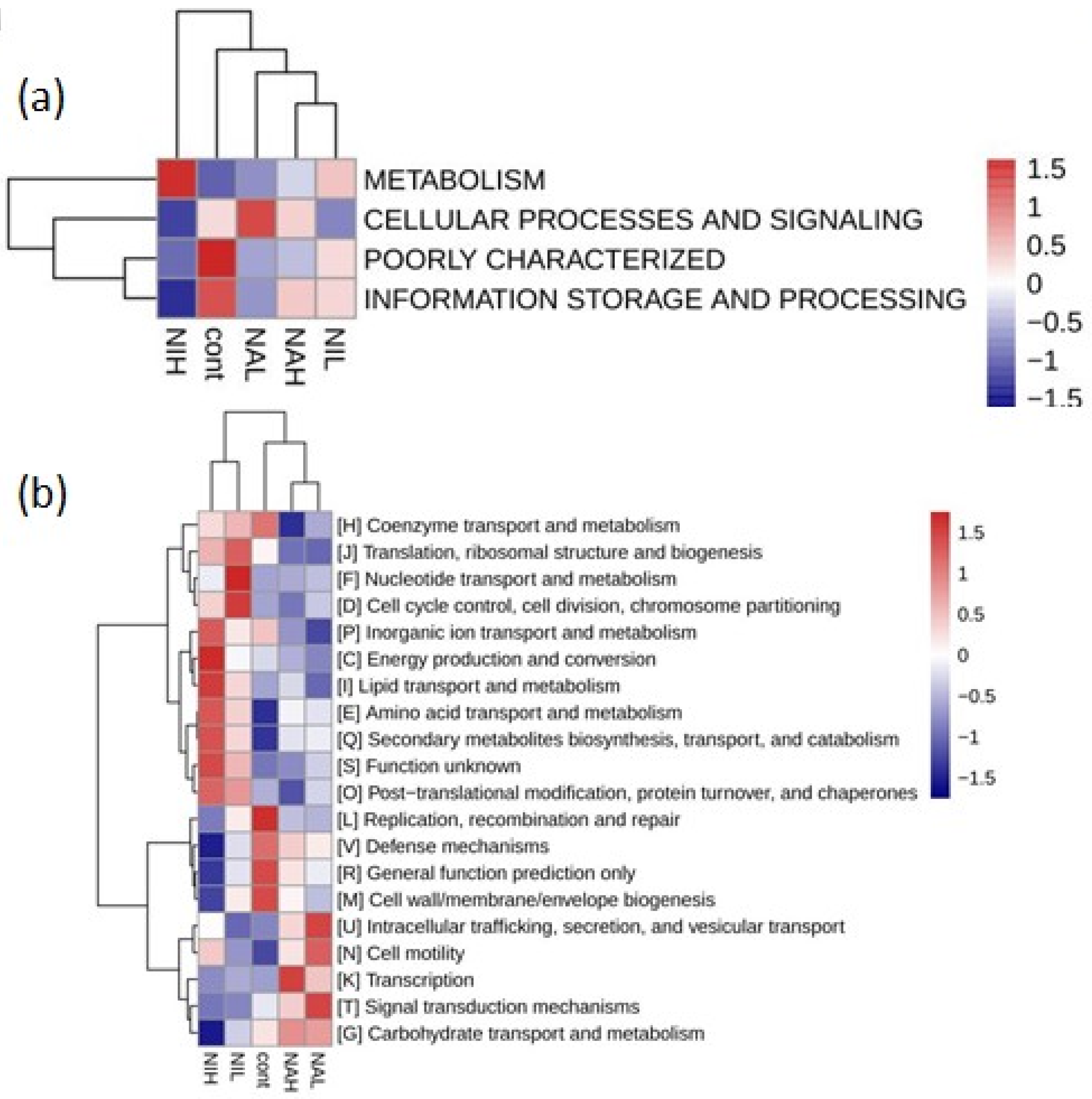
3.3. Functions of Microbial on Biofilm
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arnér, E.S.J.; Holmgren, A. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 2000, 20, 6102–6109. [Google Scholar] [CrossRef]
- Novoselov, S.V.; Gladyshev, V.N. Non-animal origin of animal thioredoxin reductases: Implications for selenocysteine evolution and evolution of protein function through carboxy-terminal extensions. Protein Sci. 2003, 2, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Wang, S.; Tang, C.; Duan, P.; Yao, L.; Tang, J.; Wong, P.K.; An, T.; Dionysiou, D.D.; Wu, Y. Protection Mechanisms of Periphytic Biofilm to Photocatalytic Nanoparticle Exposure. Environ. Sci. Technol. 2019, 53, 1585–1594. [Google Scholar] [CrossRef]
- Lenz, M.; Lens, P.N. The essential toxin: The changing perception of selenium in environmental sciences. Sci. Total Environ. 2009, 407, 3620–3633. [Google Scholar] [CrossRef]
- Chasteen, T.G.; Bentley, R. Biomethylation of Selenium and Tellurium: Microorganisms and Plants. Chem. Rev. 2002, 103, 1–26. [Google Scholar] [CrossRef]
- Hilton, J.W.; Hodson, P.V.; Slinger, S.J. The requirement and toxicity of selenium in rainbow trout (Salmo gairdneri). J. Nutr. 1980, 110, 2527–2535. [Google Scholar] [CrossRef]
- Hodson, P.V.; Hilton, J.W. The Nutritional Requirements and Toxicity to Fish of Dietary and Waterborne Selenium. Eco-Log. Bull. 1983, 35, 335–340. [Google Scholar]
- Cutter, G.A.; Bruland, K.W. The marine biogeochemistry of selenium: A re-evaluation1. Limnol. Oceanogr. 1984, 29, 1179–1192. [Google Scholar] [CrossRef] [Green Version]
- Ozturk Kurt, B.; Konukoglu, D.; Kalayci, R.; Ozdemir, S. Investigation of the Protective Role of Selenium in the Changes Caused by Chlorpyrifos in Trace Elements, Biochemical and Hematological Parameters in Rats. Biol. Trace Elem. Res. 2022, 200, 1–10. [Google Scholar] [CrossRef]
- Copeland, P.R. Making sense of nonsense: The evolution of selenocysteine usage in proteins. Genome Biol. 2005, 6, 221. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Zhong, Y.; Huang, Z.; Yang, Y. Selenium Accumulation in Unicellular Green Alga Chlorella vulgaris and Its Effects on Antioxidant Enzymes and Content of Photosynthetic Pigments. PLoS ONE 2014, 9, e112270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Li, G.; Yin, H.; An, T. Bacterial response mechanism during biofilm growth on different metal material substrates: EPS characteristics, oxidative stress and molecular regulatory network analysis. Environ. Res. 2020, 185, 109451. [Google Scholar] [CrossRef]
- Tang, J.; Zhu, N.; Zhu, Y.; Kerr, P.; Wu, Y. Distinguishing the roles of different extracellular polymeric substance fractions of a periphytic biofilm in defending against Fe2O3 nanoparticle toxicity. Environ. Sci. Nano 2017, 4, 1682–1691. [Google Scholar] [CrossRef]
- Shabbir, S.; Faheem, M.; Ali, N.; Kerr, P.G.; Wang, L.-F.; Kuppusamy, S.; Li, Y. Periphytic biofilm: An innovative approach for biodegradation of microplastics. Sci. Total Environ. 2020, 717, 137064. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Wu, Y.; Esquivel-Elizondo, S.; Sørensen, S.J.; Rittmann, B.E. How Microbial Aggregates Protect against Nanoparticle Toxicity. Trends Biotechnol. 2018, 36, 1171–1182. [Google Scholar] [CrossRef]
- Wu, Y.; Hu, Z.; Kerr, P.G.; Yang, L. A multi-level bioreactor to remove organic matter and metals, together with its associated bacterial diversity. Bioresour. Technol. 2011, 102, 736–741. [Google Scholar] [CrossRef] [PubMed]
- Battin, T.J.; Besemer, K.; Bengtsson, M.M.; Romaní, A.M.; Packmann, A.I. The ecology and biogeochemistry of stream biofilms. Nat. Rev. Microbiol. 2016, 14, 251–263. [Google Scholar] [CrossRef] [Green Version]
- Larned, S.T. A prospectus for periphyton: Recent and future ecological research. J. N. Am. Benthol. Soc. 2010, 29, 182–206. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Liu, J.; Rene, E.R. Periphytic biofilms: A promising nutrient utilization regulator in wetlands. Bioresour. Technol. 2018, 248, 44–48. [Google Scholar] [CrossRef]
- Janz, D.M.; Liber, K.; Pickering, I.J.; Wiramanaden, C.I.; Weech, S.A.; Gallego-Gallegos, M.; Driessnack, M.K.; Franz, E.D.; Goertzen, J.P.; Tse, J.J.; et al. Integrative assessment of selenium speciation, biogeochemistry, and distribution in a northern coldwater ecosystem. Integr. Environ. Assess. Manag. 2014, 10, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Zhnag, X.; Fan, W.-Y.; Yao, M.-C.; Yang, C.-W.; Sheng, G.-P. Redox state of microbial extracellular polymeric substances regulates reduction of selenite to elemental sele-nium accompanying with enhancing microbial detoxification in aquatic environments. Water Res. 2020, 172, 115538. [Google Scholar] [CrossRef]
- Mitchell, K.; Lima, A.T.; Van Cappellen, P. Philippe, Selenium in buoyant marine debris biofilm. Mar. Pollut. Bull. 2019, 149, 110562. [Google Scholar] [CrossRef]
- Tendenedzai, J.T.; Chirwa, E.M.N.; Brink, H.G. Performance Evaluation of Selenite (SeO32−) Reduction by Enterococcus spp. Catalysts 2021, 11, 1024. [Google Scholar] [CrossRef]
- Sinharoy, A.; Lens, P.N.L. Biological removal of selenate and selenite from wastewater: Options for selenium recovery as nanoparticles. Curr. Pollut. Rep. 2020, 3, 230–249. [Google Scholar] [CrossRef]
- Xiong, J.; Wang, H.; Yao, J.; He, Q.; Ma, J.; Yang, J.; Liu, C.; Chen, Y.; Huangfu, X.; Liu, H. A critical review on sulfur reduction of aqueous selenite: Mechanisms and applications. J. Hazard. Mater. 2021, 422, 126852. [Google Scholar] [CrossRef] [PubMed]
- Zonaro, E.; Lampis, S.; Turner, R.J.; Qazi, S.; Vallini, G. Biogenic selenium and tellurium nanoparticles synthesized by environmental microbial isolates effica-ciously inhibit bacterial planktonic cultures and biofilms. Front. Microbiol. 2015, 6, 584. [Google Scholar] [CrossRef] [Green Version]
- Wadhwani, S.A.; Shedbalkar, U.U.; Singh, R.; Chopade, B.A. Biogenic selenium nanoparticles: Current status and future prospects. Appl. Microbiol. Biotechnol. 2016, 100, 2555–2566. [Google Scholar] [CrossRef]
- De Feudis, M.; Massaccesi, L.; D’Amato, R.; Bussinelli, D.; Casucci, C.; Agnelli, A. Impact of Na-selenite fertilization on the microbial biomass and enzymes of a soil under corn (Zea mays L.) cultivation. Geoderma 2020, 373, 114425. [Google Scholar] [CrossRef]
- Nowak, J.; Kaklewski, K.; Klódka, D. Influence of various concentrations of selenic acid (IV) on the activity of soil enzymes. Sci. Total Environ. 2001, 291, 105–110. [Google Scholar] [CrossRef]
- Espinosa-Ortiz, E.J.; Pechaud, Y.; Lauchnor, E.; Rene, E.R.; Gerlach, R.; Peyton, B.M.; van Hullebusch, E.D.; Piet, N.L.L. Effect of selenite on the morphology and respiratory activity of Phanerochaete chrysosporium bio-films. Bioresour. Technol. 2016, 210, 138–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, L.C.; Espinosa-Ortiz, E.J.; Nancharaiah, Y.V.; van Hullebusch, E.D.; Gerlach, R.; Lens, P.N.L. Selenate removal in biofilm systems: Effect of nitrate and sulfate on selenium removal efficiency, biofilm structure and microbial community. J. Chem. Technol. Biotechnol. 2018, 93, 2380–2389. [Google Scholar] [CrossRef] [Green Version]
- Miao, L.; Wang, P.; Hou, J.; Ning, D.; Liu, Z.; Liu, S.; Li, T. Chronic exposure to CuO nanoparticles induced community structure shift and a delay inhibition of microbial functions in multi-species biofilms. J. Clean. Prod. 2020, 262, 121353. [Google Scholar] [CrossRef]
- Moore, J.D.; Stegemeier, J.P.; Bibby, K.; Marinakos, S.M.; Lowry, G.V.; Gregory, K.B. Impacts of Pristine and Transformed Ag and Cu Engineered Nanomaterials on Surficial Sediment Microbial Communities Appear Short-Lived. Environ. Sci. Technol. 2016, 50, 2641–2651. [Google Scholar] [CrossRef]
- Philippot, L.; Spor, A.; Hénault, C.; Bru, D.; Bizouard, F.; Jones, C.; Sarr, A.; Maron, P.-A. Loss in microbial diversity affects nitrogen cycling in soil. ISME J. 2013, 7, 1609–1619. [Google Scholar] [CrossRef] [PubMed]
- Newton, R.J.; Jones, S.E.; Eiler, A.; Mcmahon, K.D.; Bertilsson, S. A guide to the natural history of freshwater lake bacteria. Microbiol. Mol. Biol. Rev. MMBR 2011, 75, 14–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenk, S.; Arnds, J.; Zerjatke, K.; Musat, N.; Amann, R.; Mußmann, M. Novel groups of Gamma-Proteobacteria catalyse sulfur oxidation and carbon fixation in a coastal, intertidal sediment. Environ. Microbiol. 2011, 13, 758–774. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.L.; Smith, D.L.; Connolly, J.; McDonald, J.E.; Cox, M.J.; Joint, I.; Edwards, C.; McCarthy, A.J. Identification of Carbohydrate Metabolism Genes in the Metagenome of a Marine Biofilm Community Shown to Be Dominated by Gamma-Proteobacteria and Bacteroidetes. Genes 2010, 1, 371–384. [Google Scholar] [CrossRef]
- Ancion, P.-Y.; Lear, G.; Lewis, G.D. Three common metal contaminants of urban runoff (Zn, Cu & Pb) accumulate in freshwater biofilm and modify embedded bacterial communities. Environ. Pollut. 2010, 158, 2738–2745. [Google Scholar] [CrossRef] [PubMed]
- Bonnineau, C.; Artigas, J.; Chaumet, B.; Dabrin, A.; Faburé, J.; Ferrari, B.J.D.; Lebrun, J.D.; Margoum, C.; Mazzella, N.; Miège, C.; et al. Role of Biofilms in Contaminant Bioaccumulation and Trophic Transfer in Aquatic Ecosystems: Current State of Knowledge and Future Challenges. Rev. Environ. Contam. Toxicol. 2020, 253, 115–153. [Google Scholar] [CrossRef]
- Maroušek, J.; Hašková, S.; Zeman, R.; Váchal, J.; Vaníčková, R. Nutrient Management in Processing of Steam-Exploded Lignocellulose Phytomass. Chem. Eng. Technol. 2014, 37, 1945–1948. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, W.; Tao, C.; Adyel, T.M.; Zhao, J.; Hou, J.; Miao, L.; Zeng, Y. Effects of Selenium in Different Valences on the Community Structure and Microbial Functions of Biofilms. Water 2022, 14, 2394. https://doi.org/10.3390/w14152394
Hao W, Tao C, Adyel TM, Zhao J, Hou J, Miao L, Zeng Y. Effects of Selenium in Different Valences on the Community Structure and Microbial Functions of Biofilms. Water. 2022; 14(15):2394. https://doi.org/10.3390/w14152394
Chicago/Turabian StyleHao, Wenbin, Chunmei Tao, Tanveer M. Adyel, Junjie Zhao, Jun Hou, Lingzhan Miao, and Yuan Zeng. 2022. "Effects of Selenium in Different Valences on the Community Structure and Microbial Functions of Biofilms" Water 14, no. 15: 2394. https://doi.org/10.3390/w14152394




