Machine Learning and Multiple Imputation Approach to Predict Chlorophyll-a Concentration in the Coastal Zone of Korea
Abstract
:1. Introduction
2. Materials and Methods
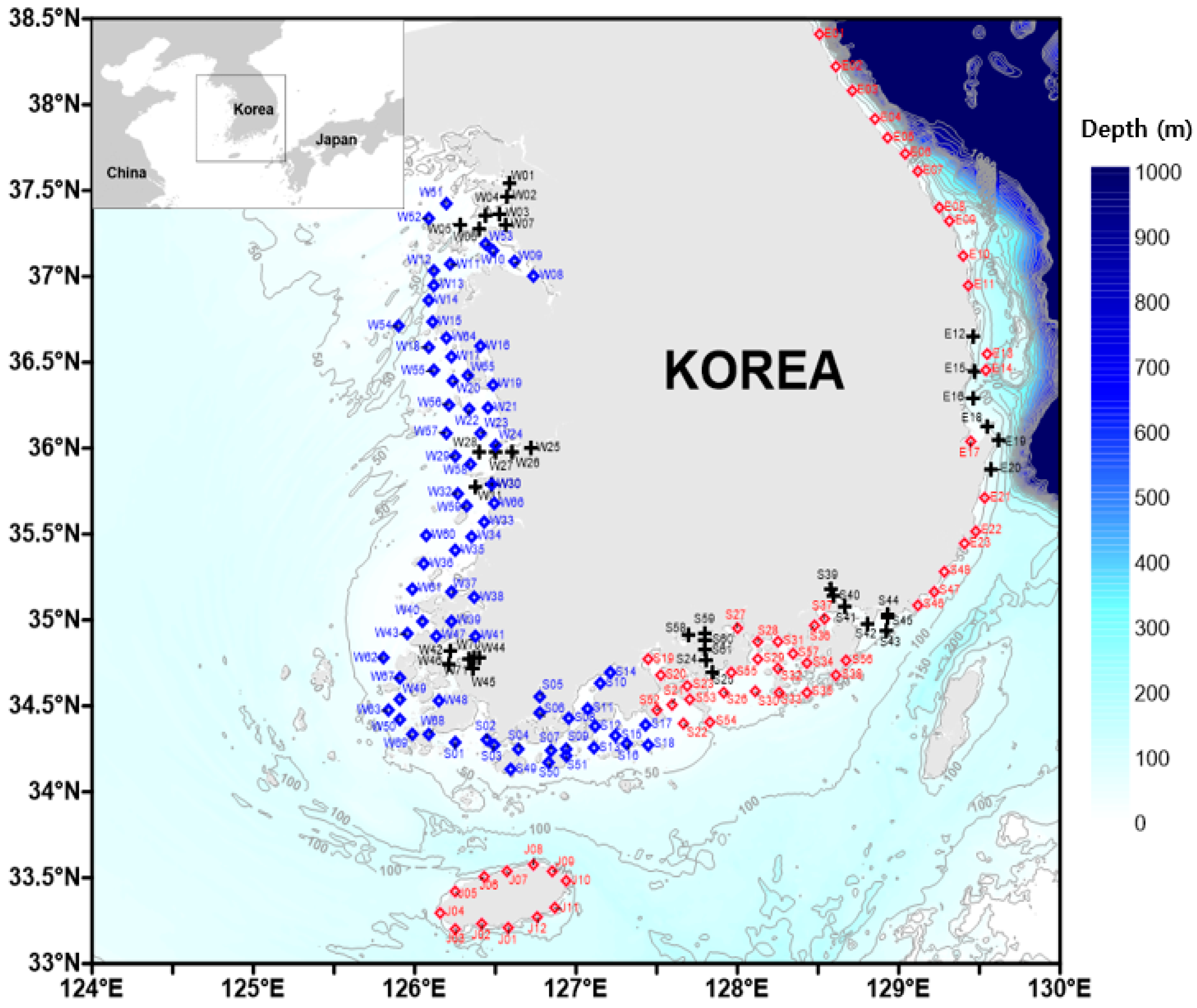
2.1. Study Area and Data
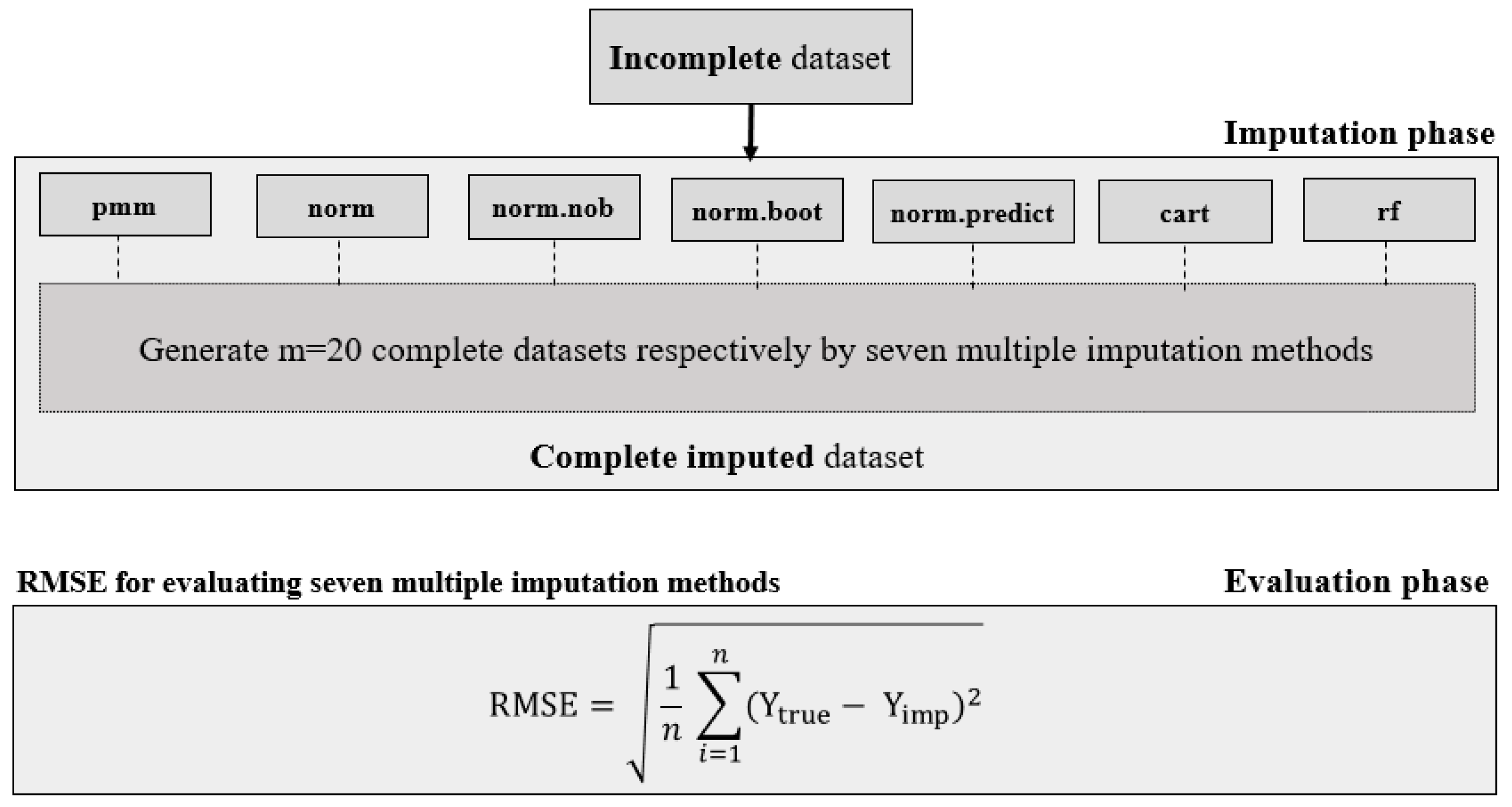
2.2. Evaluation of Multiple Imputation (MI) Methods
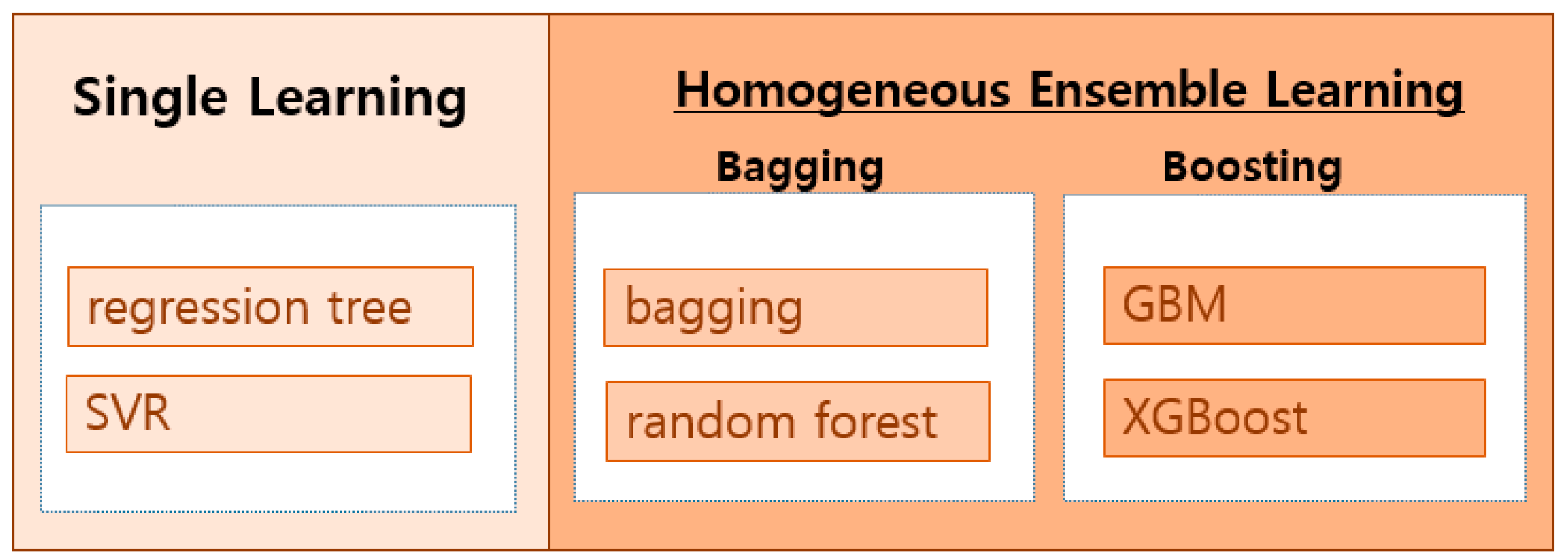
2.3. Machine Learning Algorithms
2.4. Regression Model Accuracy Metrics
3. Results
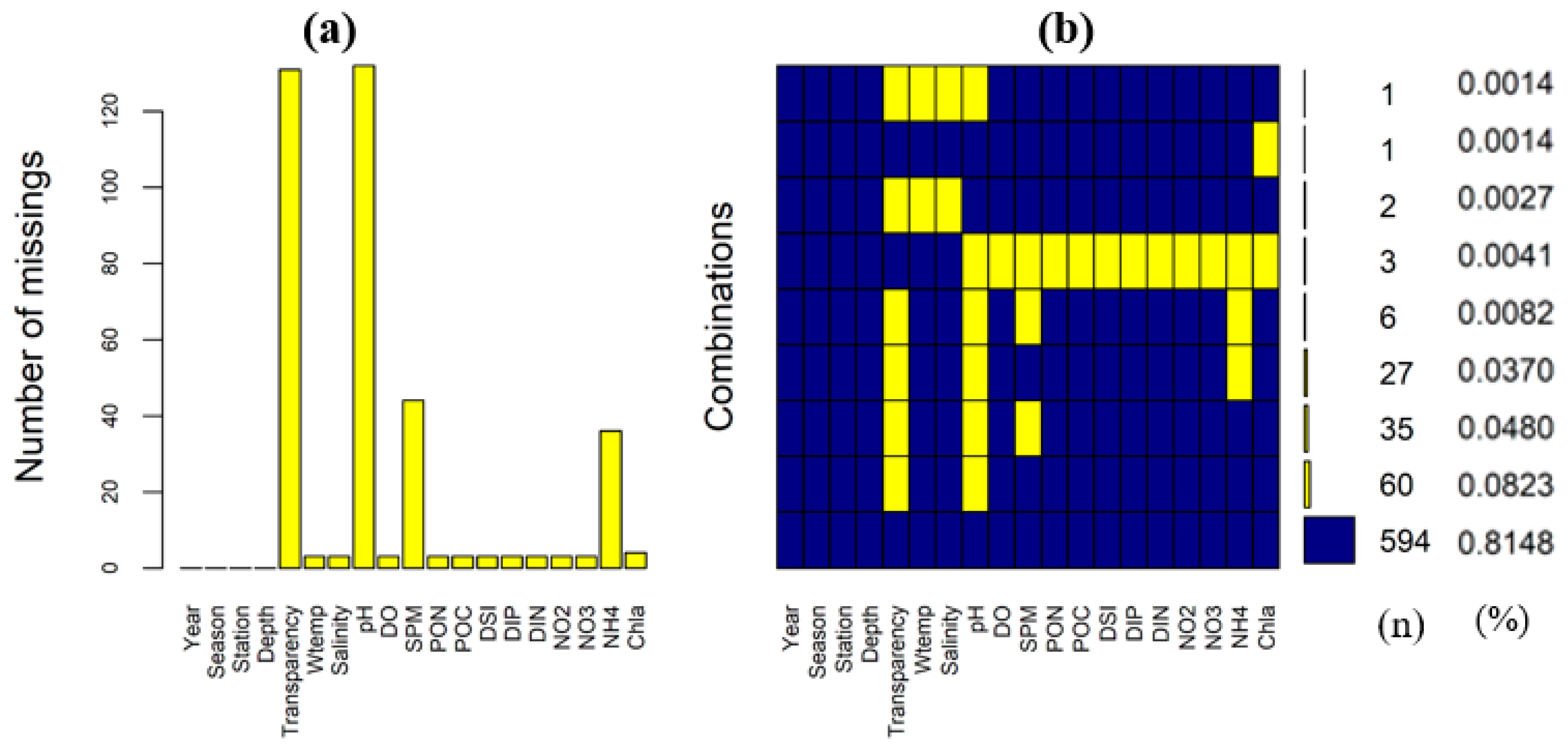
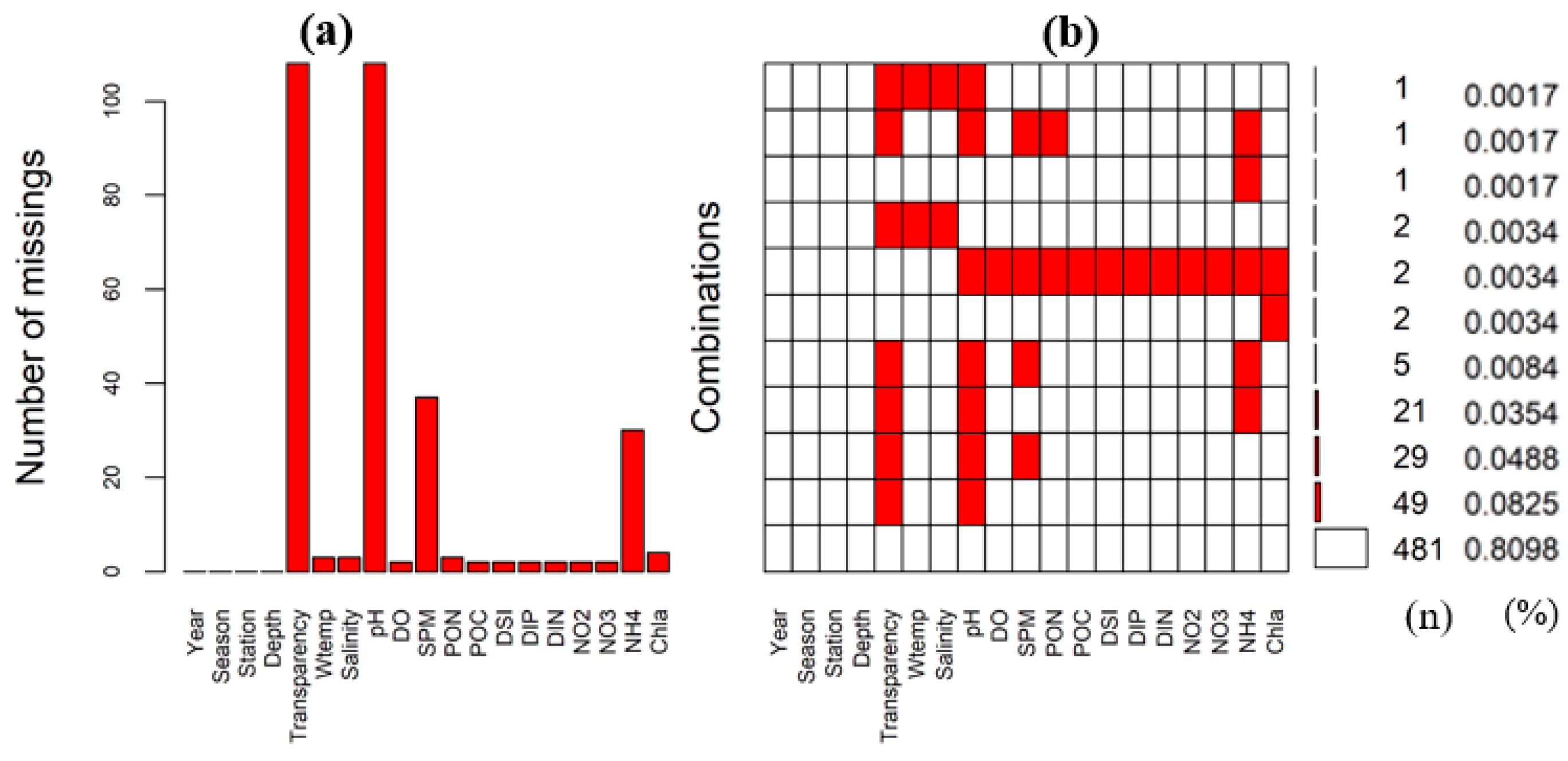
3.1. Missing Data Pattern
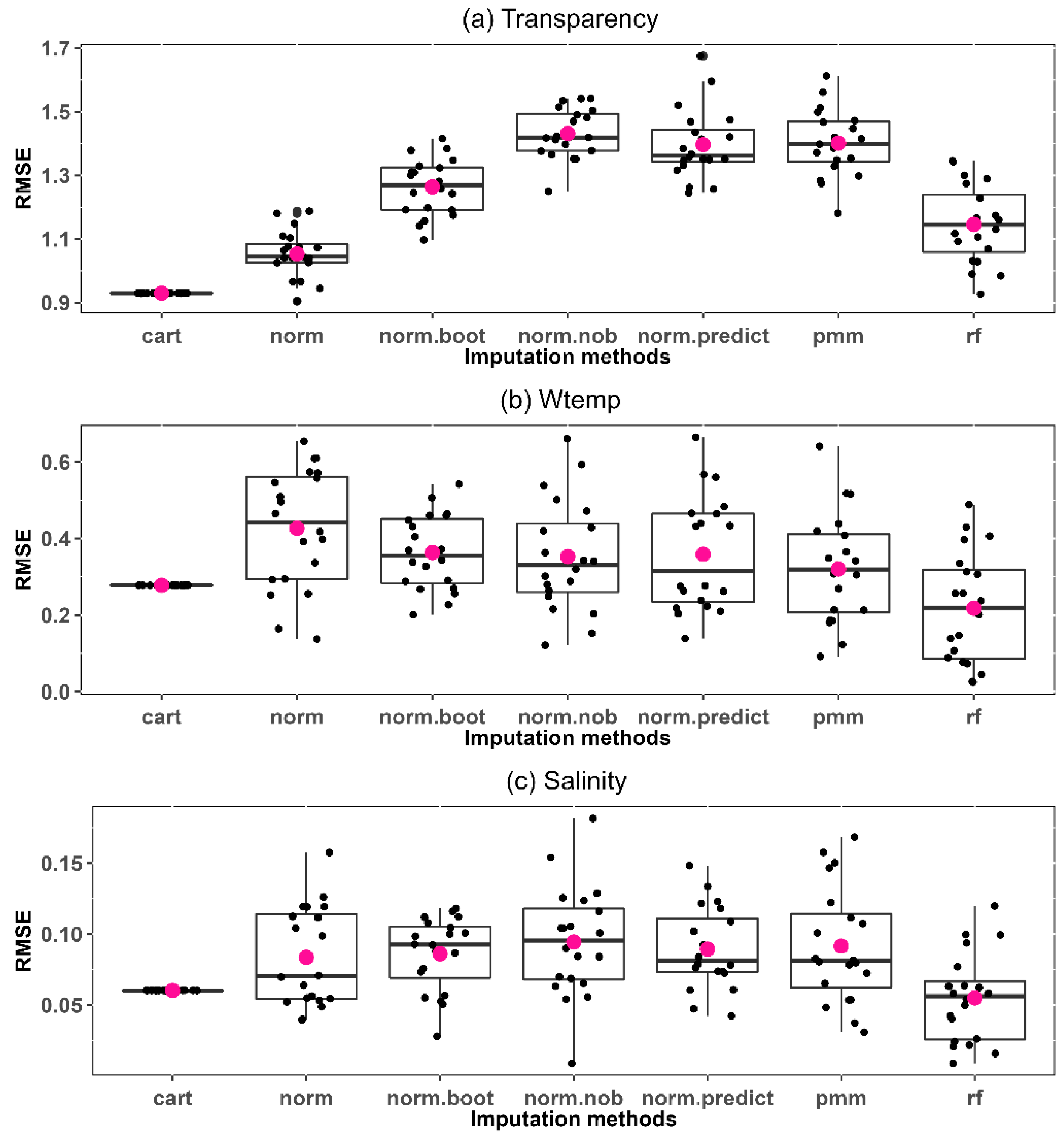
3.2. Selection of Multiple Imputation Method
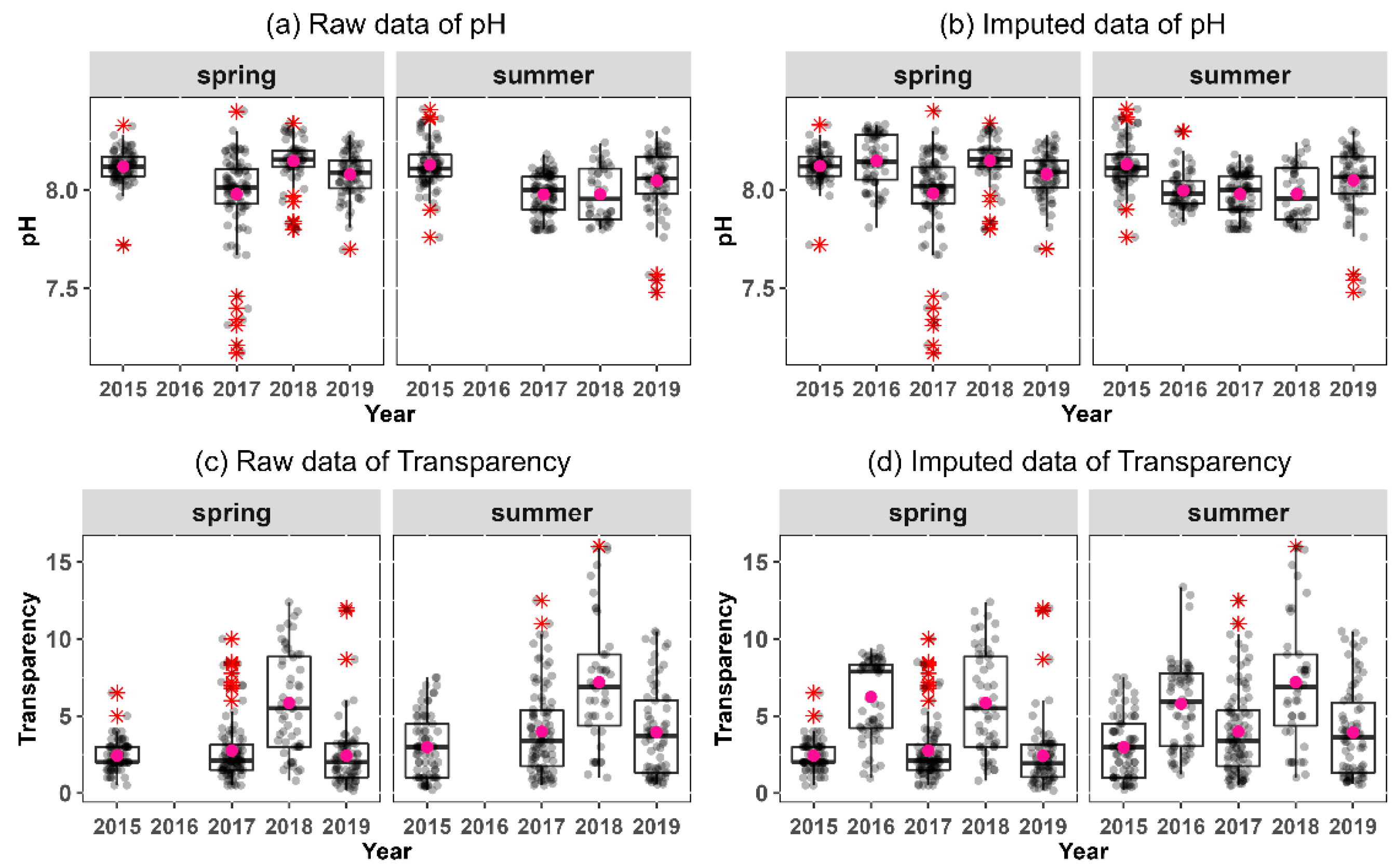
3.3. Missing Imputation by Cart Multiple Imputation and Exploratory Data Analysis
3.4. Distribution of Features before and after Imputation
3.5. Performance Evaluation of Machine Learning Model
3.6. Feature Importance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rajkomar, A.; Dean, J.; Kohane, I. Machine learning in medicine. N. Engl. J. Med. 2019, 380, 1347–1358. [Google Scholar] [CrossRef] [PubMed]
- Jordan, M.I.; Mitchell, T.M. Machine learning: Trends, perspectives, and prospects. Science 2015, 349, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Gudivada, V.; Apon, A.; Ding, J. Data quality considerations for big data and machine learning: Going beyond data cleaning and transformations. Int. J. Adv. Softw. 2017, 10, 1–20. [Google Scholar]
- Kim, E.D.; Ko, S.K.; Son, S.C.; Lee, B.T. Technical Trends of Time-Series Data Imputation. Electron. Telecommun. Trends 2021, 36, 145–153. [Google Scholar] [CrossRef]
- El-Masri, M.M.; Fox-Wasylyshyn, S.M. Missing data: An introductory conceptual overview for the novice researcher. Can. J. Nurs. Res. 2005, 37, 156–171. [Google Scholar]
- Allison, P.D. Multiple imputation for missing data: A cautionary tale. Sociol. Methods Res. 2000, 28, 301–309. [Google Scholar] [CrossRef] [Green Version]
- Patrician, P.A. Multiple imputation for missing data. Res. Nurs. Health 2002, 25, 76–84. [Google Scholar] [CrossRef]
- Emmanuel, T.; Maupong, T.; Mpoeleng, D.; Semong, T.; Mphago, B.; Tabona, O. A survey on missing data in machine learning. J. Big Data 2021, 8, 140. [Google Scholar] [CrossRef]
- Barnard, J.; Meng, X.L. Applications of multiple imputation in medical studies: From AIDS to NHANES. Stat. Methods Med. Res. 1999, 8, 17–36. [Google Scholar] [CrossRef]
- Vilas, L.G.; Spyrakos, E.; Palenzuela, J.M.T. Neural network estimation of chlorophyll a from MERIS full resolution data for the coastal waters of Galician rias (NW Spain). Remote Sens. Environ. 2011, 115, 524–535. [Google Scholar] [CrossRef]
- Park, Y.; Cho, K.H.; Park, J.; Cha, S.M.; Kim, J.H. Development of early-warning protocol for predicting chlorophyll-a concentration using machine learning models in freshwater and estuarine reservoirs, Korea. Sci. Total Environ. 2015, 502, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Hartnett, M.; Nash, S. Modelling nutrient and chlorophyll_a dynamics in an Irish brackish waterbody. Environ. Model. Softw. 2004, 19, 47–56. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.M.; Park, K.D.; Kim, I.K. Comparison of machine learning algorithms for Chl-a prediction in the middle of Nakdong River (focusing on water quality and quantity factors). J. Korean Soc. Water Wastewater 2020, 34, 277–288. [Google Scholar] [CrossRef]
- Shin, Y.; Kim, T.; Hong, S.; Lee, S.; Lee, E.; Hong, S.; Heo, T.Y. Prediction of chlorophyll-a concentrations in the Nakdong River using machine learning methods. Water 2020, 12, 1822. [Google Scholar] [CrossRef]
- Cao, Z.; Ma, R.; Duan, H.; Pahlevan, N.; Melack, J.; Shen, M.; Xue, K. A machine learning approach to estimate chlorophyll-a from Landsat-8 measurements in inland lakes. Remote Sens. Environ. 2020, 248, 111974. [Google Scholar] [CrossRef]
- Yu, P.; Gao, R.; Zhang, D.; Liu, Z.-P. Predicting coastal algal blooms with environmental factors by machine learning methods. Ecol. Indic. 2021, 123, 107334. [Google Scholar] [CrossRef]
- Amorim, F.; Rick, J.; Lohmann, G.; Wiltshire, K. Evaluation of Machine Learning Predictions of a Highly Resolved Time Series of Chlorophyll-a Concentration. Appl. Sci. 2021, 11, 7208. [Google Scholar] [CrossRef]
- Baek, Y.M.; Park, R.S. Missing Data Analysis Using R; Hannara Academy Press: Seoul, Korea, 2021; pp. 110–114. [Google Scholar]
- Rubin, D.B. An overview of multiple imputation. In Proceedings of the Survey Research Methods Section of the American Statistical Association, Princeton, NJ, USA, August 1998; Citeseer. pp. 79–84. [Google Scholar]
- Zhang, Z. Multiple imputation with multivariate imputation by chained equation (MICE) package. Ann. Transl. Med. 2016, 4, 30. [Google Scholar] [CrossRef]
- Yun, S.C. Imputation of missing values. J. Prev. Med. Public Health 2004, 37, 209–211. [Google Scholar]
- Alruhaymi, A.Z.; Kim, C.J. Why Can Multiple Imputations and How (MICE) Algorithm Work? Open J. Stat. 2021, 11, 759–777. [Google Scholar] [CrossRef]
- Kim, J.H. A Study on the Multiple Imputation of Missing Values: Focus on Fine Dust Data. Soc. Converg. Knowl. Trans. 2020, 8, 149–156. [Google Scholar] [CrossRef]
- Murray, J.S. Multiple Imputation: A Review of Practical and Theoretical Findings. Stat. Sci. 2018, 33, 142–159. [Google Scholar] [CrossRef] [Green Version]
- Flexible Imputation of Missing Data (Second Edition). Available online: https://stefvanbuuren.name/fimd/ (accessed on 5 March 2022).
- White, I.R.; Royston, P.; Wood, A.M. Multiple imputation using chained equations: Issues and guidance for practice. Stat. Med. 2011, 30, 377–399. [Google Scholar] [CrossRef] [PubMed]
- Van Buuren, S.; Groothuis-Oudshoorn, K. mice: Multivariate imputation by chained equations in R. J. Stat. Softw. 2011, 45, 1–67. [Google Scholar] [CrossRef] [Green Version]
- Azur, M.J.; Stuart, E.A.; Frangakis, C.; Leaf, P.J. Multiple imputation by chained equations: What is it and how does it work? Int. J. Methods Psychiatr. Res. 2011, 20, 40–49. [Google Scholar] [CrossRef]
- Iterative Imputation for Missing Values in Machine Learning. Available online: https://machinelearningmastery.com/iterative-imputation-for-missing-values-in-machine-learning/ (accessed on 7 March 2022).
- Noh, J.H. Machine Learning Models and Missing Data Imputation Methods in Predicting the Progression of IgA Nephropathy. Master’s Thesis, The Graduate School Seoul National University, Seoul, Korea, February 2015. [Google Scholar]
- Kang, B.K.; Park, J.S. Effect of input variable characteristics on the performance of an ensemble machine learning model for algal bloom prediction. J. Korean Soc. Water Wastewater 2021, 35, 417–424. [Google Scholar] [CrossRef]
- Kim, J.H.; Shin, J.-K.; Lee, H.; Lee, D.H.; Kang, J.-H.; Cho, K.H.; Lee, Y.-G.; Chon, K.; Baek, S.-S.; Park, Y. Improving the performance of machine learning models for early warning of harmful algal blooms using an adaptive synthetic sampling method. Water Res. 2021, 207, 117821. [Google Scholar] [CrossRef]
- Kim, Y.N.; Yoo, J.K.; Yeo, J.W.; Kho, B.S.; Hwang, I.S. History and Status of the National Marine Ecosystem Monitoring Program in Korea. Sea J. Korean Soc. Oceanogr. 2019, 24, 49–53. [Google Scholar]
- Korea Marine Environment Management Corporation (KOEM). Available online: http://koem.or.kr/ (accessed on 7 March 2022).
- Marine Environment Information Portal (MEIS). Available online: http://meis.go.kr/ (accessed on 7 March 2022).
- Package ‘Mice’. Available online: https://cran.r-project.org/web/packages/mice/mice.pdf (accessed on 7 March 2022).
- Rincy, T.N.; Gupta, R. Ensemble Learning Techniques and its Efficiency in Machine Learning: A Survey. In Proceedings of the 2nd International Conference on Data, Engineering and Applications (IDEA), Bhopal, India, 28–29 February 2020; pp. 1–6. [Google Scholar] [CrossRef]
- Schapire, R.E. The Boosting Approach to Machine Learning: An Overview. In Nonlinear Estimation and Classification; Lecture Notes in Statistics; Denison, D.D., Hansen, M.H., Holmes, C.C., Mallick, B., Yu, B., Eds.; Springer: New York, NY, USA, 2003; Volume 171, pp. 149–171. [Google Scholar]
- Yang, X.; Wang, Y.; Byrne, R.; Schneider, G.; Yang, S. Concepts of Artificial Intelligence for Computer-Assisted Drug Discovery. Chem. Rev. 2019, 119, 10520–10594. [Google Scholar] [CrossRef] [Green Version]
- Chung, D.H.; Yun, J.S.; Yang, S.M. Machine Learning for Predicting Entrepreneurial Innovativeness. Asia-Pac. J. Bus. Ventur. Entrep. 2021, 16, 73–86. [Google Scholar]
- Yuvaraj, P.; Murthy, A.R.; Iyer, N.R.; Sekar, S.; Samui, P. Support vector regression based models to predict fracture characteristics of high strength and ultra high strength concrete beams. Eng. Fract. Mech. 2013, 98, 29–43. [Google Scholar] [CrossRef]
- Nti, I.K.; Adekoya, A.F.; Weyori, B.A. A comprehensive evaluation of ensemble learning for stock-market prediction. J. Big Data 2020, 7, 1–40. [Google Scholar] [CrossRef]
- Mitchell, R.; Frank, E. Accelerating the XGBoost algorithm using GPU computing. PeerJ Comput. Sci. 2017, 3, e127–e163. [Google Scholar] [CrossRef]
- Choi, S.; Kim, C. The Empirical Evaluation of Machine Learning Models Predicting Round-Trip Time in Cellular Network. In Proceedings of the 2021 International Conference on Information and Communication Technology Convergence (ICTC), Jeju Island, Korea, 20–22 October 2021; pp. 1374–1376. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A scalable tree boosting system. In Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data mining, San Francisco, CA, USA, 13–17 August 2016. [Google Scholar]
- Chicco, D.; Warrens, M.J.; Jurman, G. The coefficient of determination R-squared is more informative than SMAPE, MAE, MAPE, MSE and RMSE in regression analysis evaluation. PeerJ Comput. Sci. 2021, 7, e623. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Rahman, M.; Haque, M.; Hasan, M.W.; Alam, M.M. Performance evaluation of SVM and GBM in predicting compressive and splitting tensile strength of concrete prepared with ceramic waste and nylon fiber. J. King Saud Univ. Eng. Sci. 2021, in press. [Google Scholar] [CrossRef]
- Kooh, M.R.R.; Thotagamuge, R.; Chau, Y.-F.C.; Mahadi, A.H.; Lim, C.M. Machine learning approaches to predict adsorption capacity of Azolla pinnata in the removal of methylene blue. J. Taiwan Inst. Chem. Eng. 2022, 132, 104134. [Google Scholar] [CrossRef]
- Chhabra, G.; Vashisht, V.; Ranjan, J. A Comparison of Multiple Imputation Methods for Data with Missing Values. Indian J. Sci. Technol. 2017, 10, 1–7. [Google Scholar] [CrossRef]
- Jadhav, A.; Pramod, D.; Ramanathan, K. Comparison of Performance of Data Imputation Methods for Numeric Dataset. Appl. Artif. Intell. 2019, 33, 913–933. [Google Scholar] [CrossRef]
- Kim, W.; Cho, W.; Choi, J.; Kim, J.; Park, C.; Choo, J. A Comparison of the Effects of Data Imputation Methods on Model Performance. In Proceedings of the 2019 21st International Conference on Advanced Communication Technology (ICACT), PyeongChang, Korea, 17–20 February 2019. [Google Scholar] [CrossRef]
- Amdevýren, H.; Demýr, N.; Kanik, A.; Keskýn, S. Use of principal component scores in multiple linear regression models for prediction of Chlorophyll-a in reservoirs. Ecol. Model. 2005, 181, 581–589. [Google Scholar] [CrossRef]
- Cho, K.H.; Kang, J.-H.; Ki, S.J.; Park, Y.; Cha, S.M.; Kim, J.H. Determination of the optimal parameters in regression models for the prediction of chlorophyll-a: A case study of the Yeongsan Reservoir, Korea. Sci. Total Environ. 2009, 407, 2536–2545. [Google Scholar] [CrossRef]
- National Institute of Fisheries Science (NIFS). Available online: https://www.nifs.go.kr/red/info_1.red (accessed on 2 April 2022).
- National Oceanic and Atmospheric Administration (NOAA). Available online: https://oceanservice.noaa.gov/facts/why_habs.html (accessed on 3 June 2022).
- Yi, H.-S.; Lee, B.; Park, S.; Kwak, K.-C.; An, K.-G. Prediction of short-term algal bloom using the M5P model-tree and extreme learning machine. Environ. Eng. Res. 2019, 24, 404–411. [Google Scholar] [CrossRef]


| Model Accuracy Metric | Formula Definition |
|---|---|
| Coefficient of determination (R-squared or R2) | |
| Mean absolute error (MAE) | |
| Root mean square error (RMSE) | |
| Spearman’s correlation coefficient (rs) |
| Feature | Unit | Min. | Max | 1st Qu. | Median | Mean | 3rd Qu. | Skewness | Kurtosis | CV |
|---|---|---|---|---|---|---|---|---|---|---|
| Transparency | m | 0.16 | 16 | 1.81 | 3 | 4.076 | 5.9 | 1.09 | 0.83 | 0.73 |
| Wtemp | °C | 9.22 | 30.28 | 15.94 | 20.25 | 20.42 | 24.74 | 0.05 | −1.22 | 0.24 |
| Salinity | psu | 16.34 | 34.86 | 31.65 | 32.22 | 32.21 | 33.12 | −2.94 | 22.92 | 0.05 |
| pH | pH | 7.17 | 8.41 | 7.98 | 8.075 | 8.059 | 8.15 | −1.23 | 4.54 | 0.02 |
| DO | mg/L | 3.53 | 12.31 | 7.11 | 7.89 | 7.811 | 8.49 | 0.09 | 0.76 | 0.13 |
| SPM | mg/L | 0.5 | 75.55 | 6.16 | 10.28 | 13.34 | 17.83 | 1.72 | 4.41 | 0.79 |
| PON | μM | 0.64 | 88.82 | 2.67 | 4.28 | 9.049 | 7.68 | 5.06 | 35.58 | 1.77 |
| POC | μM | 1.29 | 179.99 | 13.19 | 21.52 | 26.99 | 34.32 | 2.41 | 9.13 | 0.81 |
| DSi | μM | 0.03 | 59.23 | 2.85 | 5.81 | 7.09 | 8.88 | 2.81 | 12.66 | 0.91 |
| DIP | μM | 0 | 2.52 | 0.07 | 0.15 | 0.2139 | 0.3 | 3.37 | 21.08 | 1.1 |
| DIN | μM | 0.1 | 76.28 | 1.53 | 2.96 | 4.482 | 6.04 | 5.42 | 52.36 | 1.2 |
| NO2 | μM | 0 | 3.1 | 0.05 | 0.16 | 0.2706 | 0.33 | 3.3 | 14.91 | 1.33 |
| NO3 | μM | 0 | 30.58 | 0.46 | 1.22 | 2.535 | 3.5 | 3.36 | 17.89 | 1.33 |
| NH4 | μM | 0 | 17.56 | 0.39 | 1.25 | 1.59 | 2.08 | 3.53 | 19.16 | 1.18 |
| Chl-a | μg/L | 0.03 | 14.58 | 0.79 | 1.46 | 2.084 | 2.82 | 2.29 | 7.55 | 0.95 |
| Model | MAE | RMSE | Spearman’s Correlation | R2 | |
|---|---|---|---|---|---|
| Single | regression tree | 0.073 | 0.107 | 0.557 | 0.308 |
| SVR | 0.061 | 0.094 | 0.744 | 0.493 | |
| Ensemble | bagging | 0.069 | 0.099 | 0.658 | 0.413 |
| random forest | 0.063 | 0.093 | 0.731 | 0.500 | |
| GBM | 0.065 | 0.094 | 0.698 | 0.471 | |
| XGBoost | 0.062 | 0.090 | 0.720 | 0.520 | |
| Feature | Raw Value | Imputed Value | Year-Season | Station |
|---|---|---|---|---|
| Chl-a | 45.24 | 3.32 | 2015-spring | W26 |
| Salinity | 5.4 | 27.64 | 2016-spring | S45 |
| NO3 | 72.97 | 20.72 | 2016-spring | S45 |
| NH4 | 24.08 | 9.67 | 2019-spring | W59 |
| PON | 314.99 | 9.89 | 2017-summer | W50 |
| SPM | 286.5 | 9.35 | 2017-summer | W50 |
| Category | Features | References |
|---|---|---|
| Physical quality | water temperature, salinity, transparency, Secchi depth | [11,13,14,16,17,32,56] |
| Chemical quality | pH, conductivity, DO, BOD, COD, SS, TOC, silicate, phosphate, nitrogen, carbonate, TN, TP, TDN, TDP, NO3, NO2, NH3, NH4 | |
| Biological | chlorophyll-a, phytoplankton abundance, zooplankton abundance | |
| Meteorological | temperature, precipitation, wind speed, wind direction, sunlight radiation | |
| Hydrodynamic | inflow, outflow, water level, flux, water volume, discharge rate |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.-R.; Soh, H.Y.; Kwak, M.-T.; Han, S.-H. Machine Learning and Multiple Imputation Approach to Predict Chlorophyll-a Concentration in the Coastal Zone of Korea. Water 2022, 14, 1862. https://doi.org/10.3390/w14121862
Kim H-R, Soh HY, Kwak M-T, Han S-H. Machine Learning and Multiple Imputation Approach to Predict Chlorophyll-a Concentration in the Coastal Zone of Korea. Water. 2022; 14(12):1862. https://doi.org/10.3390/w14121862
Chicago/Turabian StyleKim, Hae-Ran, Ho Young Soh, Myeong-Taek Kwak, and Soon-Hee Han. 2022. "Machine Learning and Multiple Imputation Approach to Predict Chlorophyll-a Concentration in the Coastal Zone of Korea" Water 14, no. 12: 1862. https://doi.org/10.3390/w14121862
APA StyleKim, H.-R., Soh, H. Y., Kwak, M.-T., & Han, S.-H. (2022). Machine Learning and Multiple Imputation Approach to Predict Chlorophyll-a Concentration in the Coastal Zone of Korea. Water, 14(12), 1862. https://doi.org/10.3390/w14121862







