Moroccan Lagoon Microbiomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Water Sampling
2.2. 16S/18S Amplicon Sequencing
2.3. Preparation of Shotgun Libraries for Metagenomic Samples and Sequencing
2.4. Data Processing
| Filter Size | # Reads | Total Length (Mb) | Average Read Length (bp) | ||
|---|---|---|---|---|---|
| Marchica | 2014 | 0.22 μm | 132,322 | 80.1 | 269 |
| 2015 | 0.22 μm | 76,482 | 46.6 | 271 | |
| Oualidia | 2014 | 0.22 μm | 179,448 | 108.2 | 268 |
| 2015 | 0.22 μm | 146,886 | 89.2 | 270 | |
| Total | 535,138 | 324.1 | |||
| Samples | Input | Filtered | DenoisedF | DenoisedR | Merged | Nonchim |
|---|---|---|---|---|---|---|
| OSD2414 (Marchica) | 66,161 | 28,157 | 27,383 | 27,412 | 23,687 | 19,662 |
| OSD2415 (Marchica) | 38,241 | 18,570 | 18,124 | 18,198 | 16,957 | 16,355 |
| OSD4714 (Venice) | 44,664 | 21,137 | 20,199 | 20,354 | 17,763 | 15,331 |
| OSD4715 (Venice) | 60,996 | 31,535 | 30,156 | 30,205 | 26,166 | 23,088 |
| OSD8114 (Ria Formosa) | 53,394 | 22,538 | 22,059 | 21,935 | 18,895 | 16,205 |
| OSD8115 (Ria Formosa) | 90,023 | 45,355 | 44,652 | 44,584 | 41,038 | 37,323 |
| OSD9114 (Oualidia) | 89,724 | 43,757 | 41,329 | 41,707 | 34,329 | 29,064 |
| OSD9115 (Oualidia) | 73,443 | 33,878 | 32,946 | 32,968 | 28,967 | 23,401 |
| OSD9314 (ElJadida) | 41,323 | 20,930 | 19,889 | 20,052 | 16,612 | 13,680 |
| OSD9315 (ElJadida) | 62,923 | 31,790 | 31,088 | 31,164 | 27,840 | 25,242 |
| OSD9414 (Saidia Marina) | 61,932 | 28,805 | 27,557 | 27,544 | 22,009 | 18,190 |
| OSD9415 (Saidia Marina) | 48,501 | 20,674 | 20,169 | 20,156 | 18,152 | 16,940 |
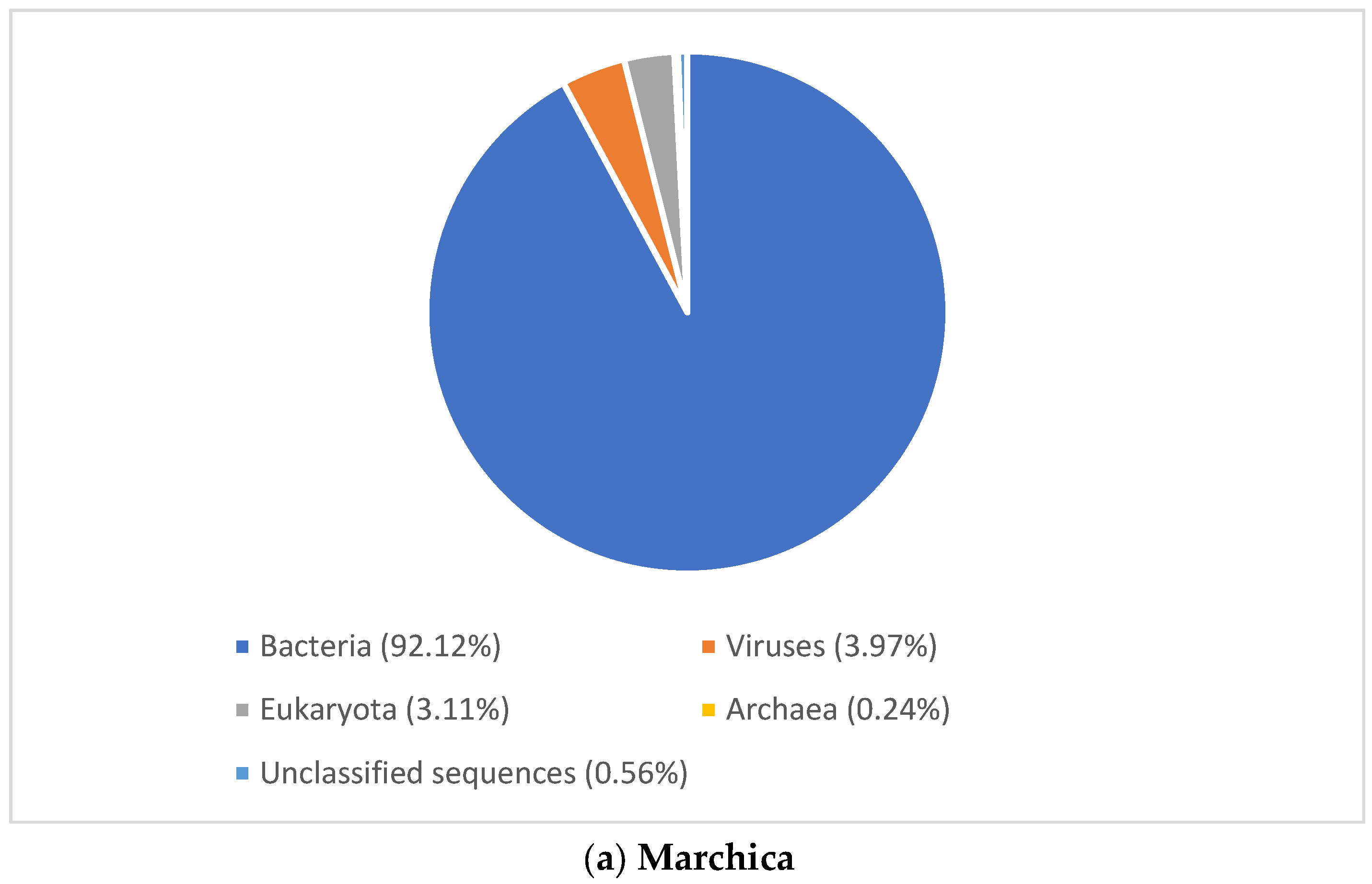
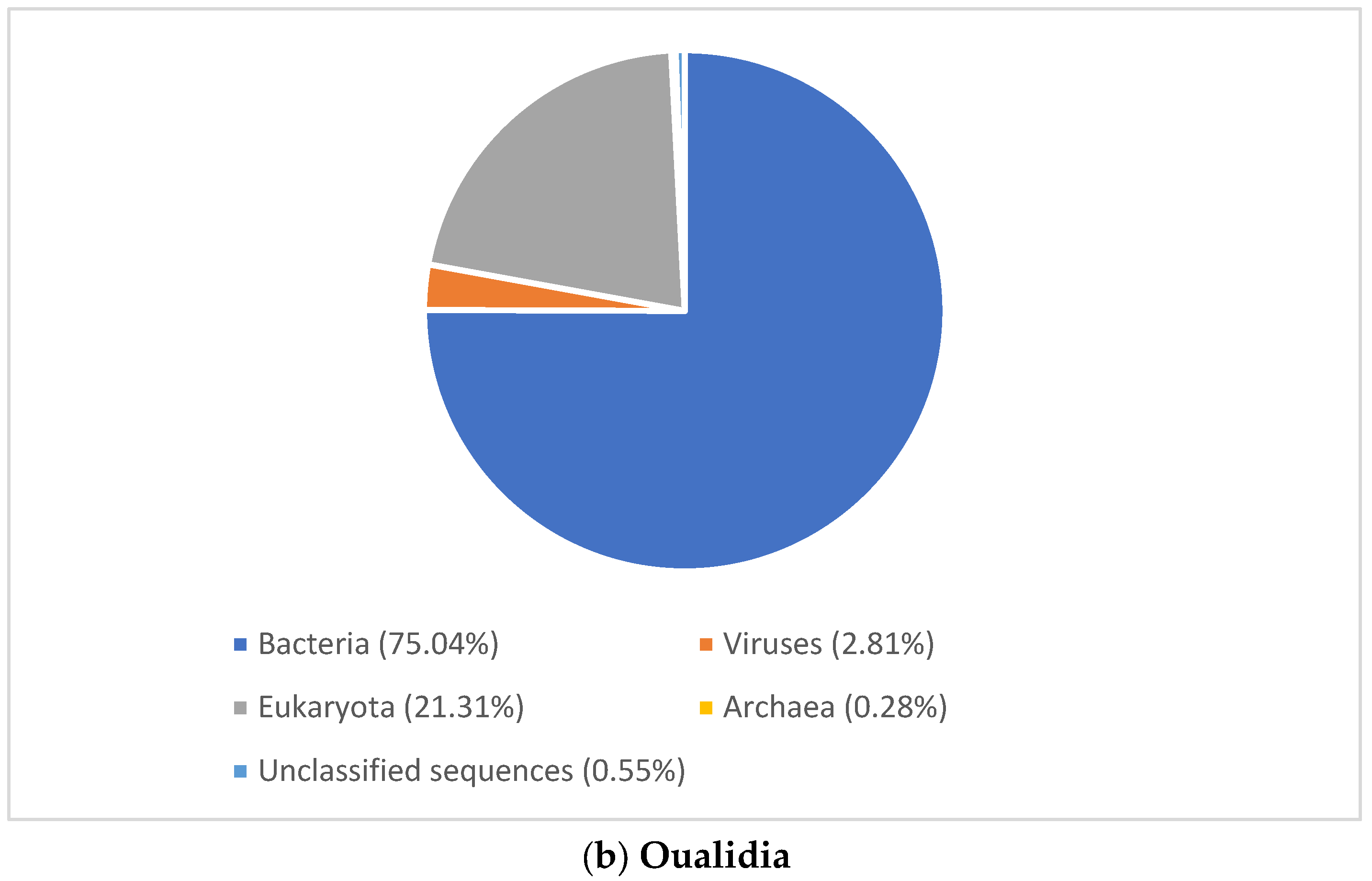
3. Results
3.1. Physicochemical Characteristics of the Lagoon Sampling Sites
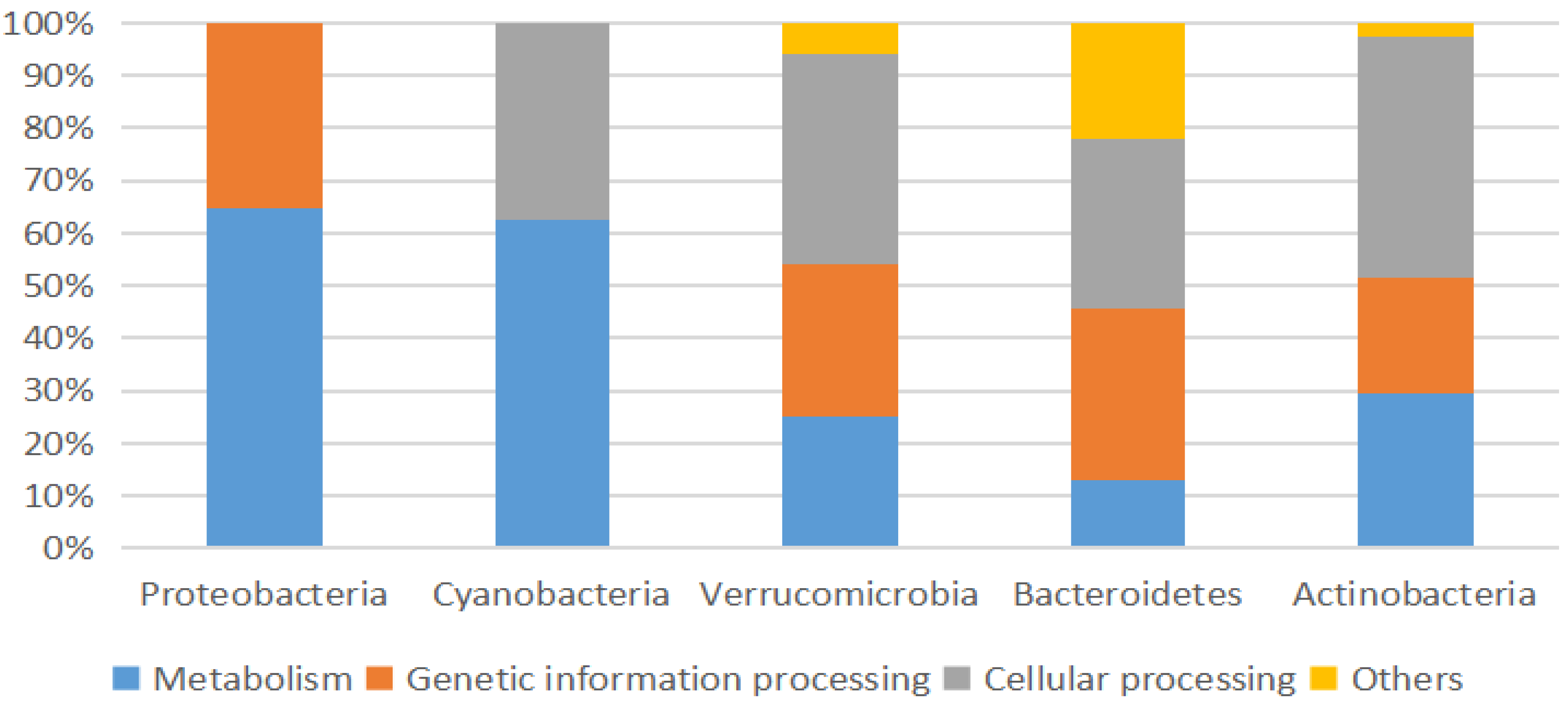
3.2. Microbial Community Structure

| Marchica 2014 | Oualidia 2014 | ||||
| Taxa | #Hits | % | Taxa | #Hits | % |
| Actinobacteria | 4793 | 7.42 | Actinobacteria | 11,670 | 13.98 |
| Bacteroidetes | 15,896 | 24.62 | Bacteroidetes | 19,660 | 23.55 |
| Cyanobacteria | 22,178 | 34.35 | Cyanobacteria | 9721 | 11.65 |
| Planctomycetes | 2190 | 3.39 | Planctomycetes | 1904 | 2.28 |
| Proteobacteria | 16,310 | 25.26 | Proteobacteria | 24,559 | 29.42 |
| Verrucomicrobia | 2712 | 4.20 | Verrucomicrobia | 13,202 | 15.82 |
| Euryarchaeota | 3 | 0 | Euryarchaeota | 19 | 0.02 |
| Marchica 2015 | Oualidia 2015 | ||||
| Taxa | #Hits | % | Taxa | #Hits | % |
| Actinobacteria | 4408 | 11.75 | Actinobacteria | 5613 | 7.80 |
| Bacteroidetes | 6173 | 16.46 | Bacteroidetes | 31,312 | 43.49 |
| Cyanobacteria | 4751 | 12.67 | Cyanobacteria | 384 | 0.53 |
| Planctomycetes | 1217 | 3.25 | Planctomycetes | 142 | 0.20 |
| Proteobacteria | 20,109 | 53.62 | Proteobacteria | 21,011 | 29.18 |
| Verrucomicrobia | 656 | 1.75 | Verrucomicrobia | 10,740 | 14.92 |
| Euryarchaeota | 10 | 0.03 | Euryarchaeota | 1 | 0 |
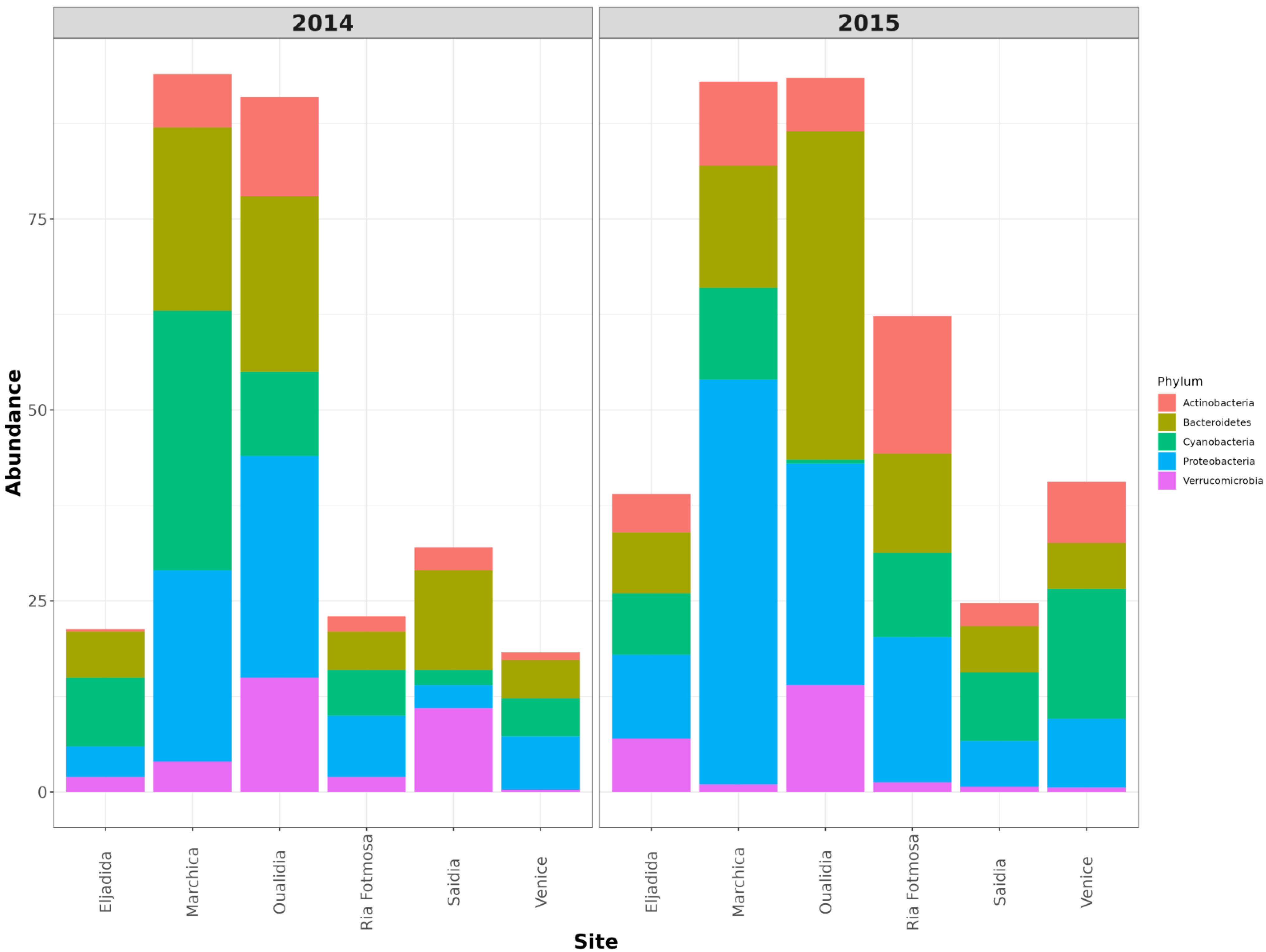
| Dataset | Observed Richness | ACE | Chao1 | Shannon | Simpson |
|---|---|---|---|---|---|
| Marchica Bv4v5 (OSD24, 2014) | 462 | 677.66 | 656.16 | 4.18 | 0.86 |
| Marchica Bv4v5 (OSD24, 2015) | 316 | 490.39 | 497.5 | 4.13 | 0.87 |
| Oualidia Bv4v5 (OSD91, 2014) | 1144 | 1591.09 | 1605.32 | 6.13 | 0.94 |
| Oualidia Bv4v5 (OSD91, 2015) | 704 | 1053.14 | 1022.78 | 4.59 | 0.89 |
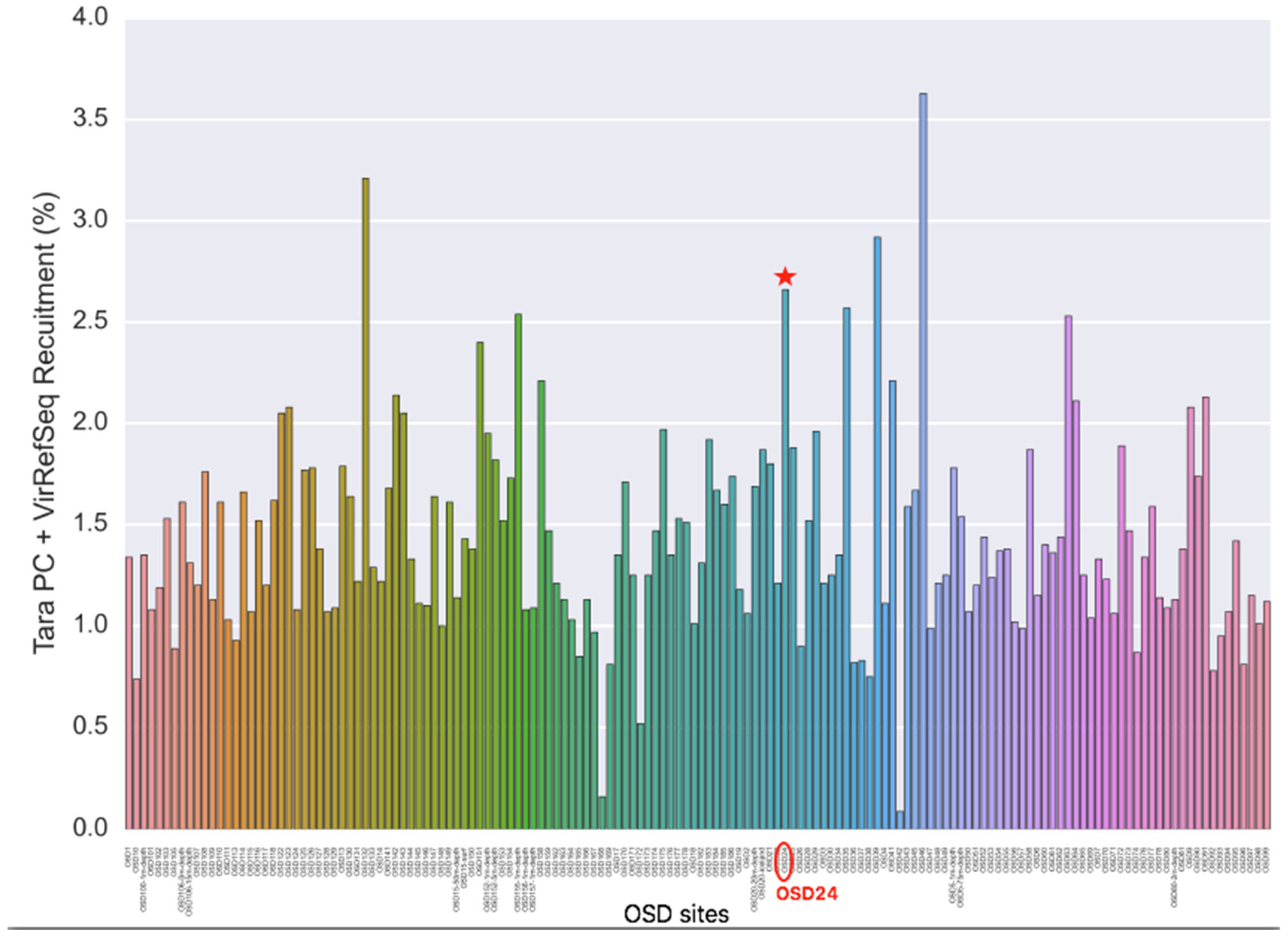
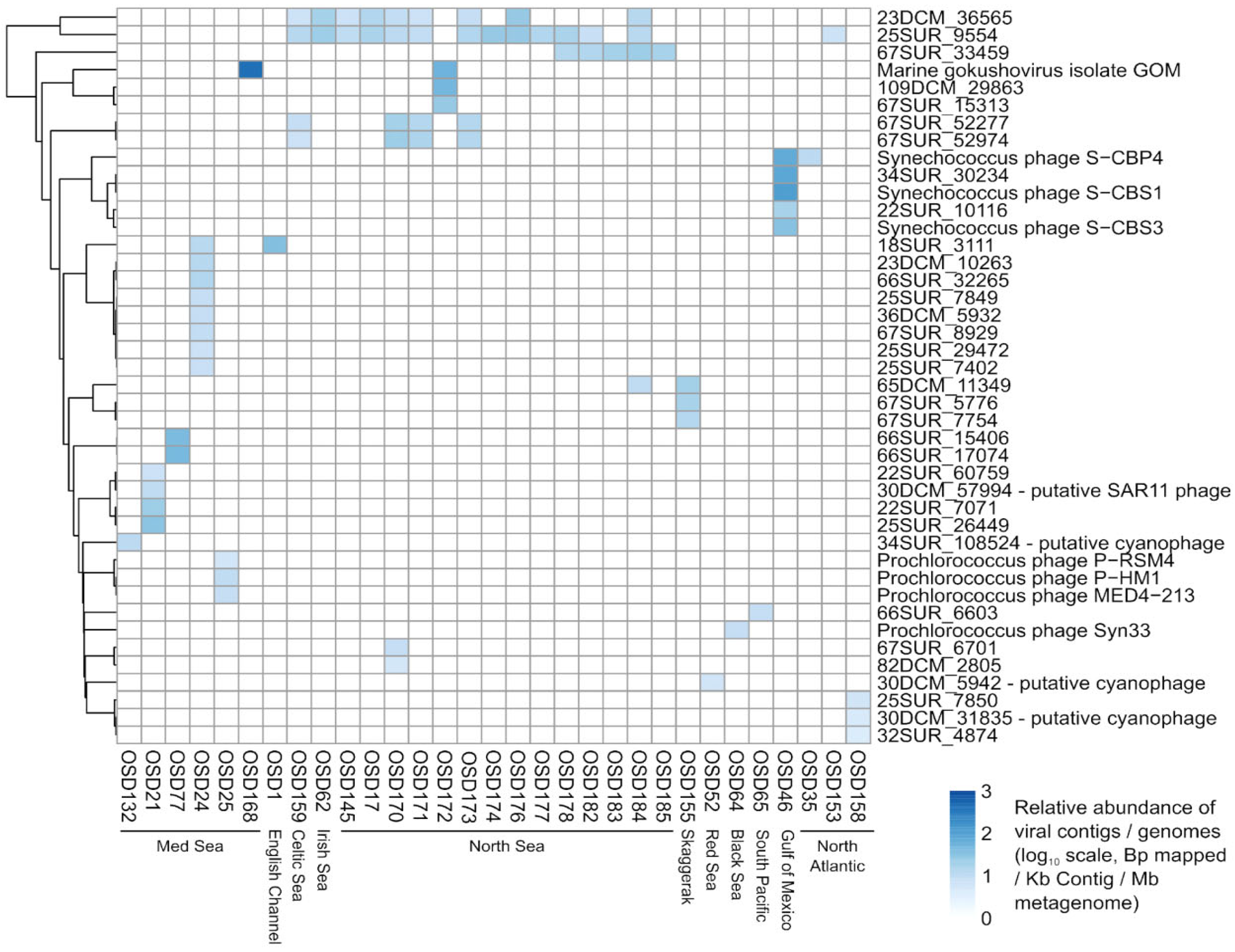
3.2.1. Viral Signature
3.2.2. Proteobacteria
3.2.3. Cyanobacteria
3.2.4. Bacteroidetes
3.2.5. Verrucomicrobia
3.2.6. Actinobacteria
3.2.7. Eukaryotes
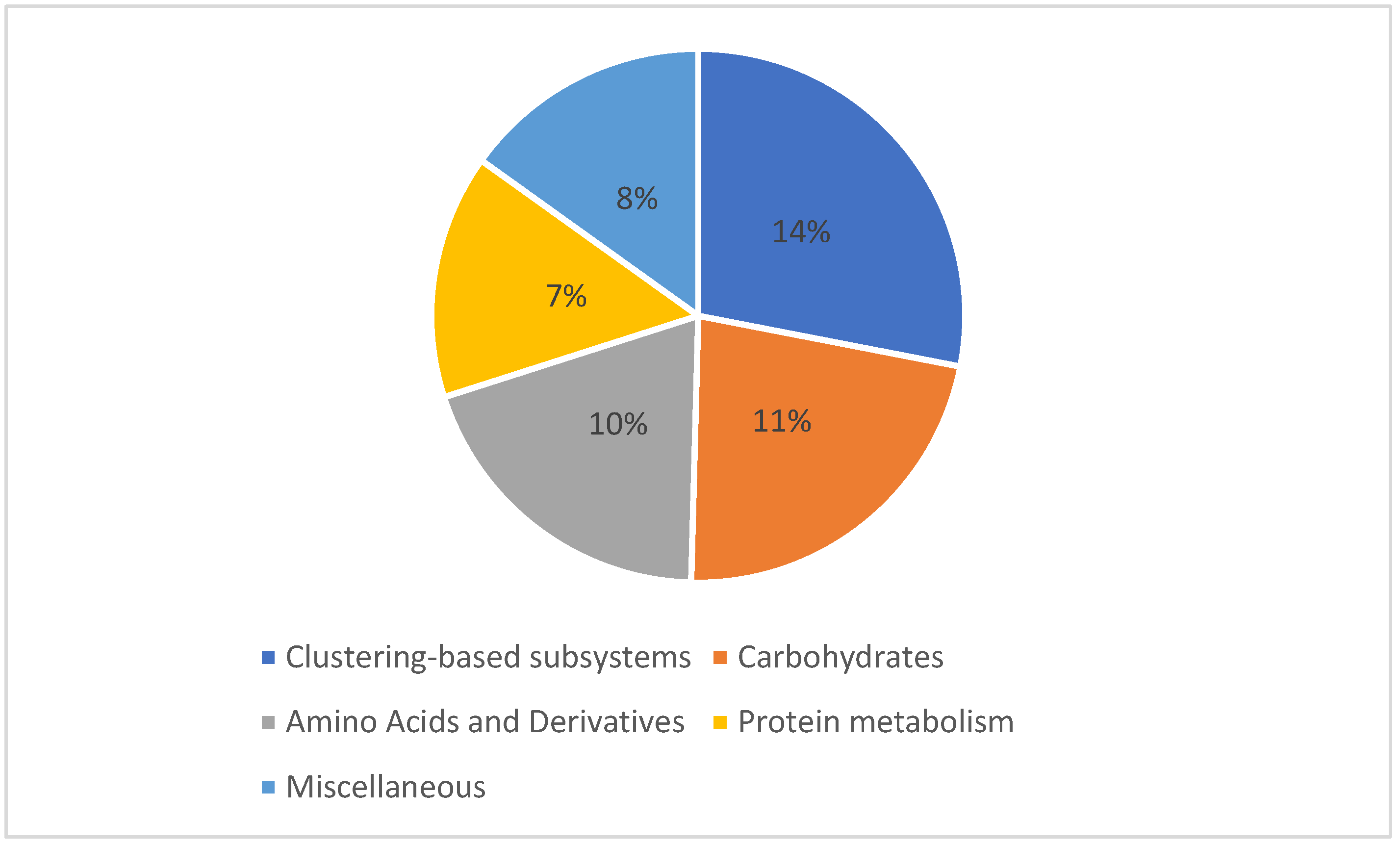
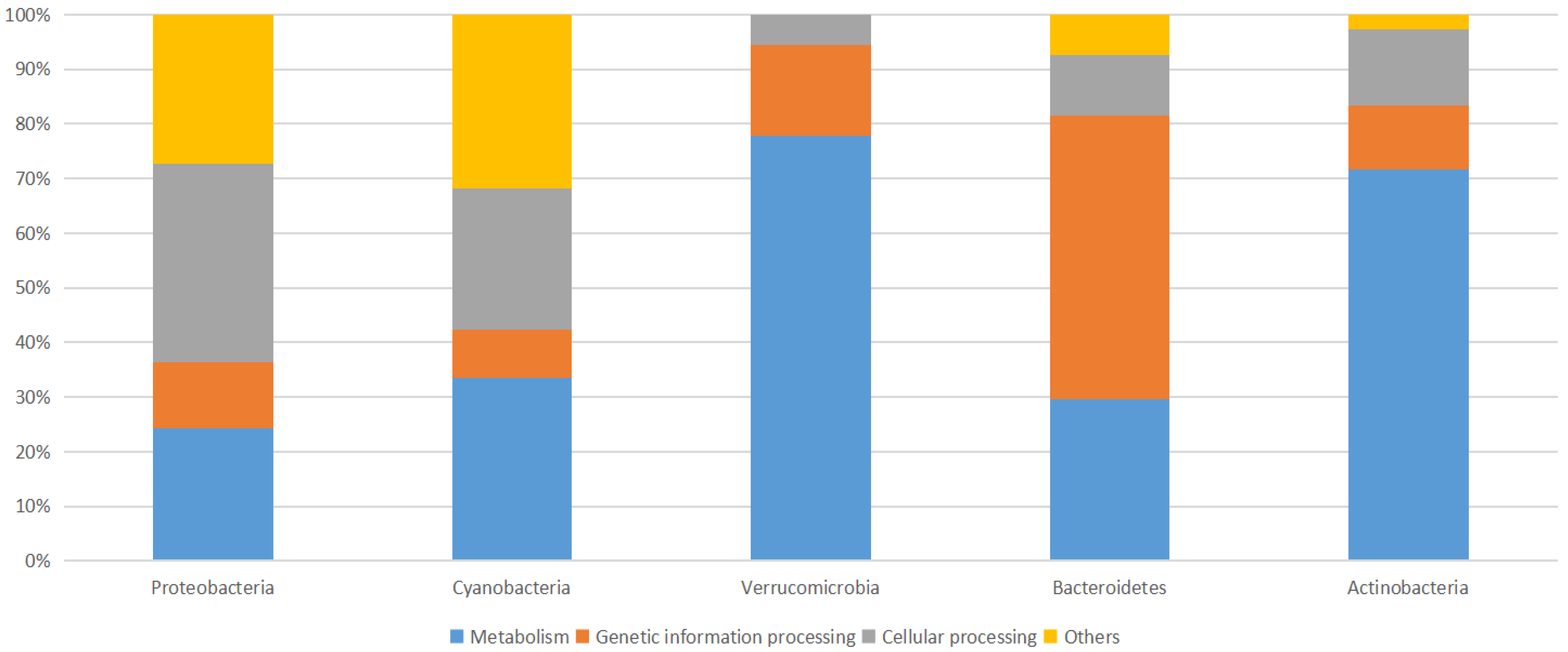
3.3. Microbial Communities Functional Analysis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sánchez-Baracaldo, P.; Bianchini, G.; Wilson, J.D.; Knoll, A.H. Cyanobacteria and Biogeochemical Cycles through Earth History. Trends Microbiol. 2022, 30, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Amaral-Zettler, L.; Artigas, L.F.; Baross, J.; LokaBharathi, P.A.; Boetius, A.; Chandramohan, D.; Herndl, G.; Kogure, K.; Neal, P.; Pedros-Alio, C.; et al. A Global Census of Marine Microbes; Wiley-Blackwell: Hoboken, NJ, USA, 2010. [Google Scholar]
- Venter, J.C.; Remington, K.; Heidelberg, J.F.; Halpern, A.L.; Rusch, D.; Eisen, J.A.; Wu, D.; Paulsen, I.; Nelson, K.E.; Nelson, W.; et al. Environmental Genome Shotgun Sequencing of the Sargasso Sea. Science 2004, 304, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Sunagawa, S.; Coelho, L.P.; Chaffron, S.; Kultima, J.R.; Labadie, K.; Salazar, G.; Djahanschiri, B.; Zeller, G.; Mende, D.R.; Alberti, A.; et al. Structure and Function of the Global Ocean Microbiome. Science 2015, 348, 1261359. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.; Lovell, C.R. Microbial Surface Colonization and Biofilm Development in Marine Environments. Microbiol. Mol. Biol. Rev. 2016, 80, 91–138. [Google Scholar] [CrossRef]
- Liu, S.; Ren, H.; Shen, L.; Lou, L.; Tian, G.; Zheng, P.; Hu, B. PH Levels Drive Bacterial Community Structure in Sediments of the Qiantang River as Determined by 454 Pyrosequencing. Front. Microbiol. 2015, 6, 285. [Google Scholar] [CrossRef]
- Ghai, R.; Hernandez, C.M.; Picazo, A.; Mizuno, C.M.; Ininbergs, K.; Díez, B.; Valas, R.; DuPont, C.L.; McMahon, K.D.; Camacho, A.; et al. Metagenomes of Mediterranean Coastal Lagoons. Sci. Rep. 2012, 2, 490. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Knight, R. Global Patterns in Bacterial Diversity. Proc. Natl. Acad. Sci. USA 2007, 104, 11436–11440. [Google Scholar] [CrossRef]
- Wang, S.; Yan, Z.; Wang, P.; Zheng, X.; Fan, J. Comparative Metagenomics Reveals the Microbial Diversity and Metabolic Potentials in the Sediments and Surrounding Seawaters of Qinhuangdao Mariculture Area. PLoS ONE 2020, 15, e0234128. [Google Scholar] [CrossRef]
- Ruiz, F.; Abad, M.; Olías, M.; Galán, E.; González, I.; Aguilá, E.; Hamoumi, N.; Pulido, I.; Cantano, M. The Present Environmental Scenario of the Nador Lagoon (Morocco). Environ. Res. 2006, 102, 215–229. [Google Scholar] [CrossRef]
- El Madani, F.; Chiaar, A.; Chafi, A. Phytoplankton Composition and Abundance Assessment in the Nador Lagoon (Mediterranean Coast of Morocco). Acta Bot. Croat. 2011, 70, 269–288. [Google Scholar] [CrossRef]
- Natij, L.; Damsiri, Z.; Khalil, K.; Loudiki, M.; Ettahiri, O.; Elkalay, K. Phytoplankton Abundance and Diversity in the Coastal Waters of Oualidia Lagoon, South Moroccan Atlantic in Relation to Environmental Variables. Int. J. Adv. Res. 2014, 2, 1022–1032. [Google Scholar]
- Kopf, A.; Bicak, M.; Kottmann, R.; Schnetzer, J.; Kostadinov, I.; Lehmann, K.; Fernandez-Guerra, A.; Jeanthon, C.; Rahav, E.; Ullrich, M.; et al. The Ocean Sampling Day Consortium. GigaScience 2015, 4, 27. [Google Scholar] [CrossRef] [PubMed]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every Base Matters: Assessing Small Subunit RRNA Primers for Marine Microbiomes with Mock Communities, Time Series and Global Field Samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Huse, S.M.; Mark Welch, D.B.; Voorhis, A.; Shipunova, A.; Morrison, H.G.; Eren, A.M.; Sogin, M.L. VAMPS: A Website for Visualization and Analysis of Microbial Population Structures. BMC Bioinform. 2014, 15, 41. [Google Scholar] [CrossRef]
- Huse, S.M.; Dethlefsen, L.; Huber, J.A.; Welch, D.M.; Relman, D.A.; Sogin, M.L. Exploring Microbial Diversity and Taxonomy Using SSU RRNA Hypervariable Tag Sequencing. PLoS Genet. 2008, 4, e1000255. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. MetaSPAdes: A New Versatile Metagenomic Assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef]
- Mikheenko, A.; Saveliev, V.; Gurevich, A. MetaQUAST: Evaluation of Metagenome Assemblies. Bioinformatics 2016, 32, 1088–1090. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Truong, D.T.; Franzosa, E.A.; Tickle, T.L.; Scholz, M.; Weingart, G.; Pasolli, E.; Tett, A.; Huttenhower, C.; Segata, N. MetaPhlAn2 for Enhanced Metagenomic Taxonomic Profiling. Nat. Methods 2015, 12, 902–903. [Google Scholar] [CrossRef] [PubMed]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; et al. The Metagenomics RAST Server—A Public Resource for the Automatic Phylogenetic and Functional Analysis of Metagenomes. BMC Bioinform. 2008, 9, 386. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Brum, J.R.; Ignacio-Espinoza, J.C.; Roux, S.; Doulcier, G.; Acinas, S.G.; Alberti, A.; Chaffron, S.; Cruaud, C.; de Vargas, C.; Gasol, J.M.; et al. Patterns and Ecological Drivers of Ocean Viral Communities. Science 2015, 348, 1261498. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Aknaf, A.; Akodad, M.; Layachi, M.; El Madani, F.; Jaddar, A.; Mesfioui, A.; Baghour, M. Study of the Spatial and Temporal Variation of Physical-Chemical Parameters Characterizing the Quality of Surface Waters of the Lagoon Marchica–North-East Morocco. J. Mater. Environ. Sci. 2017, 8, 3216–3225. [Google Scholar]
- Padan, E.; Bibi, E.; Ito, M.; Krulwich, T.A. Alkaline PH Homeostasis in Bacteria: New Insights. Biochim. Biophys. Acta 2005, 1717, 67–88. [Google Scholar] [CrossRef]
- Kortz, A.R.; Magurran, A.E. Increases in Local Richness (α-Diversity) Following Invasion Are Offset by Biotic Homogenization in a Biodiversity Hotspot. Biol. Lett. 2019, 15, 20190133. [Google Scholar] [CrossRef]
- Magurran, A.E. Measuring Biological Diversity; John Wiley & Sons: Hoboken, NJ, USA, 2003. [Google Scholar]
- Simpson, E.H. Measurement of Diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Kim, B.-R.; Shin, J.; Guevarra, R.; Lee, J.H.; Kim, D.W.; Seol, K.-H.; Lee, J.-H.; Kim, H.B.; Isaacson, R. Deciphering Diversity Indices for a Better Understanding of Microbial Communities. J. Microbiol. Biotechnol. 2017, 27, 2089–2093. [Google Scholar] [CrossRef]
- Yilmaz, P.; Yarza, P.; Rapp, J.Z.; Glöckner, F.O. Expanding the World of Marine Bacterial and Archaeal Clades. Front. Microbiol. 2016, 6, 1524. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S. The Phylogeny of Proteobacteria: Relationships to Other Eubacterial Phyla and Eukaryotes. FEMS Microbiol. Rev. 2000, 24, 367–402. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.; Lovell, C.R. Bacterial Primary Colonization and Early Succession on Surfaces in Marine Waters as Determined by Amplified RRNA Gene Restriction Analysis and Sequence Analysis of 16S RRNA Genes. Appl. Environ. Microbiol. 2000, 66, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Yang, J.; Liu, L.; Zhang, W.; Amalfitano, S. Bacterioplankton Community Shifts Associated with Epipelagic and Mesopelagic Waters in the Southern Ocean. Sci. Rep. 2015, 5, 12897. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Steele, J.A.; Caporaso, J.G.; Steinbrück, L.; Reeder, J.; Temperton, B.; Huse, S.; McHardy, A.C.; Knight, R.; Joint, I.; et al. Defining Seasonal Marine Microbial Community Dynamics. ISME J. 2012, 6, 298–308. [Google Scholar] [CrossRef]
- Hartsock, A.; Shapleigh, J.P. Physiological Roles for Two Periplasmic Nitrate Reductases in Rhodobacter Sphaeroides 2.4.3 (ATCC 17025). J. Bacteriol. 2011, 193, 6483–6489. [Google Scholar] [CrossRef]
- Vincent, P.; Pignet, P.; Talmont, F.; Bozzi, L.; Fournet, B.; Guezennec, J.; Jeanthon, C.; Prieur, D. Production and Characterization of an Exopolysaccharide Excreted by a Deep-Sea Hydrothermal Vent Bacterium Isolated from the Polychaete Annelid Alvinella Pompejana. Appl. Environ. Microbiol. 1994, 60, 4134–4141. [Google Scholar] [CrossRef]
- Morris, R.M.; Rappé, M.S.; Connon, S.A.; Vergin, K.L.; Siebold, W.A.; Carlson, C.A.; Giovannoni, S.J. SAR11 Clade Dominates Ocean Surface Bacterioplankton Communities. Nature 2002, 420, 806–810. [Google Scholar] [CrossRef]
- Solano-Gallego, L.; Kidd, L.; Trotta, M.; Marco, M.D.; Caldin, M.; Furlanello, T.; Breitschwerdt, E.B. Febrile Illness Associated with Rickettsia Conorii Infection in Dogs from Sicily. Emerg. Infect. Dis. 2006, 12, 1985–1988. [Google Scholar] [CrossRef]
- Introduction to the Cyanobacteria. Available online: https://ucmp.berkeley.edu/bacteria/cyanointro.html (accessed on 27 February 2022).
- Bullerjahn, G.S.; Post, A.F. Physiology and Molecular Biology of Aquatic Cyanobacteria. Front. Microbiol. 2014, 5, 359. [Google Scholar] [CrossRef]
- Cooke, G.D.; Welch, E.B.; Peterson, S.; Nichols, S.A. Restoration and Management of Lakes and Reservoirs, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2005; ISBN 978-0-429-18923-4. [Google Scholar]
- Fernández-Gómez, B.; Richter, M.; Schüler, M.; Pinhassi, J.; Acinas, S.G.; González, J.M.; Pedrós-Alió, C. Ecology of Marine Bacteroidetes: A Comparative Genomics Approach. ISME J. 2013, 7, 1026–1037. [Google Scholar] [CrossRef] [PubMed]
- GLOBAL: Marine Census Publishes Latest Findings. Available online: https://www.universityworldnews.com/post.php?story=20100903170738623 (accessed on 27 February 2022).
- Gómez-Pereira, P.R.; Fuchs, B.M.; Alonso, C.; Oliver, M.J.; van Beusekom, J.E.E.; Amann, R. Distinct Flavobacterial Communities in Contrasting Water Masses of the North Atlantic Ocean. ISME J. 2010, 4, 472–487. [Google Scholar] [CrossRef] [PubMed]
- Mann, A.J.; Hahnke, R.L.; Huang, S.; Werner, J.; Xing, P.; Barbeyron, T.; Huettel, B.; Stüber, K.; Reinhardt, R.; Harder, J.; et al. The Genome of the Alga-Associated Marine Flavobacterium Formosa Agariphila KMM 3901T Reveals a Broad Potential for Degradation of Algal Polysaccharides. Appl. Environ. Microbiol. 2013, 79, 6813–6822. [Google Scholar] [CrossRef]
- Cardman, Z.; Arnosti, C.; Durbin, A.; Ziervogel, K.; Cox, C.; Steen, A.D.; Teske, A. Verrucomicrobia Are Candidates for Polysaccharide-Degrading Bacterioplankton in an Arctic Fjord of Svalbard. Appl. Environ. Microbiol. 2014, 80, 3749–3756. [Google Scholar] [CrossRef]
- Bünger, W.; Jiang, X.; Müller, J.; Hurek, T.; Reinhold-Hurek, B. Novel Cultivated Endophytic Verrucomicrobia Reveal Deep-Rooting Traits of Bacteria to Associate with Plants. Sci. Rep. 2020, 10, 8692. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, C.; Samudrala, R.; Anderson, I.; Hedlund, B.P.; Petroni, G.; Michailova, N.; Pinel, N.; Overbeek, R.; Rosati, G.; Staley, J.T. Genes for the Cytoskeletal Protein Tubulin in the Bacterial Genus Prosthecobacter. Proc. Natl. Acad. Sci. USA 2002, 99, 17049–17054. [Google Scholar] [CrossRef] [PubMed]
- Dunfield, P.F.; Yuryev, A.; Senin, P.; Smirnova, A.V.; Stott, M.B.; Hou, S.; Ly, B.; Saw, J.H.; Zhou, Z.; Ren, Y.; et al. Methane Oxidation by an Extremely Acidophilic Bacterium of the Phylum Verrucomicrobia. Nature 2007, 450, 879–882. [Google Scholar] [CrossRef]
- Hou, S.; Makarova, K.S.; Saw, J.H.W.; Senin, P.; Ly, B.V.; Zhou, Z.; Ren, Y.; Wang, J.; Galperin, M.Y.; Omelchenko, M.V.; et al. Complete Genome Sequence of the Extremely Acidophilic Methanotroph Isolate V4, Methylacidiphilum Infernorum, a Representative of the Bacterial Phylum Verrucomicrobia. Biol. Direct 2008, 3, 26. [Google Scholar] [CrossRef]
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of Actinobacteria: Tracing the Evolutionary History of an Ancient Phylum. Microbiol. Mol. Biol. Rev. 2007, 71, 495–548. [Google Scholar] [CrossRef]
- Shivlata, L.; Satyanarayana, T. Thermophilic and Alkaliphilic Actinobacteria: Biology and Potential Applications. Front. Microbiol. 2015, 6, 1014. [Google Scholar] [CrossRef]
- Pinto, I.; Calisto, R.; Serra, C.R.; Lage, O.M.; Antunes, S.C. Bacterioplankton Community as a Biological Element for Reservoirs Water Quality Assessment. Water 2021, 13, 2836. [Google Scholar] [CrossRef]
- Pinheiro, R.O.; de Souza Salles, J.; Sarno, E.N.; Sampaio, E.P. Mycobacterium Leprae-Host-Cell Interactions and Genetic Determinants in Leprosy: An Overview. Future Microbiol. 2011, 6, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Hewson, I.; Paerl, R.W.; Tripp, H.J.; Zehr, J.P.; Karl, D.M. Metagenomic Potential of Microbial Assemblages in the Surface Waters of the Central Pacific Ocean Tracks Variability in Oceanic Habitat. Limnol. Oceanogr. 2009, 54, 1981–1994. [Google Scholar] [CrossRef]
- Schloerke, B.; Cook, D.; Larmarange, J.; Briatte, F.; Marbach, M.; Thoen, E.; Elberg, A.; Crowley, J. Extension to “ggplot2” [R Package GGally Version 2.0.0]. 2020. Available online: https://ggobi.github.io/ggally/index.html (accessed on 27 March 2022).
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A Communal Catalogue Reveals Earth’s Multiscale Microbial Diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef]
- Matoir, M.; Belabed, A.; Najih, M.; Kada, O.; Rezzoume, N. Surrounding influence on the Ecological state of the lagoon of Marchica. J. Mater. Environ. Sci. 2015, 1260–1265. [Google Scholar]
- Riouchi, O.; Madani, F.E.; Abadie, E.; Skalli, A.; Baghour, M. The Spatio-Temporal Evolution of the Genus Nitzschia Longissima at the Level of the Lagoon in Nador, Morocco. E3S Web Conf. 2021, 234, 00081. [Google Scholar] [CrossRef]
- Hassou, N.; Boujaber, N.; Oumaskour, K.; Lakhdar, F.; Benba, J.; Benhaddou, Z.; Assoobhei, O.; Etahiri, S. Nutrients Variation in the Coastal Waters of Oualidia Lagoon, Morocco. Relation to the Contribution of Watershed and Hydro-Chemical Characteristics. Int. J. Adv. Res. 2016, 4, 1697–1706. [Google Scholar] [CrossRef]
- Chagas, G.G.; Suzuki, M.S. Seasonal Hydrochemical Variation in a Tropical Coastal Lagoon (Açu Lagoon, Brazil). Braz. J. Biol. 2005, 65, 597–607. [Google Scholar] [CrossRef][Green Version]
- Damsiri, Z.; Natij, L.; Khalil, K.; Loudiki, M.; Rabouille, C.; Ettahiri, O.; Bougadir, B.; Elkalay, K. Spatio-Temporal Nutrients Variability in the Oualidia Lagoon (Atlantic Moroccan Coast). Int. J. Curr. Adv. Res. 2014, 2, 609–618. [Google Scholar]
- Aknaf, A.; Akodad, M.; Moumen, A.; Chekroun, K.B.; Elhamouti, C.; Bailal, A.; Baghour, M. Impact of the New Pass on the Eutrophication of the Lagoon Marchica: Study of the Two Sites Bou Areg and Mohandis. J. Mater. Environ. Sci. 2015, 6, 2939–2943. [Google Scholar]
- Giovannoni, S.J. SAR11 Bacteria: The Most Abundant Plankton in the Oceans. Annu. Rev. Mar. Sci. 2017, 9, 231–255. [Google Scholar] [CrossRef] [PubMed]
- MacConnachie, K.; Tishkowski, K. Boutonneuse Fever. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Zinger, L.; Amaral-Zettler, L.A.; Fuhrman, J.A.; Horner-Devine, M.C.; Huse, S.M.; Welch, D.B.M.; Martiny, J.B.H.; Sogin, M.; Boetius, A.; Ramette, A. Global Patterns of Bacterial Beta-Diversity in Seafloor and Seawater Ecosystems. PLoS ONE 2011, 6, e24570. [Google Scholar] [CrossRef] [PubMed]
- Juhmani, A.-S.; Vezzi, A.; Wahsha, M.; Buosi, A.; Pascale, F.D.; Schiavon, R.; Sfriso, A. Diversity and Dynamics of Seaweed Associated Microbial Communities Inhabiting the Lagoon of Venice. Microorganisms 2020, 8, 1657. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.-H.; Kang, I.; Yang, S.-J.; Cho, J.-C. Characterization of Spatial Distribution of the Bacterial Community in the South Sea of Korea. PLoS ONE 2017, 12, e0174159. [Google Scholar] [CrossRef]
- Schneider, K.; van der Werf, W.; Cendoya, M.; Mourits, M.; Navas-Cortés, J.A.; Vicent, A.; Oude Lansink, A. Impact of Xylella Fastidiosa Subspecies Pauca in European Olives. Proc. Natl. Acad. Sci. USA 2020, 117, 9250–9259. [Google Scholar] [CrossRef]
- Partensky, F.; Blanchot, J.; Vaulot, D. DifferentiaI Distribution and Ecology of Prochlorococcus and Synechococcus in Oceanic Waters: A Review. Bull. Inst. Océanographique Monaco 1999, 19, 457–475. [Google Scholar]
- Kim, Y.; Jeon, J.; Kwak, M.S.; Kim, G.H.; Koh, I.; Rho, M. Photosynthetic Functions of Synechococcus in the Ocean Microbiomes of Diverse Salinity and Seasons. PLoS ONE 2018, 13, e0190266. [Google Scholar] [CrossRef]
- Lürling, M.; Mello, M.M.E.; van Oosterhout, F.; de Senerpont Domis, L.; Marinho, M.M. Response of Natural Cyanobacteria and Algae Assemblages to a Nutrient Pulse and Elevated Temperature. Front. Microbiol. 2018, 9, 1851. [Google Scholar] [CrossRef]
- Brouwer, P.; Bräutigam, A.; Buijs, V.A.; Tazelaar, A.O.E.; van der Werf, A.; Schlüter, U.; Reichart, G.-J.; Bolger, A.; Usadel, B.; Weber, A.P.M.; et al. Metabolic Adaptation, a Specialized Leaf Organ Structure and Vascular Responses to Diurnal N2 Fixation by Nostoc Azollae Sustain the Astonishing Productivity of Azolla Ferns without Nitrogen Fertilizer. Front. Plant Sci. 2017, 8, 442. [Google Scholar] [CrossRef]
- Niestępski, S.; Harnisz, M.; Korzeniewska, E.; Osińska, A. Markers Specific to Bacteroides Fragilis Group Bacteria as Indicators of Anthropogenic Pollution of Surface Waters. Int. J. Environ. Res. Public Health 2020, 17, 7137. [Google Scholar] [CrossRef]
- Rusch, D.B.; Halpern, A.L.; Sutton, G.; Heidelberg, K.B.; Williamson, S.; Yooseph, S.; Wu, D.; Eisen, J.A.; Hoffman, J.M.; Remington, K.; et al. The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 2007, 5, e77. [Google Scholar] [CrossRef] [PubMed]
- Biers, E.J.; Sun, S.; Howard, E.C. Prokaryotic Genomes and Diversity in Surface Ocean Waters: Interrogating the Global Ocean Sampling Metagenome. Appl. Environ. Microbiol. 2009, 75, 2221–2229. [Google Scholar] [CrossRef] [PubMed]
- Signori, C.N.; Thomas, F.; Enrich-Prast, A.; Pollery, R.C.G.; Sievert, S.M. Microbial Diversity and Community Structure across Environmental Gradients in Bransfield Strait, Western Antarctic Peninsula. Front. Microbiol. 2014, 5, 647. [Google Scholar] [CrossRef] [PubMed]
- Luria, C.M.; Ducklow, H.W.; Amaral-Zettler, L.A. Marine Bacterial, Archaeal and Eukaryotic Diversity and Community Structure on the Continental Shelf of the Western Antarctic Peninsula. Aquat. Microb. Ecol. 2014, 73, 107–121. [Google Scholar] [CrossRef]
- Brown, M.V.; Philip, G.K.; Bunge, J.A.; Smith, M.C.; Bissett, A.; Lauro, F.M.; Fuhrman, J.A.; Donachie, S.P. Microbial Community Structure in the North Pacific Ocean. ISME J. 2009, 3, 1374–1386. [Google Scholar] [CrossRef]
- Gárate -Lizárraga, I.; Sevilla -Torres, G.; Álvarez -Añorve, M.; Aguirre -Bahena, F.; Violante -González, J.; Rojas -Herrera, A. First Record of a Red Tide Caused by Gyrodinium Instriatum (Dinophyceae: Gymnodiniales) in Bahía de Acapulco, Guerrero. CICIMAR Oceánides 2013, 28, 43–47. [Google Scholar] [CrossRef]
- Teo, C.L.; Jamaluddin, H.; Zain, N.A.M.; Idris, A. Biodiesel Production via Lipase Catalysed Transesterification of Microalgae Lipids from Tetraselmis Sp. Renew. Energy 2014, 68, 1–5. [Google Scholar] [CrossRef]
- Chappell, P.; Whitney, L.; Haddock, T.; Menden-Deuer, S.; Roy, E.; Wells, M.; Jenkins, B. Thalassiosira Spp. Community Composition Shifts in Response to Chemical and Physical Forcing in the Northeast Pacific Ocean. Front. Microbiol. 2013, 4, 273. [Google Scholar] [CrossRef]
- Scofield, V.; Jacques, S.M.S.; Guimarães, J.R.D.; Farjalla, V.F. Potential Changes in Bacterial Metabolism Associated with Increased Water Temperature and Nutrient Inputs in Tropical Humic Lagoons. Front. Microbiol. 2015, 6, 310. [Google Scholar] [CrossRef]
- Krause, E.; Wichels, A.; Giménez, L.; Lunau, M.; Schilhabel, M.B.; Gerdts, G. Small Changes in PH Have Direct Effects on Marine Bacterial Community Composition: A Microcosm Approach. PLoS ONE 2012, 7, e47035. [Google Scholar] [CrossRef]
- Singh, J.S.; Kumar, A.; Rai, A.N.; Singh, D.P. Cyanobacteria: A Precious Bio-Resource in Agriculture, Ecosystem, and Environmental Sustainability. Front. Microbiol. 2016, 7, 529. [Google Scholar] [CrossRef] [PubMed]
- Bajpai, V.K.; Shukla, S.; Kang, S.-M.; Hwang, S.K.; Song, X.; Huh, Y.S.; Han, Y.-K. Developments of Cyanobacteria for Nano-Marine Drugs: Relevance of Nanoformulations in Cancer Therapies. Mar. Drugs 2018, 16, 179. [Google Scholar] [CrossRef] [PubMed]
- Zadjelovic, V.; Chhun, A.; Quareshy, M.; Silvano, E.; Hernandez-Fernaud, J.R.; Aguilo-Ferretjans, M.M.; Bosch, R.; Dorador, C.; Gibson, M.I.; Christie-Oleza, J.A. Beyond Oil Degradation: Enzymatic Potential of Alcanivorax to Degrade Natural and Synthetic Polyesters. Environ. Microbiol. 2020, 22, 1356–1369. [Google Scholar] [CrossRef] [PubMed]
- Antoniou, E.; Fodelianakis, S.; Korkakaki, E.; Kalogerakis, N. Biosurfactant Production from Marine Hydrocarbon-Degrading Consortia and Pure Bacterial Strains Using Crude Oil as Carbon Source. Front. Microbiol. 2015, 6, 274. [Google Scholar] [CrossRef]
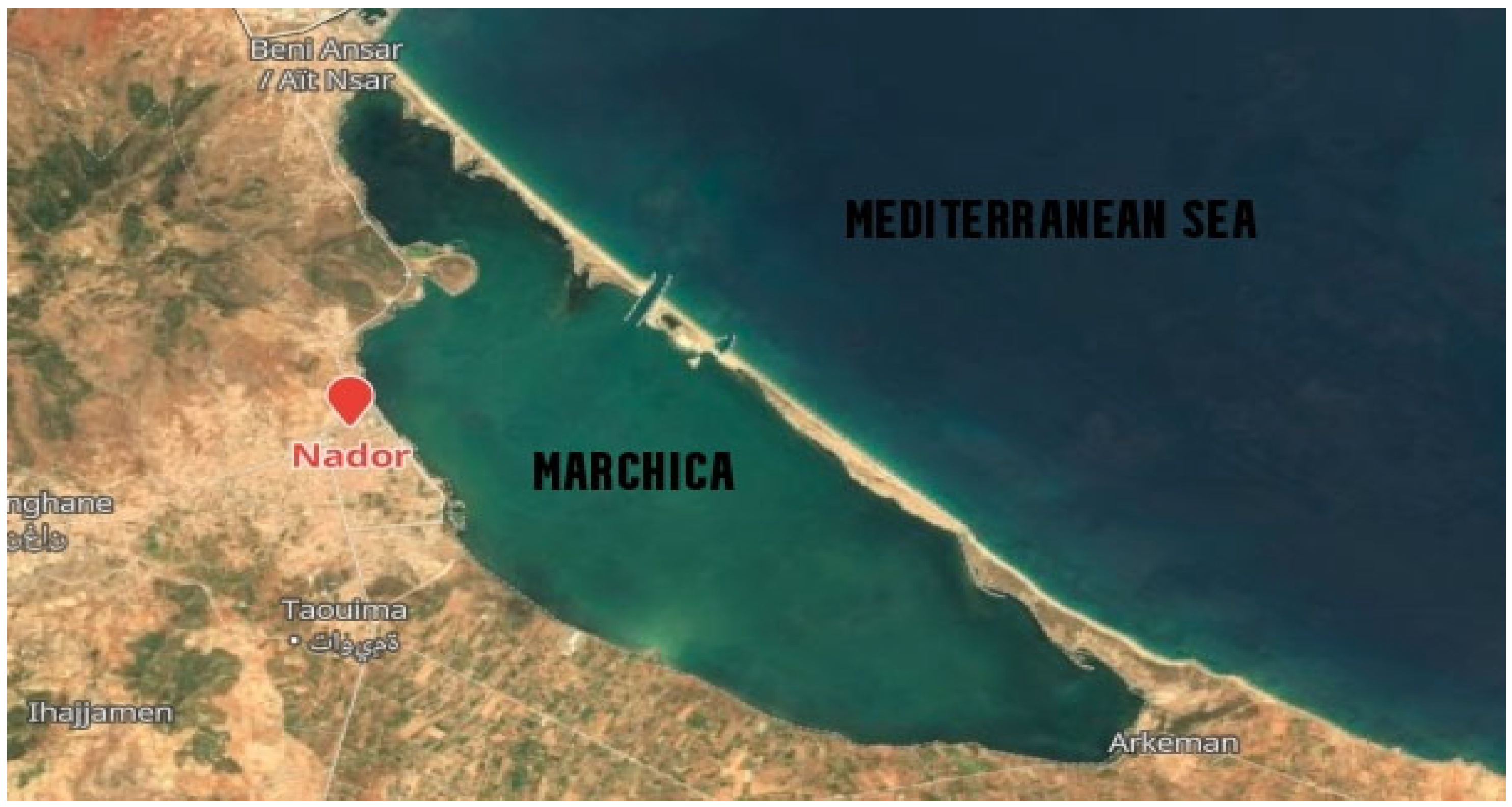


| Units | Marchica | Oualidia | ||||
|---|---|---|---|---|---|---|
| 2014 | 2015 | 2014 | 2015 | |||
| Parameters | Temperature | Celsius | 26.5 | 27.2 | 21 | 20 |
| Salinity | ppt | 35.98 | 35.96 | 27.24 | 29.41 | |
| Electrical conductivity | mS | 53.7 | 54.2 | 39.6 | 45.1 | |
| pH | <7.6 to 8.4> | 8.16 | 8.74 | 8.36 | 8.16 | |
| Dissolved Oxygen | mg/L | 8.33 | 8.50 | 7 | 7.5 | |
| Phosphate | mg/L | 0.03 | 0.04 | 0 | 0 | |
| Turbidity | NTU | 3.62 | 3.60 | 3 | 3.2 | |
| Nitrates | mg/L | 12.5 | 4.8 | 10.79 | 2.68 | |
| Nitrites | mg/L | <0.3 | 0.11 | <0.001 | <0.05 | |
| Oualidia Lagoon | Marchica Lagoon | ||||||
|---|---|---|---|---|---|---|---|
| Taxa | Genus | Species | Strain | #Hits (bp) | % | #Hits (bp) | % |
| Alphaproteobacteria | Anaplasma | marginale | Maries | 3,521,109 | 8.28 | 2,149,110 | 2.21 |
| Alphaproteobacteria | Rickettsia | conorii | Malish 7 | 3,079,827 | 7.24 | 1,016,387 | 10.49 |
| Alphaproteobacteria | Anaplasma | phagocytophilum | HZ | 1,683,742 | 3.96 | 3,623,917 | 3.74 |
| Gammaproteobacteria | Xylella | fastidiosa | Temecula1 | 1,501,655 | 3.53 | 3,857,299 | 3.98 |
| Gammaproteobacteria | Xenorhabdus | nematophila | 1_9_0_6_1 | 1,117,290 | 2.62 | 4,068,450 | 4.19 |
| Alphaproteobacteria | Orientia | tsutsugamushi | Boryong | 829,248 | 1.95 | 3,639,647 | 3.75 |
| Gammaproteobacteria | Escherichia | colia | UMN026 | 490,326 | 1.15 | 1,533,000 | 1.58 |
| Betaproteobacteria | Burkholderia | rhizoxinica | 4_5_4 | 430,554 | 1.01 | 1,112,721 | 1.15 |
| Alphaproteobacteria | Brucella | canis | 2_3_3_6_5 | 283,501 | 0.66 | 592,700 | 0.61 |
| Alphaproteobacteria | Erythrobacter | litoralis | HTCC2594 | 277,397 | 0.65 | 725,363 | 0.75 |
| Betaproteobacteria | Ralstonia | solanacearum | PSI07 | 241,328 | 0.56 | 614,227 | 0.63 |
| Alphaproteobacteria | Rhizobium | sp | N_G_R_2_3_4 | 217,740 | 0.51 | 305,658 | 0.31 |
| Gammaproteobacteria | Shigella | dysenteriae | Sd197 | 124,696 | 0.29 | 346,084 | 0.35 |
| Gammaproteobacteria | Marinobacter | aquaeolei | VT8 | 664,804 | 1.56 | 2,030,270 | 2.09 |
| Cyanobacteria | Prochlorococcus | marinus | 9_3_1_3 | 1,387,027 | 0.3 | 3,261,370 | 3.36 |
| Cyanobacteria | Nostoc | azollae | 0_7_0_8 | 1,340,033 | 3.15 | 5,328,515 | 5.5 |
| Cyanobacteria | Synechococcus | sp | C_C_9_6_0_5 | 406,828 | 0.95 | 3,636,034 | 3.75 |
| Cyanobacteria | Synechococcus | sp | JA_2_1_3 | 287,486 | 0.67 | 1,310,559 | 1.35 |
| Bacteroidetes | Candidatus -Amoebophilus | asiaticus | 5_a_2 | 2,748,208 | 6.46 | 10,026,340 | 10.34 |
| Bacteroidetes | Bacteroides | fragilis | 9_3_4_3 | 956,572 | 2.25 | 2,783,907 | 2.87 |
| Verrucomicrobia | Methylacidiphilum | infernorum | V4 | 655,181 | 1.54 | 1,576,020 | 1.62 |
| Verrucomicrobia | Akkermansia | muciniphila | 8_3_5 | 517,529 | 1.21 | 571,583 | 0.58 |
| Actinobacteria | Rothia | dentocariosa | 1_7_9_3_1 | 2,513,106 | 5.91 | 3,917,261 | 4.04 |
| Actinobacteria | Mycobacterium | leprae | TN | 1,072,610 | 2.52 | 1,013,872 | 1.04 |
| Actinobacteria | Tropheryma | whipplei | 2_7 | 1,010,398 | 2.37 | 1,929,685 | 1.99 |
| Actinobacteria | Rhodococcus | jostii | RHA1 | 199,761 | 0.47 | 277,060 | 0.28 |
| Actinobacteria | Catenulispora | acidiphila | 4_4_9_2_8 | 194,558 | 0.45 | 614,181 | 0.63 |
| Marchica Lagoon | Oualidia Lagoon | |||||||
|---|---|---|---|---|---|---|---|---|
| 2014 | 2015 | 2014 | 2015 | |||||
| Taxa | #Hits | % | #Hits | % | #Hits | % | #Hits | % |
| Dinophyta | 4233 | 37.7 | 22,505 | 62.09 | 7617 | 22.7 | 1723 | 5.4 |
| Ochrophyta | 2725 | 24.2 | 6128 | 16.9 | 10,848 | 32.4 | 13,302 | 42.2 |
| Chlorophyta | 777 | 6.9 | 939 | 2.5 | 1827 | 5.4 | 6806 | 21.6 |
| Marchica Lagoon | Oualidia Lagoon | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2014 | 2015 | 2014 | 2015 | ||||||||
| Taxa | Genus | #Hits % | #Hits % | Taxa | Genus | #Hits | %#Hits % | ||||
| Dinophyta | Gyrodinium | 2844 | 25.3 | 12,160 | 33.5 | Dinophyta | Pelagodinium | 4051 | 12.1 | 20 | 0.06 |
| Ochrophyta | Pseudo-Nitzschia | 1776 | 15.8 | 752 | 2 | Ochrophyta | Thalassiosira | 2862 | 8.5 | 7979 | 25.3 |
| Chlorophyta | Tetraselmis | 503 | 4.4 | 61 | 0.1 | Chlorophyta | Ostreococcus | 8 | 0.02 | 5240 | 16.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaouni, B.; Idrissi Azami, A.; Essayeh, S.; Arrafiqui, E.H.; Bailal, A.; Raoui, S.; Amzazi, S.; Twaddle, A.; El Hamouti, C.; Boukhatem, N.; et al. Moroccan Lagoon Microbiomes. Water 2022, 14, 1715. https://doi.org/10.3390/w14111715
Chaouni B, Idrissi Azami A, Essayeh S, Arrafiqui EH, Bailal A, Raoui S, Amzazi S, Twaddle A, El Hamouti C, Boukhatem N, et al. Moroccan Lagoon Microbiomes. Water. 2022; 14(11):1715. https://doi.org/10.3390/w14111715
Chicago/Turabian StyleChaouni, Bouchra, Abdellah Idrissi Azami, Soumaya Essayeh, El Houcine Arrafiqui, Abdelhakim Bailal, Sanae Raoui, Saaïd Amzazi, Alan Twaddle, Chahrazade El Hamouti, Noureddine Boukhatem, and et al. 2022. "Moroccan Lagoon Microbiomes" Water 14, no. 11: 1715. https://doi.org/10.3390/w14111715
APA StyleChaouni, B., Idrissi Azami, A., Essayeh, S., Arrafiqui, E. H., Bailal, A., Raoui, S., Amzazi, S., Twaddle, A., El Hamouti, C., Boukhatem, N., Timinouni, M., El Otmani, F., Chahboune, R., Barrijal, S., El Homani, A., Nejjari, C., Zaid, E. H., Hamamouch, N., Bakkali, F., ... Ghazal, H. (2022). Moroccan Lagoon Microbiomes. Water, 14(11), 1715. https://doi.org/10.3390/w14111715








