Abstract
In classical microbiology, developing a high-efficiency bacterial consortium is a great challenge for faster biodegradation of petroleum contaminants. In this study, a systematic experimental and mathematical procedure was adopted to establish a bacterial consortium for the effective biodegradation of heavy oil constituents. A total of 27 bacterial consortia were established as per orthogonal experiments, using 8 petroleum-degrading bacterial strains. These bacteria were closer phylogenetic relatives of Brevundimonas sp. Tibet-IX23 (Y1), Bacillus firmus YHSA15, B. cereus MTCC 9817, B. aquimaris AT8 (Y2, Y6 and Y7), Pseudomonas alcaligenes NBRC (Y3), Microbacterium oxydans CV8.4 (Y4), Rhodococcus erythropolis SBUG 2052 (Y5), and Planococcus sp. Tibet-IX21 (Y8), and were used in different combinations. Partial correlation analysis and a general linear model hereafter were applied to investigate interspecific relationships among different strains and consortia. The Y1 bacterial species showed a remarkable synergy, whereas Y3, Y4, and Y6 displayed a strong antagonism in all consortia. Inoculation ratios of different strains significantly influenced biodegradation. An optimal consortium was constructed with Y1, Y2, Y5, Y7, and Y8, which revealed maximum degradation of 11.238 mg/mL OD600 for oil contaminants. This study provides a line of evidence that a functional consortium can be established by mathematical models for improved bioremediation of petroleum-contaminated environment.
1. Introduction
A large quantity of petroleum-derived chemicals may enter the environment during crude oil exploration, processing, storage, and transportation. Every year, approximately 600,000 tons of crude oil spills damage aquatic and terrestrial ecosystems worldwide [1]. The adverse effects of petroleum contamination have been reported in terms of water and soil quality deterioration, a decline in biodiversity, and health complications [2,3].
The classical biodegradation approaches are commonly adopted schemes for the removal of petroleum contaminants from the environment, particularly in developing countries [4]. Existing knowledge suggests that biodegradation can be accelerated by the addition of nutrients or the introduction of specific microbial strains as competent degraders [5,6,7]. These microbial strains are initially cultivated in the laboratory but then applied to contaminated media for subsequent cleanup. These cultivable hydrocarbon-degrading bacteria usually belong to Pseudomonas, Rhodococcus, Acinetobacter, Bacillus, and Micrococcus genera [8,9,10,11]. Since crude oil is complex, containing different types of organic compounds, a single microbial strain is often incapable of degrading a wide range of contaminants. This is mainly caused by the low viability, adaptability, and biodegradability of the applied strain under diverse environmental conditions [12,13]. In natural settings, biodegradation involves a succession of species that carry out catabolic processes, leading to enhanced degradation of pollutants.
The primary aim of establishing a functional microbial consortium is to develop similar conditions in a given environment where multiple bacteria are exploited synergistically to promote biodegradation of petroleum contaminants. Principally, in a consortium, multiple strains have different physiological properties that offer diverse degradation abilities [14,15,16]. However, a bottleneck here is that not all of the strains exhibit positive interactions/synergy, but they could also compete with each other and act antagonistically [17,18]. Hence, an ideal consortium comprises synergistic strains that work together and improve biodegradation. To this end, the understanding of microbial interspecific relationships and interactions between strains and contaminants is an essential criterion [19].
Usually, a bacterial consortium is developed by mixing individual strains in an equal inoculation dosage [20,21]. Nevertheless, the initial inoculation dosage influences degradation efficiency [22], because it results in differentiated microbial communities and functions in the micro-ecosystem [23]. In the past, response surface methodology (RSM) has been used to optimize inoculation dosages and other operational conditions [24]. However, each variable in RSM is independent and yields little to no information about the interactions among strains [25].
In this study, a systematic, experimental, and mathematical procedure was adopted to establish and optimize a petroleum-degrading bacterial consortium. By using 8 petroleum-degrading strains, 27 bacterial consortia were developed in multiple combinations. Then, the interactions between bacterial strains were investigated, and a consortium with maximum degradation efficiency was developed. The proposed methodology and the constructed consortium showed the potential for enhancing the biological treatment of petroleum contamination in a conventional setup.
2. Materials and Methods
2.1. Bacteria Isolation and Culture Medium
Petroleum-degrading bacteria were isolated from activated sludge by enrichment cultivation. Firstly, sludge was sampled from a petroleum wastewater treatment plant in Liaohe Petrochemical Company, China (coordinates: 122.09998, 41.15971). Then, a sterilized inorganic salt (IS) medium was prepared as a minimal growth media [26]. Approximately, 0.1 g of Liaohe heavy oil was added into 100 mL of IS medium as a sole carbon source. Thereon, 100 mL of IS medium and 3 mL of sludge supernatant were added into a 250 mL Erlenmeyer flask, shaking for 5 days at an agitation speed of 180 rpm at 30 °C. Then, 5 mL of the mixture was transferred to a fresh sterilized IS medium for further cultivation. Repetitive sub-cultivation was performed four times in the IS medium by increasing oil contents from 0.1 g/L to 0.4 g/L. The culture, which was diluted with different gradients (10−1, 10−2, 10−3, 10−4, and 10−5), was inoculated onto lysogeny broth (LB) agar medium. Finally, phenotypically distinct colonies were obtained from the plates and stored in a refrigerator at −4 °C for biochemical and molecular characterization.
2.2. Identification of Isolates
Petroleum-degrading isolates were identified and characterized according to the standard protocols in the Identification Manual of Systematic Bacteriology [27]. At first, biochemical characterization was performed by doing Gram staining [28], methyl red test [29], starch hydrolysis test [30], Simmons’ citrate test [31], gelatin hydrolysis test [32], and catalase test [33]. Then, 16S rRNA gene analysis was performed for the species identification. Briefly, the total DNA of the isolates was extracted using the CTAB method [34]. 16S rRNA genes were amplified using the domain-specific bacteria primer, Bac27_F, and universal reverse primer Uni_1492R [35]. The PCR products were sequenced with the primer 27F at Shanghai Meiji Biomedical Technology Co., Ltd. (Shanghai, China). The 16S rRNA gene sequences were analyzed using the BLAST tool available at National Center for Biotechnology Information (NCBI) [36]. Phylogenetic trees were constructed based on partial 16S rRNA gene sequences (1000bp) by MEGA ver.5.0 (ASU, Phoenix, AZ, USA) with a neighbor-joining method [37].
2.3. Bacterial Growth Curves and Oil Degradation
The isolated strains were incubated at 30 °C for 3 days on a shaker (180 rpm). Their growth curves were obtained by measuring optical density (OD600 nm) on a UVmini-1240 UV–Visible spectrophotometer (Shimadzu, Tokyo, Japan) daily. Briefly, 10 mL strains with OD600 of 0.8 were inoculated into 100 mL of IS medium in 250 mL Erlenmeyer flasks, which were supplemented with 0.1 g of Liaohe heavy oil as a sole carbon source. Along with control treatment, these flasks were put on a shaker at 180 rpm for 7 days (30 °C). The residual oil was extracted from each sample, and the oil content was detected by an oil-content analyzer (OIL-480, China Invent Instrument, Beijing, China) as per standard method HJ 637-2018 [38]. Oil degradation efficiency (ƞ %) was calculated according to Equation (1).
where W1 and W0 are the oil contents in the control sample (added 5 mL deionized water) and treated sample (mg/L), respectively.
The oil removal amount μ (mg/mLOD600) was calculated by Equation (2).
where M0 is the weight of total oil in a medium (100 mg); ƞ is the oil degradation efficiency determined by Equation (1) (%); V and A are the inoculation volume (mL) and OD600 of each strain (0.8), respectively.
The SARA fractions (saturates, aromatics, resins, asphaltenes) of the oil were analyzed using an AcceleSep system (Agela Technologies, Tianjin, China) according to the standard SY/T 5119-2008 [39].
2.4. Establishing Bacterial Consortia in an Orthogonal Experiment
Eight strains were selected to construct bacterial consortia. The inoculated combinations were designed by orthogonal experiment using the software Minitab v17 (Minitab Inc., State College, PA, USA). Eight isolates were selected as the factors of the orthogonal experiments, and each factor had three levels, i.e., initial inoculation volume of 1, 1.5, and 2 mL (Table S1). A total of 27 consortia were established, and each consortium was inoculated into 100 mL of IS medium added with 0.1 g Liaohe heavy oil. All samples were incubated under 30 °C for 7 days on a shaker at 180 rpm. The oil removal amount μ (mg/mLOD600), used to evaluate the oil removal efficiencies of the bacterial consortia, was calculated by Equation (3).
where M0 is the weight of total oil in a medium (0.1 g); ƞ is the oil degradation efficiency determined by Equation (1) (%); j is the total of the strains in bacterial consortia; Vi and Ai are the inoculation volume (mL) and OD600 of the i-th strain (0.8), respectively.
2.5. Mathematical Analysis of Microbial Interspecific Relationship
Based on the results of the orthogonal experiment, partial correlation analysis was carried out to identify the correlation between strains and oil removal performance. This was performed by using Statistical Product and Service Solutions (SPSS) (IBM China Company, Ltd., Beijing, China). Moreover, a general linear model shown in Equation (4) was also applied, using SPSS to analyze the interactions between each strain during oil biodegradation [40].
where Y is the petroleum removal amount; Xi is the inoculation volume of the i-th strain; βi is the regression coefficients; β0 is a random error.
3. Results and Discussion
3.1. Bacteria Isolation and Identification
In this research, 14 petroleum-degrading bacterial strains were isolated by enrichment cultivation and isolation. Among them, eight strains (Y1–Y8) displayed successful growth on IS medium in the presence of Liaohe heavy oil as a sole carbon source. The results of the catalase test indicated that all strains can secrete catalase; furthermore, two of the isolates (Y1 and Y3) were Gram negative (Table S2). Methyl red test showed that Y1, Y2, and Y4 can produce organic acid by glucose fermentation. The substrate utilization test indicated that Y2 and Y6-Y8 can utilize starch; Y2, Y4, and Y8 can utilize citrate; Y4, Y5, Y7, and Y8, can utilize gelatin. The gene sequencing indicated that isolated strains were closely related to Breuvndimonas sp. strain Tibet-IX23, Bacillus firmus strain YHSA15, B. aquimaris strain AT8, Pseudomonas alcaligenes strain NBRC, Microbacterium oxydans strain CV8.4, Rhodococcus erythropolis strain SBUG 2052, Planococcus cereus strain MTCC 9817, and Planococcus sp. Tibet-IX21. (Table 1). A phylogenic tree displaying similarities among these strains is presented in Figure 1.

Table 1.
Closest relatives of the 16S rRNA gene sequences of the isolated strains.
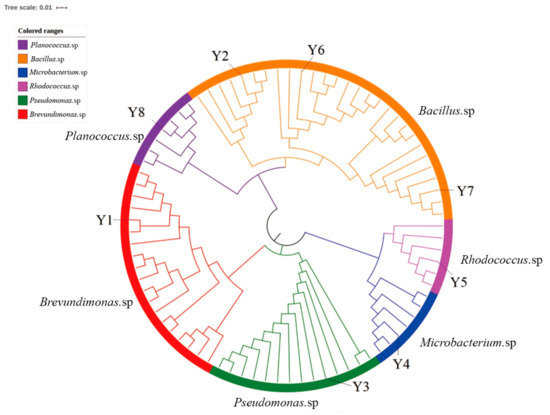
Figure 1.
The phylogenetic tree based on 16S rRNA gene sequences of the isolates and related species. Neighbor-joining analysis with 1000 bootstrap replicates was used to infer tree topology. The bar represents 0.1% sequence divergence. Sequenced data showing the location of selected isolated strain.
Many species belonging to the genera Brevundimonas, Bacillus, Pseudomonas, Rhodococcus, Microbacterium, and Planococcus were previously isolated from petroleum contaminated sites and exhibited the ability of petroleum contaminants degradation [41]. For instance, Microbacterium hydrocarbonoxydans and Microbacterium oleivorans were reported as crude oil-degrading bacteria [42]. Likewise, Planococcus sp. strain ZD22 could degrade benzene and its derivatives [43]. Thus, strains Y1–Y8 are alternative bacterial resources for the bioremediation of petroleum-contaminated environments.
3.2. Bacterial Growth Curves and Oil Degradation
The results of OD600 were used to obtain the growth curves for each bacterial strain (Figure 2). The lag phase lasted over 18 h for Y1 and Y5, but it was less than 15 h for other bacteria. However, a long period of acclimation displayed a faster growth for Y1 and Y5 in the exponential phase. Thus, these strains were collected in the exponential phase and then inoculated for oil degradation experiments. The oil degradation efficiency of each strain was recorded at least 35% individually, and the total oil removal per OD600 was around 4.37 to 5.75 mg/mL OD600 (Figure 3). A relatively better oil degradation efficiency (45.8%) on the 7th day was recorded for Y6 and Y8. Due to the complex composition of crude oil, low oil degradation efficiency using a single strain is generally reported. Yemisi, et al. [44] isolated two strains of Bacillus sp. and Pseudomonas sp. with total petroleum hydrocarbon degradation rates of 42% and 49%, respectively. Accordingly, the strain of Brevundimonas sp. isolated from oil-contaminated seawater displayed a diesel oil biodegradation rate of 45% [45]. Application of two strains of Rhodococcus sp.—namely, CD 167 and CD 130, on petroleum-contaminated soil showed a total petroleum hydrocarbon removal rate of 38.4% and 29.8%, respectively [46]. All these studies used diesel oil or crude oil to evaluate the biodegradation efficiency of strains. In comparison, heavy crude oil used in our research contained higher refractory components. The isolates Y1–Y8 having over 35% of oil removal efficiency are acceptable.
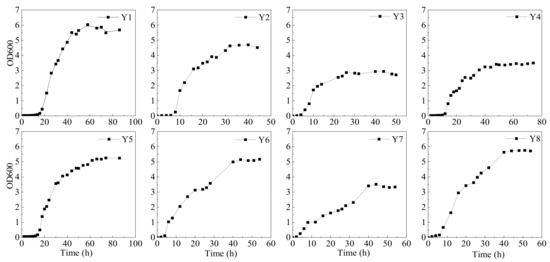
Figure 2.
The growth curves obtained for each strain in IS medium with heavy oil as the sole carbon source.
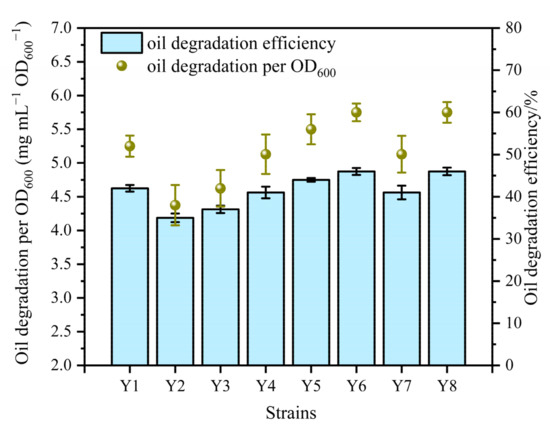
Figure 3.
Degradation efficiencies efficiency (%) and amount (mg/mL OD600) of the strains Y1–Y8.
The SARA fractions were studied for differentially treated Liaohe heavy oil in the presence of different bacterial strains before and after biodegradation experiments (Figure 4). In the beginning, the SARA fraction of Liaohe heavy oil was recorded as 33.5% for saturates, 32.2% for aromatics, 17.5% for resins, and 17.8% for asphaltene. After biodegradation, the saturates’ content in all samples reduced remarkably. The highest reduction rate of saturates content (from 33.5% to 19.5%) was observed for Y8. The saturates are readily biodegradable components, as reported previously [47]. The content of aromatics and resins was decreased for Y1, indicating its ability to degrade these compounds. However, Y2, Y3, Y6, and Y8 showed a biodegradation potential for asphaltene (Figure 4). Different strains exhibited different degradation characteristics and resulted in varying oil degradation efficiency. Therefore, the construction of a bacterial consortium could promote the biodegradation of complex petroleum contaminants.
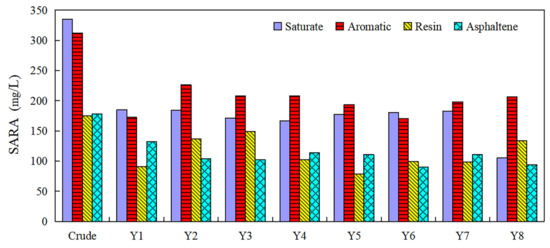
Figure 4.
SARA fractions extracted from the initial heavy oil sample and samples after biodegradation.
3.3. Oil Degradation in an Orthogonal Experiment for Consortia Establishment
The abovementioned 8 strains were used to establish 27 consortia (C1–C27) as per the orthogonal experiment. Their performances in oil degradation amounts are shown in Table 2. Results illustrated that the oil degradation amounts of the established consortia were less than 3.5 mg/mL at OD600. This was even lower than the individual strains (8.75~11.5 mg/mL OD600), thus indicating a mutual inhibition effect among these strains. These results are consistent with an earlier study in which the degradation efficiency of alkane and aromatics was reduced from 55.3% to 39.0% in the presence of bacterial consortium, compared with the individual bacteria [48].

Table 2.
Oil degradation amount by consortium constructed with orthogonal experiment.
3.4. Interspecific Relationship Analysis
To identify the interspecific relationship, partial correlation analysis was performed to evaluate the relevance and significance of each strain to oil biodegradation (Table 3). As shown in Table 3, a positive correlation relationship (PAR > 0) was only found with Y1, thus indicating a significant synergy of this strain with the others for oil degradation. As of today, little is known about the physiology of this strain particularly, which makes it difficult to speculate the underlying reasons/mechanisms of the synergy. Nevertheless, members of Brevundimonas genus have been found to work effectively in a consortium with other strains for enhanced oil degradation [49]. Although negative correlations were found in Y2, Y3, Y4, Y5, Y6, Y7, and Y8 (PAR < 0), p-values of Y2, Y5, Y7, and Y8 were higher than 0.05, suggesting that correlations of these strains to oil biodegradation were not significant. The minus PAR and low (<0.05) p-value of Y3, Y4, and Y6 indicated significant negative correlations. To further investigate the antagonistic correlations, a general linear model was used to analyze oil removal amount at each inoculation volume (1.0, 1.5, and 2.0 mL) for Y3, Y4, and Y6 with other strains (Figure 5). Oil removal per OD600 decreased with an increase in inoculation volume for Y3 and Y4 among each group. This confirmed that these strains were mutually antagonistic and should not coexist within the same treatment. For Y6, the oil removal amount showed first a decrease and then slightly increased. This indicated that a competitive relationship exists between Y6 and the other strains, thus suggesting antagonism potential in the consortia.

Table 3.
The result of partial correlation analysis.
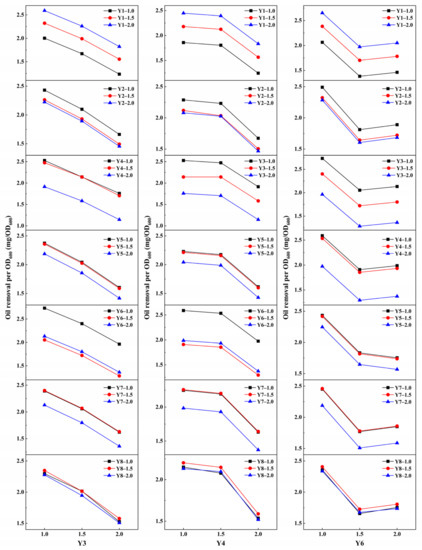
Figure 5.
Analysis of the interspecific relationships between isolates (Y3, Y4, and Y6) at different dosages.
In the presence of Y1, a variety of oil components could be degraded simultaneously, thus improving the overall oil remediation efficiency. A similar study previously reported a synergistic relationship among Brevundimonas, Pseudomonas, Nitratireductor, and Acinetobacter, which enhanced the degradation of oil [13]. The synergy mechanisms between petroleum degraders may be complex. It is likely that one species removes the toxic metabolites, but others improve biodegradation. Further, there might be the second species able to degrade oil-derived compounds that were partially degraded by the first species [50]. The antagonism of Y3 and Y4 to other strains could have lowered the oil degradation efficiency. When the antagonistic strains are co-cultured, some may produce a specific metabolite or may change environmental conditions, inhibiting the growth of others. Additionally, due to the competition for limited nutrients and space, the functions of all species cannot be fully exerted. The antagonism between certain microorganisms has often been reported. Islam et al. [51] found that antagonism between Pseudomonas aeruginosa and Bacillus cereus in a wastewater-fed microbial fuel cell inhibited cell growth and power generation.
3.5. Optimization of Petroleaum-Degrading Consortium
Based on the results of interspecific relationship analysis, petroleum-degrading consortia was optimized. Approximately, 2 mL of Y1 was inoculated to optimize the consortia as per significant synergy, as reported above (Section 3.4). However, Y3, Y4, and Y6 were not included, because they showed strong antagonism. The bacterial consortia were reconstructed with inoculation dosage conditions as follows: Y1 at 2 mL; Y2, Y5, Y7, and Y8 varied from 1 to 2 mL. A total of nine consortia were established according to the orthogonal experiment using Minitab 17 software (Minitab Inc., State College, PA, USA), and their oil degradation amount was evaluated accordingly (Table 4). The oil degradation for these nine consortia was recorded in the range of 9.98 to 11.29 mg/mL OD600, which showed 3~4 times higher degradation than the consortia C1-C27 (Table 2).

Table 4.
Optimization of petroleum-degrading consortia.
Finally, a comparative experimental study was carried out to confirm these results. Among isolated strains, the highest oil degradation efficiency was observed for Y1 (5.25 mg/mL OD600), Y6 (5.75 mg/mL OD600), and Y8 (5.75 mg/mL OD600). These bacteria were then selected to construct a real-time consortium in an equal-proportion inoculation ratio. This consortium showed 9.38 mg/mL OD600 of oil degradation in 7 days under the same experimental conditions. Compared with the conventional consortia construction method, the consortium N2 (Y1: Y2: Y5: Y7: Y8 at 2:1:1.5:2:1.5) showed 11.238 mg/mL OD600 of oil degradation, thus increasing the oil degradation efficiency by 10%. These results suggest that a systematic, experimental, and mathematical methodology can help us understand the synergy, antagonism, and inoculation dosages of petroleum-degrading strains.
4. Conclusions
This study established a systematic experimental and mathematical methodology to construct high-efficiency petroleum-degrading bacterial consortia. The methodology successfully determined the synergy and antagonism between strains, which helped optimize the inoculation dosages of strains in consortia. The constructed consortium was able to degrade 11.875 mg/mL OD600 of heavy oil and could increase the degradation efficiency by 10% when compared with the conventional construction methods. This methodology is a promising strategy to construct functional consortia and improve the biological treatment of petroleum contaminants.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/w13223311/s1, Table S1: Range and levels of independent variables, Table S2: Biochemical activities of eight isolated strains.
Author Contributions
Conceptualization, J.D.; methodology, S.G.; software, H.N.; formal analysis, M.G.E.-D.; writing—original draft preparation, B.W.; writing—review and editing, Q.W.; supervision, M.A.; project administration, J.L.; funding acquisition, C.C. All authors have read and agreed to the published version of the manuscript.
Funding
National Key R&D Program of China (2019YFC1806201-01 and 2018YFC1801903-01) supported the sample collection. Science Foundation of China University of Petroleum-Beijing (No. 2462018BJB001 and 2462020YXJJ035) supported data analysis and manuscript writing.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Acknowledgments
This work was supported in part by the National Key R&D Program of China (2019YFC1806201-01 and 2018YFC1801903-01) and the Science Foundation of China University of Petroleum-Beijing (No. 2462018BJB001 and 2462020YXJJ035).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kvenvolden, K.A.; Cooper, C.K. Natural Seepage of Crude Oil into the Marine Environment. Geo Marine Lett. 2003, 23, 140–146. [Google Scholar] [CrossRef]
- Du, W.; Wan, Y.; Zhong, N.; Fei, J.; Zhang, Z.; Chen, L.; Hao, J. Status Quo of Soil Petroleum Contamination and Evolution of Bioremediation. Pet. Sci. 2011, 8, 502–514. [Google Scholar] [CrossRef]
- Hussain, F.; Hussain, I.; Khan, A.H.A.; Muhammad, Y.S.; Iqbal, M.; Soja, G.; Reichenauer, T.G.; Zeshan; Yousaf, S. Combined Application of Biochar, Compost, and Bacterial Consortia with Italian Ryegrass Enhanced Phytoremediation of Petroleum Hydrocarbon Contaminated Soil. Environ. Exp. Bot. 2018, 153, 80–88. [Google Scholar] [CrossRef]
- Lovely, D.R. Cleaning Up with Genomics: Application of Molecular Biology to Bioremediation. Soil Sediment Contam. 2004, 13, 193–194. [Google Scholar] [CrossRef]
- Gao, J.; Ming, J.; Xu, M.; Fu, X.; Duan, L.-F.; Xu, C.-C.; Gao, Y.; Xue, J.-L.; Xiao, X.-F. Isolation and Characterization of a High-Efficiency Marine Diesel Oil-Degrading Bacterium. Pet. Sci. 2021, 18, 641–653. [Google Scholar] [CrossRef]
- Eze, M. Metagenome Analysis of a Hydrocarbon-Degrading Bacterial Consortium Reveals the Specific Roles of BTEX Biodegraders. Genes 2021, 12, 98. [Google Scholar] [CrossRef]
- Shi, K.; Zhang, Q.; Xue, J.; Chen, X.; Chen, Y.; Qiao, Y.; Yang, Y.; Sun, J. Study on the Degradation Performance and Bacterial Community of Bioaugmentation in Petroleum-Pollution Seawater. J. Environ. Chem. Eng. 2020, 8, 103900. [Google Scholar] [CrossRef]
- Al-Saleh, E.; Drobiova, H.; Obuekwe, C. Predominant Culturable Crude Oil-Degrading Bacteria in the Coast of Kuwait. Int. Biodeterior. Biodegrad. 2009, 63, 400–406. [Google Scholar] [CrossRef]
- Crisafi, F.; Giuliano, L.; Yakimov, M.M.; Azzaro, M.; Denaro, R. Isolation and Degradation Potential of a Cold-Adapted Oil/PAH-Degrading Marine Bacterial Consortium from Kongsfjorden (Arctic Region). Rend. Lince 2016, 27, 261–270. [Google Scholar] [CrossRef]
- Pramila, M.; Manikandan, S.; Anju, K.; Kannan, M.M.; Hong, S.; Maruthamuthu, S.; Subramanian, K. Electrochemical Decolorization and Degradation of Turquoise Blue G (TBG) by Pre-Adapted Petroleum Degrading Bacteria. Sep. Purif. Technol. 2014, 132, 719–727. [Google Scholar] [CrossRef]
- Rajasekar, A.; Anandkumar, B.; Maruthamuthu, S.; Ting, Y.-P.; Rahman, P.K.S.M. Characterization of Corrosive Bacterial Consortia Isolated from Petroleum-Product-Transporting Pipelines. Appl. Microbiol. Biotechnol. 2009, 85, 1175–1188. [Google Scholar] [CrossRef]
- Herrero, M.; Stuckey, D.C. Bioaugmentation and its Application in Wastewater Treatment: A Review. Chemosphere 2015, 140, 119–128. [Google Scholar] [CrossRef]
- Ma, M.; Zheng, L.; Yin, X.; Gao, W.; Han, B.; Li, Q.; Zhu, A.; Chen, H.; Yang, H. Reconstruction and Evaluation of Oil-Degrading Consortia Isolated from Sediments of Hydrothermal Vents in the South Mid-Atlantic Ridge. Sci. Rep. 2021, 11, 1456. [Google Scholar] [CrossRef]
- Al-Kindi, S.; Abed, R.M.M. Effect of Biostimulation Using Sewage Sludge, Soybean Meal, and Wheat Straw on Oil Degradation and Bacterial Community Composition in a Contaminated Desert Soil. Front. Microbiol. 2016, 7, 240. [Google Scholar] [CrossRef]
- Gurav, R.; Lyu, H.; Ma, J.; Tang, J.; Liu, Q.; Zhang, H. Degradation of N-Alkanes and PAHs from the Heavy Crude Oil Using Salt-Tolerant Bacterial Consortia and Analysis Catabolic Genes. Environ. Sci. Pollut. Res. 2017, 24, 11392–11403. [Google Scholar] [CrossRef]
- M’Rassi, A.G.; Bensalah, F.; Gury, J.; Duran, R. Isolation and Characterization of Different Bacterial Strains for Bioremediation of N-Alkanes and Polycyclic Aromatic Hydrocarbons. Environ. Sci. Pollut. Res. 2015, 22, 15332–15346. [Google Scholar] [CrossRef]
- Chandra, R.; Bharagava, R.N.; Kapley, A.; Purohit, H.J. Isolation and Characterization of Potential Aerobic Bacteria Capable for Pyridine Degradation in Presence of Picoline, Phenol and Formaldehyde as Co-Pollutants. World J. Microbiol. Biotechnol. 2009, 25, 2113–2119. [Google Scholar] [CrossRef]
- Kato, S.; Haruta, S.; Cui, Z.J.; Ishii, M.; Igarashi, Y. Stable Coexistence of Five Bacterial Strains as a Cellulose-Degrading Community. Appl. Environ. Microbiol. 2005, 71, 7099–7106. [Google Scholar] [CrossRef]
- Mikesková, H.; Novotný, Č.; Svobodová, K. Interspecific Interactions in Mixed Microbial Cultures in a Biodegradation Perspective. Appl. Microbiol. Biotechnol. 2012, 95, 861–870. [Google Scholar] [CrossRef]
- Dai, X.; Lv, J.; Yan, G.; Chen, C.; Guo, S.; Fu, P. Bioremediation of Intertidal Zones Polluted by Heavy Oil Spilling Using Immobilized Laccase-Bacteria Consortium. Bioresour. Technol. 2020, 309, 123305. [Google Scholar] [CrossRef]
- Hussain, F.; Tahseen, R.; Arslan, M.; Iqbal, S.; Afzal, M. Removal of Hexadecane by Hydroponic Root Mats in Partnership with Alkane-Degrading Bacteria: Bacterial Augmentation Enhances System’s Performance. Int. J. Environ. Sci. Technol. 2019, 16, 4611–4620. [Google Scholar] [CrossRef]
- Shabir, G.; Arslan, M.; Fatima, K.; Amin, I.; Khan, Q.M.; Afzal, M. Effects of Inoculum Density on Plant Growth and Hydrocarbon Degradation. Pedosphere 2016, 26, 774–778. [Google Scholar] [CrossRef]
- Jin, D.F.; Hu, H.; Liu, D.F.; Ding, H.T.; Jia, X.M.; Zhao, Y.H. Optimization of a Bacterial Consortium for Nitrobenzene Degradation. Water Sci. Technol. 2012, 65, 795–801. [Google Scholar] [CrossRef]
- Zaroual, Z.; Chaair, H.; Essadki, A.; El Ass, K.; Azzi, M. Optimizing the Removal of Trivalent Chromium by Electrocoagulation Using Experimental Design. Chem. Eng. J. 2009, 148, 488–495. [Google Scholar] [CrossRef]
- Zhao, C.; Wen, D.; Zhang, Y.; Zhang, J.; Tang, X. Experimental and Mathematical Methodology on the Optimization of Bacterial Consortium for the Simultaneous Degradation of Three Nitrogen Heterocyclic Compounds. Environ. Sci. Technol. 2012, 46, 6205–6213. [Google Scholar] [CrossRef]
- Wang, J.; Yan, G.; An, M.; Liu, J.; Zhang, H.; Chen, Y. Study of a Plugging Microbial Consortium using Crude Oil as Sole Carbon Source. Pet. Sci. 2008, 5, 367–374. [Google Scholar] [CrossRef]
- Dong, X.Z.; Cai, M.Y. Identification Manual of Systematic Bacteriology; Science Press: Beijing, China, 2001; pp. 364–398. [Google Scholar]
- Coico, R. Gram Staining. Curr. Protoc. Microbiol. 2006, 3, A–3C. [Google Scholar] [CrossRef]
- Cowan, S.T. Micromethod for the Methyl Red Test. J. Gen. Microbiol. 1953, 9, 101–109. [Google Scholar] [CrossRef][Green Version]
- Dunican, L.K.; Seeley, H.W. Starch Hydrolysis by Streptococcus Equinus. J. Bacteriol. 1962, 83, 264–269. [Google Scholar] [CrossRef]
- Simmons, J.S. A Culture Medium for Differentiating Organisms of Typhoid-Colon Aerogenes Groups and for Isolation of Certain Fungi. J. Infect. Disease 1926, 39, 209–214. Available online: http://www.jstor.org/stable/30083347 (accessed on 15 June 2021). [CrossRef]
- McDade, J.J.; Weaver, R.H. Rapid Methods for the Detection of Gelatin Hydrolysis. J. Bacteriol. 1959, 77, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Taylor, W.I.; Achanzar, D. Catalase Test as an Aid to the Identification of Enterobacteriaceae. Appl. Microbiol. 1972, 24, 58–61. [Google Scholar] [CrossRef] [PubMed]
- Simon, E.; Hoffmann, J. Extraction of High Molecular Weight DNA from Molluscs. Trends Genet. 1993, 9, 407. [Google Scholar] [CrossRef]
- Cappello, S.; Russo, D.; Santisi, S.; Calogero, R.; Gertler, C.; Crisafi, F.; De Domenico, M.; Yakimov, M.M. Presence of Hydrocarbon-Degrading Bacteria in the Gills of Mussel Mytilus Galloprovincialis in a Contaminated Environment: A Mesoscale Simulation Study. Chem. Ecol. 2012, 28, 239–252. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.S.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Gai, H.; Zhang, X.; Chen, S.; Wang, C.; Xiao, M.; Huang, T.; Wang, J.; Song, H. An Improved Tar–Water Separation Process of Low–Rank Coal Conversion Wastewater for Increasing the Tar Yield and Reducing the Oil Content in Wastewater. Chem. Eng. J. 2020, 383, 123229. [Google Scholar] [CrossRef]
- Lin, B.; Wang, J.; Huang, Q.; Chi, Y. Effects of Potassium Hydroxide on the Catalytic Pyrolysis of Oily Sludge for High-Quality Oil Product. Fuel 2017, 200, 124–133. [Google Scholar] [CrossRef]
- Nueda, M.J.; Conesa, A.; Westerhuis, J.A.; Hoefsloot, H.C.J.; Smilde, A.K.; Talón, M.; Ferrer, A. Discovering Gene Expression Patterns in Time Course Microarray Experiments by ANOVA–SCA. Bioinformatics 2007, 23, 1792–1800. [Google Scholar] [CrossRef]
- Mahjoubi, M.; Jaouani, A.; Guesmi, A.; Ben Amor, S.; Jouini, A.; Cherif, H.; Najjari, A.; Boudabous, A.; Koubaa, N.; Cherif, A. Hydrocarbonoclastic Bacteria Isolated from Petroleum Contaminated Sites in Tunisia: Isolation, Identification and Characterization of the Biotechnological Potential. New Biotechnol. 2013, 30, 723–733. [Google Scholar] [CrossRef]
- Schippers, A.; Bosecker, K.; Spröer, C.; Schumann, P. Microbacterium oleivorans sp. nov. and Microbacterium hydrocarbonoxydans sp. nov., Novel Crude-Oil-Degrading Gram-Positive Bacteria. Int. J. Syst. Evol. Microbiol. 2005, 55, 655–660. [Google Scholar] [CrossRef]
- Li, H.; Liu, Y.H.; Luo, N.; Zhang, X.Y.; Luan, T.G.; Hu, J.M.; Wang, Z.Y.; Wu, P.C.; Chen, M.J.; Lu, J.Q. Biodegradation of Benzene and its Derivatives by a Psychrotolerant and Moderately Haloalkaliphilic Planococcus sp. Strain ZD22. Res. Microbiol. 2006, 157, 629–636. [Google Scholar] [CrossRef]
- Obafemi, Y.; Taiwo, O.S.; Omodara, O.J.; Dahunsi, O.S.; Oranusi, S. Biodegradation of Crude Petroleum by Bacterial Consortia from Oil-Contaminated Soils in Ota, Ogun State, South-Western, Nigeria. Environ. Technol. Innov. 2018, 12, 230–242. [Google Scholar] [CrossRef]
- Wang, X.; Wang, X.; Liu, M.; Zhou, L.; Gu, Z.; Zhao, J. Bioremediation of Marine Oil Pollution by Brevundimonas diminuta: Effect of Salinity and Nutrients. Desalination Water Treat. 2015, 57, 19768–19775. [Google Scholar] [CrossRef]
- Pacwa-Płociniczak, M.; Czapla, J.; Płociniczak, T.; Piotrowska-Seget, Z. The Effect of Bioaugmentation of Petroleum-Contaminated Soil with Rhodococcus Erythropolis Strains on Removal of Petroleum from Soil. Ecotoxicol. Environ. Saf. 2019, 169, 615–622. [Google Scholar] [CrossRef]
- Beškoski, V.P.; Gojgic-Cvijovic, G.; Ilic, M.; Miletic, S.; Šolević, T.; Vrvic, M. Ex Situ Bioremediation of a Soil Contaminated by Mazut (Heavy Residual Fuel Oil)—A Field Experiment. Chemosphere 2011, 83, 34–40. [Google Scholar] [CrossRef]
- Wang, J.N.; Shi, Y.Y.; Zheng, L.Y.; Wang, Z.; Cai, Z.; Liu, J. Isolation and Identification of Petroleum Degradation Bacteria and Interspecific Interactions among Four Bacillus Strains. Environ. Sci. 2015, 36, 2245–2251. [Google Scholar] [CrossRef]
- Diallo, M.; Vural, C.; Cay, H.; Ozdemir, G. Enhanced Biodegradation of Crude Oil in Soil by a Developed Bacterial Consortium and Indigenous Plant Growth Promoting Bacteria. J. Appl. Microbiol. 2021, 130, 1192–1207. [Google Scholar] [CrossRef]
- Schink, B. Synergistic Interactions in the Microbial World. Antonie van Leeuwenhoek 2002, 81, 257–261. [Google Scholar] [CrossRef]
- Islam, M.A.; Karim, A.; Mishra, P.; Dubowski, J.J.; Yousuf, A.; Sarmin, S.; Khan, M.R. Microbial Synergistic Interactions Enhanced Power Generation in Co-Culture Driven Microbial Fuel Cell. Sci. Total. Environ. 2020, 738, 140138. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).