Characterization of Antibiotic Resistance and Metal Homeostasis Genes in Midwest USA Agricultural Sediments
Abstract
1. Introduction
2. Materials and Methods
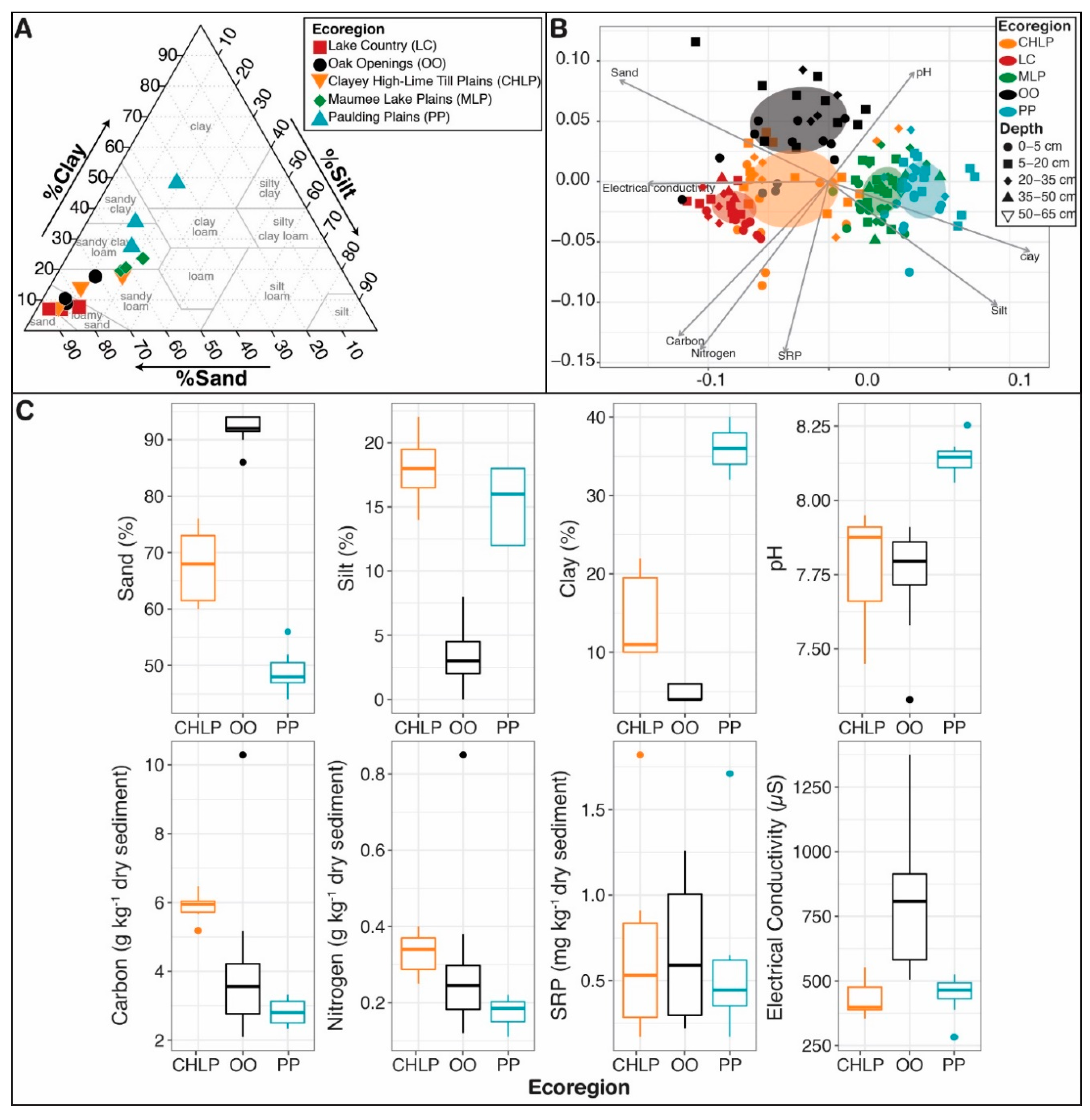
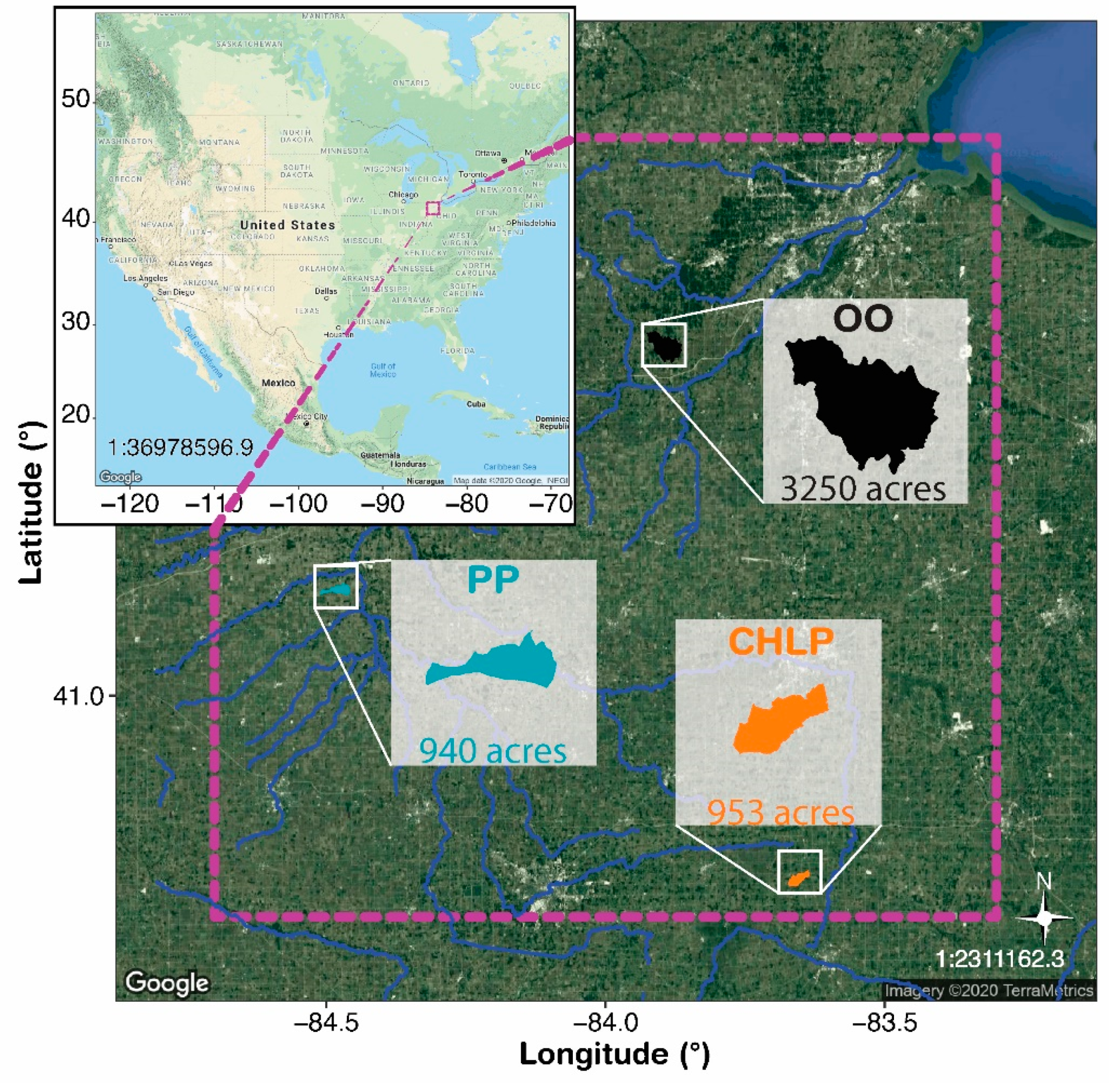
2.1. Study Area and Sample Collection
2.2. Genomic DNA Extraction & Analyses
2.3. Analysis of Antibiotics and Metals
2.4. Data Anlaysis
3. Results and Discussion
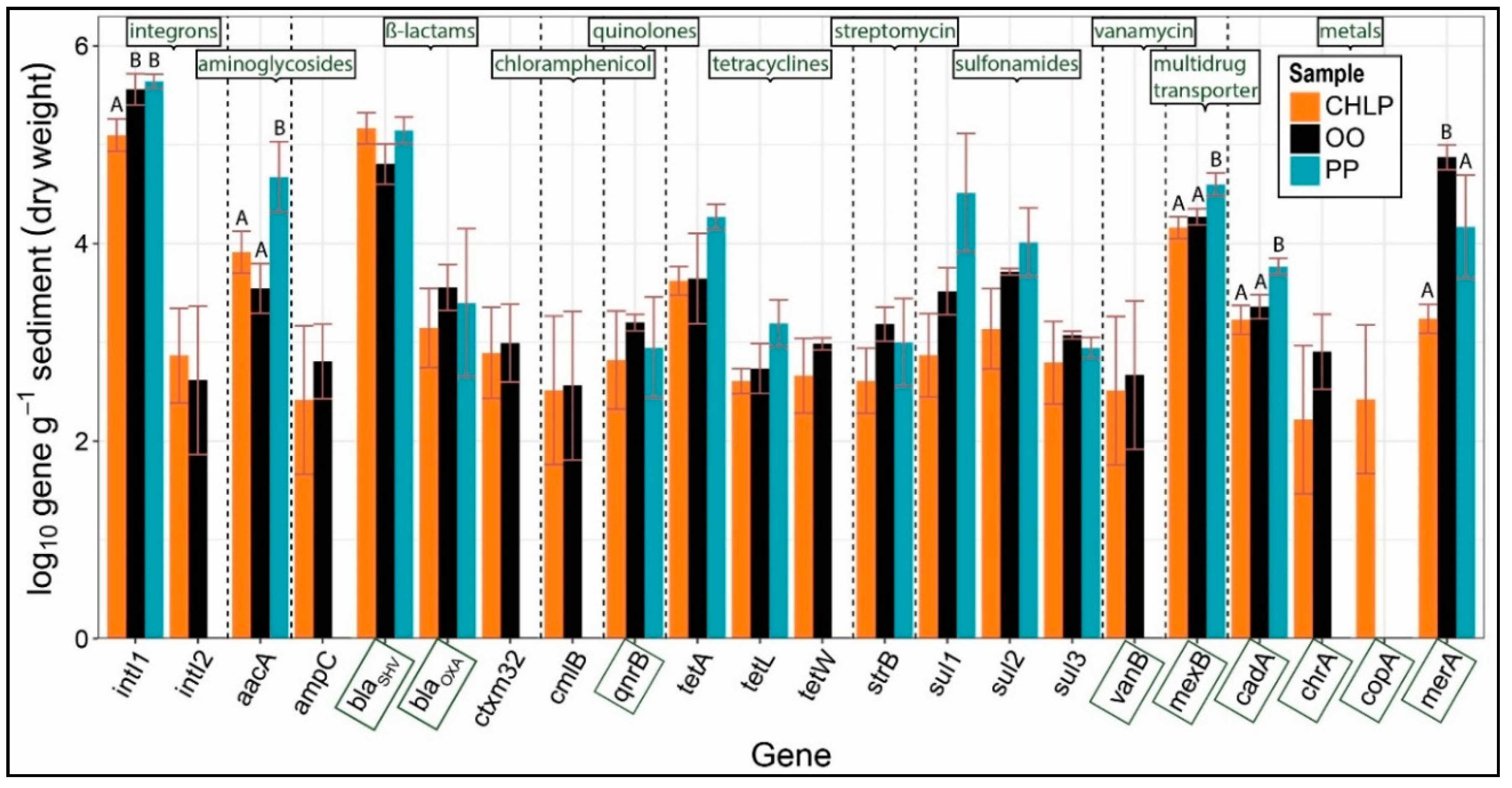
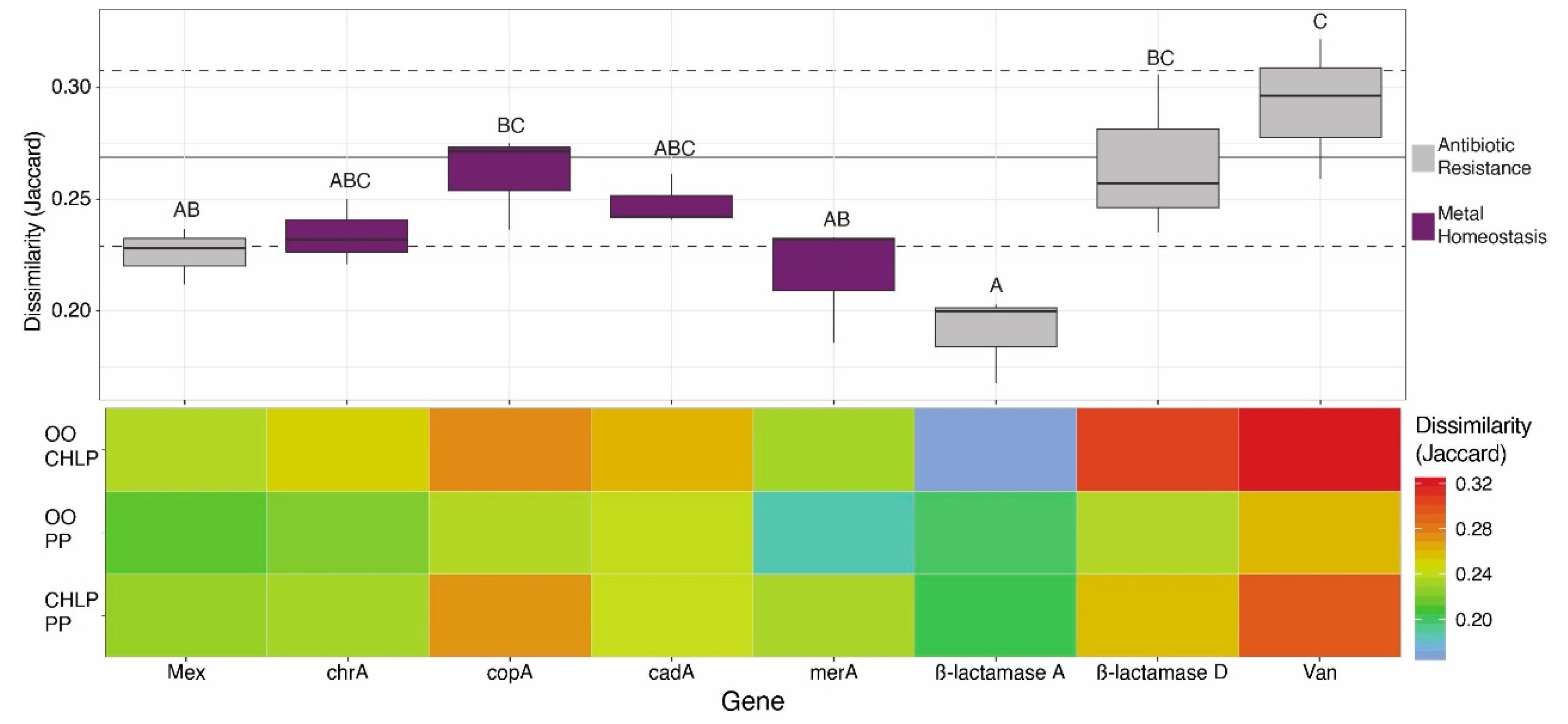
3.1. Antibiotoic Resistance and Metal Homeostasis Genes Present at Natural Levels
3.2. Few Antibiotics, Natural Concentrations of Metals
3.3. Future Research Needs for Agricultural Contributions to the Spread of Antibiotic Resistance
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B
Antibiotic Extraction and Quantification
| Method 1 | Method 2 | ||
|---|---|---|---|
| Time (min) | % B | Time (min) | % B |
| 0.0 | 0 | 0.0 | 0 |
| 5.5 | 100 | 0.5 | 0 |
| 7.5 | 100 | 4.0 | 40 |
| 8.0 | 0 | 7.0 | 100 |
| 20.0 | 0 | 9.0 | 100 |
| - | - | 10.0 | 0 |
| - | - | 20.0 | 0 |
| Analyte | Parent Ion (m/z) | Product Ion (m/z) | CE (V) | Quantification or Confirmation |
|---|---|---|---|---|
| Sulfonamides | ||||
| sulfapyridine | 250.10 | 156.00 | 17 | quantification |
| 250.10 | 108.05 | 25 | Confirmation | |
| sulfadiazine | 251.05 | 156.00 | 15 | quantification |
| 251.05 | 108.05 | 24 | Confirmation | |
| sulfamethoxazole | 254.05 | 92.10 | 29 | quantification |
| 254.05 | 108.00 | 24 | Confirmation | |
| sulfamethazine | 279.05 | 186.00 | 17 | quantification |
| 279.05 | 156.00 | 20 | Confirmation | |
| sulfachloropyridazine | 285.00 | 156.06 | 15 | quantification |
| 285.00 | 92.05 | 35 | Confirmation | |
| sulfadimethoxine | 311.10 | 156.06 | 21 | quantification |
| 311.10 | 92.05 | 35 | Confirmation | |
| 13C6-sulfamethoxazole | 260.05 | 98.10 | 32 | quantification |
| (internal standard) | 260.05 | 114.10 | 27 | Confirmation |
| 13C6-sulfamethazine | 285.05 | 186.00 | 22 | quantification |
| (surrogate) | 285.05 | 123.00 | 20 | Confirmation |
| Fluoroquinolones | ||||
| norfloxacin | 320.10 | 276.10 | 17 | quantification |
| 320.10 | 302.10 | 21 | Confirmation | |
| ciprofloxacin | 332.10 | 231.05 | 35 | quantification |
| 332.10 | 314.10 | 21 | Confirmation | |
| enrofloxacin | 360.10 | 245.10 | 25 | quantification |
| 360.10 | 316.15 | 19 | Confirmation | |
| ofloxacin | 362.10 | 261.10 | 28 | quantification |
| 362.10 | 318.10 | 19 | Confirmation | |
| clinafloxacin | 366.10 | 348.00 | 20 | Confirmation |
| (internal standard) | 366.10 | 305.00 | 22 | quantification |
| nalidixic acid | 233.15 | 187.00 | 27 | Confirmation |
| (surrogate) | 233.15 | 104.05 | 40 | quantification |
| Tetracyclines | ||||
| Tetracycline | 445.10 | 410.10 | 19 | quantification |
| 445.10 | 427.05 | 11 | confirmation | |
| doxycycline | 445.10 | 321.05 | 31 | quantification |
| 445.10 | 428.15 | 18 | confirmation | |
| oxytetracycline | 461.10 | 426.10 | 17 | quantification |
| 461.10 | 443.10 | 12 | confirmation | |
| chlortetracycline | 479.05 | 462.10 | 20 | quantification |
| & degradation products | 479.05 | 444.10 | 17 | confirmation |
| 481.05 | 464.10 | 20 | quantification | |
| 481.05 | 446.10 | 30 | confirmation | |
| demeclocycline | 465.10 | 448.05 | 20 | quantification |
| (surrogate) | 465.10 | 430.05 | 17 | confirmation |
| Macrolides | ||||
| erythromycin | 734.4 | 158.15 | 35 | quantification |
| 734.4 | 576.35 | 15 | confirmation | |
| erythromycin-H2O | 716.45 | 158.15 | 35 | quantification |
| 716.45 | 558.35 | 15 | confirmation | |
| roxithromycin | 837.45 | 158.10 | 35 | quantification |
| 837.45 | 679.45 | 20 | confirmation | |
| Tylosin | 916.45 | 174.10 | 40 | quantification |
| 916.45 | 772.45 | 30 | confirmation | |
| 13C2-erythromycin | 736.40 | 160.15 | 35 | quantification |
| 736.40 | 578.35 | 20 | confirmation | |
| 13C2-erythromycin-H2O | 718.40 | 160.15 | 35 | quantification |
| 718.40 | 560.35 | 20 | confirmation | |
| Non-categorized | ||||
| Carbadox | 263.10 | 130.05 | 22 | quantification |
| 263.10 | 231.05 | 13 | confirmation | |
| Trimethoprim | 291.10 | 230.10 | 23 | quantification |
| 291.10 | 123.05 | 24 | confirmation | |
| Lincomycin | 407.30 | 126.10 | 35 | quantification |
| 407.30 | 359.20 | 18 | confirmation | |
| Simeton | 198.20 | 68.10 | 33 | quantification |
| (internal standard) | 198.20 | 100.10 | 27 | confirmation |
References
- Chee-Sanford, J.C.; Mackie, R.I.; Koike, S.; Krapac, I.G.; Lin, Y.F.; Yannarell, A.C.; Maxwell, S.; Aminov, R.I. Fate and Transport of Antibiotic Residues and Antibiotic Resistance Genes Following Land Application of Manure Waste. J. Environ. Qual. 2009, 38, 1086–1108. [Google Scholar] [CrossRef]
- Ghosh, S.; LaPara, T.M. The Effects of Subtherapeutic Antibiotic use in Farm Animals on the Proliferation and Persistence of Antibiotic Resistance among Soil Bacteria. ISME J. 2007, 1, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Rothrock, M.J.; Keen, P.L.; Cook, K.L.; Durso, L.M.; Franklin, A.M.; Dungan, R.S. How should we be Determining Background and Baseline Antibiotic Resistance Levels in Agroecosystem Research? J. Environ. Qual. 2016, 45, 420–431. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.A.; Stedtfeld, R.D.; Wang, Q.; Cole, J.R.; Hashsham, S.A.; Looft, T.; Zhu, Y.G.; Tiedje, J.M. Clusters of Antibiotic Resistance Genes Enriched Together Stay Together in Swine Agriculture. mBio 2016, 7, 2214. [Google Scholar] [CrossRef]
- Winkworth-Lawrence, C.; Lange, K. Antibiotic Resistance Genes in Freshwater Biofilms may Reflect Influences from High-Intensity Agriculture. Microb. Ecol. 2016, 72, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Frey, S.K.; Topp, E.; Khan, I.U.; Ball, B.R.; Edwards, M.; Gottschall, N.; Sunohara, M.; Lapen, D.R. Quantitative Campylobacter Spp., Antibiotic Resistance Genes, and Veterinary Antibiotics in Surface and Ground Water Following Manure Application: Influence of Tile Drainage Control. Sci. Total Environ. 2015, 532, 138–153. [Google Scholar] [CrossRef] [PubMed]
- Chee-Sanford, J.C.; Aminov, R.I.; Krapac, I.J.; Garrigues-Jeanjean, N.; Mackie, R.I. Occurrence and Diversity of Tetracycline Resistance Genes in Lagoons and Groundwater Underlying Two Swine Production Facilities. Appl. Environ. Microbiol. 2001, 67, 1494–1502. [Google Scholar] [CrossRef]
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic Resistance is Ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef]
- Kristiansson, E.; Fick, J.; Janzon, A.; Grabic, R.; Rutgersson, C.; Weijdegård, B.; Söderström, H.; Larsson, D.J. Pyrosequencing of Antibiotic-Contaminated River Sediments Reveals High Levels of Resistance and Gene Transfer Elements. PLoS ONE 2011, 6, e17038. [Google Scholar] [CrossRef]
- Martinez, J.L. Environmental Pollution by Antibiotics and by Antibiotic Resistance Determinants. Environ. Pollut. 2009, 157, 2893–2902. [Google Scholar] [CrossRef]
- Gillings, M.R. Lateral Gene Transfer, Bacterial Genome Evolution, and the Anthropocene. Ann. N. Y. Acad. Sci. 2017, 1389, 20–36. [Google Scholar] [CrossRef] [PubMed]
- LaPara, T.M.; Burch, T.R.; McNamara, P.J.; Tan, D.T.; Yan, M.; Eichmiller, J.J. Tertiary-Treated Municipal Wastewater is a Significant Point Source of Antibiotic Resistance Genes into Duluth-Superior Harbor. Environ. Sci. Technol. 2011, 45, 9543–9549. [Google Scholar] [CrossRef] [PubMed]
- Li, A.D.; Li, L.G.; Zhang, T. Exploring Antibiotic Resistance Genes and Metal Resistance Genes in Plasmid Metagenomes from Wastewater Treatment Plants. Front. Microbiol. 2015, 6, 1025. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Shen, Y.; Liang, P.; Zhou, J.; Yang, Y.; Huang, X. Multiple Antibiotic Resistance Genes Distribution in Ten Large-Scale Membrane Bioreactors for Municipal Wastewater Treatment. Bioresour. Technol. 2016, 222, 100–106. [Google Scholar] [CrossRef]
- Negreanu, Y.; Pasternak, Z.; Jurkevitch, E.; Cytryn, E. Impact of Treated Wastewater Irrigation on Antibiotic Resistance in Agricultural . Soils Environ. Sci. Technol. 2012, 46, 4800–4808. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Wright, M.S.; Stepanauskas, R.; McArthur, J.V. Co-Selection of Antibiotic and Metal Resistance. Trends Microbiol. 2006, 14, 176–182. [Google Scholar] [CrossRef]
- Ji, X.; Shen, Q.; Liu, F.; Ma, J.; Xu, G.; Wang, Y.; Wu, M. Antibiotic Resistance Gene Abundances Associated with Antibiotics and Heavy Metals in Animal Manures and Agricultural Soils Adjacent to Feedlots in Shanghai; China. J. Hazard. Mater. 2012, 235, 178–185. [Google Scholar] [CrossRef]
- Hu, H.; Wang, J.; Li, J.; Shi, X.; Ma, Y.; Chen, D.; He, J. Long-Term Nickel Contamination Increases the Occurrence of Antibiotic Resistance Genes in Agricultural Soils. Environ. Sci. Technol. 2016, 51, 790–800. [Google Scholar] [CrossRef]
- Chen, J.; Li, J.; Zhang, H.; Shi, W.; Liu, Y. Bacterial heavy-metal and antibiotic resistance genes in a copper Tailing Dam Area in Northern China. Front. Microbiol. 2019, 10, 1916. [Google Scholar] [CrossRef]
- Bosch, N.S.; Allan, J.D.; Selegean, J.P.; Scavia, D. Scenario-Testing of Agricultural Best Management Practices in Lake Erie Watersheds. J. Great Lakes Res. 2013, 39, 429–436. [Google Scholar] [CrossRef]
- Powell, G.E.; Ward, A.D.; Mecklenburg, D.E.; Draper, J.; Word, W. Special Section: Drainage Ditches—Two-Stage Channel Systems: Part 2, Case Studies. J. Soil Water Conserv. 2007, 62, 286–296. [Google Scholar]
- King, K.W.; Williams, M.R.; LaBarge, G.A.; Smith, D.R.; Reutter, J.M.; Duncan, E.W.; Pease, L.A. Addressing Agricultural Phosphorus Loss in Artificially Drained Landscapes with 4R Nutrient Management Practices. J. Soil Water Conserv. 2018, 73, 35–47. [Google Scholar] [CrossRef]
- Jayakaran, A.D.; Mecklenburg, D.E.; Witter, J.D.; Ward, A.D.; Powell, G.E. Fluvial Processes in Agricultural Ditches in the North Central Region of the United States and Implications for Their Management. In Agricultural Drainage Ditches: Mitigation Wetlands for the 21st Century; Moore, M.T., Kroger, R., Eds.; Research Signpost: Karala, India, 2010; pp. 195–222. [Google Scholar]
- Kröger, R.; Moore, M.T.; Locke, M.A.; Cullum, R.F.; Steinriede, R.W.; Testa, S.; Bryant, C.T.; Cooper, C.M. Evaluating the Influence of Wetland Vegetation on Chemical Residence Time in Mississippi Delta Drainage Ditches. Agric. Water Manag. 2009, 96, 1175–1179. [Google Scholar] [CrossRef]
- Inwood, S.E.; Tank, J.L.; Bernot, M.J. Patterns of Denitrification Associated with Land use in 9 Midwestern Headwater Streams. J. N. Am. Benthol. Soc. 2005, 24, 227–245. [Google Scholar] [CrossRef]
- Omernik, J.M. Ecoregions of the Conterminous United States. Map Suppl. 1987, 77, 118–125. [Google Scholar] [CrossRef]
- Brooker, M.R.; Bohrer, G.; Mouser, P.J. Variations in Potential CH4 Flux and CO2 Respiration from Freshwater Wetland Sediments that Differ by Microsite Location, Depth and Temperature. Ecol. Eng. 2014, 72, 84–94. [Google Scholar] [CrossRef]
- Sandberg, K.D.; Ishii, S.; LaPara, T.M. A Microfluidic Quantitative Polymerase Chain Reaction Method for the Simultaneous Analysis of Dozens of Antibiotic Resistance and Heavy Metal Resistance Genes. Environ. Sci. Technol. Lett. 2017, 5, 20–25. [Google Scholar] [CrossRef]
- Cong, J.; Lu, H.; Li, D.; Zhang, Y.; Cong, J.; Liu, X.; Xu, H.; Li, Y.; Deng, Y. Analyses of the Influencing Factors of Soil Microbial Functional Gene Diversity in Tropical Rainforest Based on GeoChip 5.0. Genom. Data 2015, 5, 397–398. [Google Scholar] [CrossRef]
- Kerrigan, J.F.; Sandberg, K.D.; Engstrom, D.R.; LaPara, T.M.; Arnold, W.A. Sedimentary Record of Antibiotic Accumulation in Minnesota Lakes. Sci. Total Environ. 2018, 621, 970–979. [Google Scholar] [CrossRef]
- Ma, X.; Zhao, C.; Gao, Y.; Liu, B.; Wang, T.; Yuan, T.; Hale, L.; Nostrand, J.D.V.; Wan, S.; Zhou, J. Divergent Taxonomic and Functional Responses of Microbial Communities to Field Simulation of Aeolian Soil Erosion and Deposition. Mol. Ecol. 2017, 26, 4186–4196. [Google Scholar] [CrossRef]
- Zhang, J.; Gao, Q.; Zhang, Q.; Wang, T.; Yue, H.; Wu, L.; Shi, J.; Qin, Z.; Zhou, J.; Zuo, J. Bacteriophage–prokaryote Dynamics and Interaction within Anaerobic Digestion Processes Across Time and Space. Microbiome 2017, 5, 57. [Google Scholar] [CrossRef]
- Dignam, B.E.; O’Callaghan, M.; Condron, L.M.; Kowalchuk, G.A.; Van Nostrand, J.D.; Zhou, J.; Wakelin, S.A. Effect of Land use and Soil Organic Matter Quality on the Structure and Function of Microbial Communities in Pastoral Soils: Implications for Disease Suppression. PLoS ONE 2018, 13, e0196581. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2.0–10. Available online: http://CRAN.R-project.org/package=vegan (accessed on 3 September 2020).
- De Caceres, M.; Jansen, F.; De Caceres, M.M. Package ‘Indicspecies’. Relationship between Species and Groups of Sites. R Package Version. Available online: https://cran.r-project.org/web/packages/indicspecies/index.html (accessed on 3 September 2020).
- Wickham, H. Ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Pennec, E.; Slowikowski, K. Ggwordcloud: A Word Cloud Geom for’Ggplot2’. R Package Version 0.3.0. 2018. Available online: https://cran.r-project.org/package=ggwordcloud (accessed on 3 September 2020).
- Kahle, D.; Wickham, H. Ggmap: Spatial Visualization with Ggplot2. R J. 2013, 5, 144–161. [Google Scholar] [CrossRef]
- Goldstein, C.; Lee, M.D.; Sanchez, S.; Hudson, C.; Phillips, B.; Register, B.; Grady, M.; Liebert, C.; Summers, A.O.; White, D.G.; et al. Incidence of Class 1 and 2 Integrases in Clinical and Commensal Bacteria from Livestock, Companion Animals, and Exotics. Antimicrob. Agents Chemother. 2001, 45, 723–726. [Google Scholar] [CrossRef] [PubMed]
- Niehus, R.; Mitri, S.; Fletcher, A.G.; Foster, K.R. Migration and Horizontal Gene Transfer Divide Microbial Genomes into Multiple Niches. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef]
- Kerrigan, J.F.; Sandberg, K.D.; Engstrom, D.R.; LaPara, T.M.; Arnold, W.A. Small and Large-Scale Distribution of Four Classes of Antibiotics in Sediment: Association with Metals and Antibiotic Resistance Genes. Environ. Sci. Process. Impacts 2018, 20, 1167–1179. [Google Scholar] [CrossRef]
- Sandberg, K.D.; LaPara, T.M. The Fate of Antibiotic Resistance Genes and Class 1 Integrons Following the Application of Swine and Dairy Manure to Soils. FEMS Microbiol. Ecol. 2016, 92. [Google Scholar] [CrossRef]
- Burch, T.R.; Sadowsky, M.J.; LaPara, T.M. Fate of Antibiotic Resistance Genes and Class 1 Integrons in Soil Microcosms Following the Application of Treated Residual Municipal Wastewater Solids. Environ. Sci. Technol. 2014, 48, 5620–5627. [Google Scholar] [CrossRef]
- Gillings, M.R. DNA as a Pollutant: The Clinical Class 1 Integron. Curr. Pollut. Rep. 2018, 4, 49–55. [Google Scholar] [CrossRef]
- Xie, J.; He, Z.; Liu, X.; Liu, X.; Van Nostrand, J.D.; Deng, Y.; Wu, L.; Zhou, J.; Qiu, G. GeoChip-Based Analysis of the Functional Gene Diversity and Metabolic Potential of Microbial Communities in Acid Mine Drainage. Appl. Environ. Microbiol. 2011, 77, 991–999. [Google Scholar] [CrossRef] [PubMed]
- Paula, F.S.; Rodrigues, J.L.M.; Zhou, J.; Wu, L.; Mueller, R.C.; Mirza, B.S.; Bohannan, B.J.M.; Nüsslein, K.; Deng, Y.; Tiedje, J.M.; et al. Land use Change Alters Functional Gene Diversity, Composition and Abundance in Amazon Forest Soil Microbial Communities. Mol. Ecol. 2014, 23, 2988–2999. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Mukherjee, M.M.; Varela, M.F. Modulation of Bacterial Multidrug Resistance Efflux Pumps of the Major Facilitator Superfamily. Int. J. Bacteriol. 2013, 2013, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Von Wintersdorff, C.J.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.; Wolffs, P.F. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef] [PubMed]
- De Mandal, S.; Mathipi, V.; Muthukumaran, R.B.; Gurusubramanian, G.; Lalnunmawii, E.; Kumar, N.S. Amplicon Sequencing and Imputed Metagenomic Analysis of Waste Soil and Sediment Microbiome Reveals Unique Bacterial Communities and their Functional Attributes. Environ. Monit. Assess. 2019, 191, 778. [Google Scholar] [CrossRef]
- Brown, D.; Demeter, M.; Turner, R.J. Prevalence of Multidrug Resistance Efflux Pumps (MDREPs) in Environmental Communities. In Microbial Diversity in the Genomic Era; Das, S., Dash, H.R., Eds.; Academic Press: Amsterdam, The Netherlands, 2019; pp. 545–557. [Google Scholar]
- Shi, K.; Cao, M.; Li, C.; Huang, J.; Zheng, S.; Wang, G. Efflux Proteins MacAB Confer Resistance to Arsenite and Penicillin/Macrolide-Type Antibiotics in Agrobacterium Tumefaciens 5A. World J. Microb. Biot. 2019, 35, 115. [Google Scholar] [CrossRef]
- Meyer, M.T.; Lee, E.A.; Ferrell, G.M.; Bumgarner, J.E.; Varns, J. Evaluation of Offline Tandem and Online Solid-Phase Extraction with Liquid Chromatography/Electrospray Ionization-Mass Spectrometry for Analysis of Antibiotics in Ambient Water and Comparison to an Independent Method; United States Geological Survey: Hunter Mill, VA, USA, 2007. [Google Scholar]
- Kast, J.B.; Long, C.M.; Muenich, R.L.; Martin, J.F.; Kalcic, M.M. Manure Management at Ohio Confined Animal Feeding Facilities in the Maumee River Watershed. J. Great Lakes Res. 2019, 45, 116–1170. [Google Scholar] [CrossRef]
- He, Z.L.; Yang, X.E.; Stoffella, P.J. Trace Elements in Agroecosystems and Impacts on the Environment. J. Trace Elem. Med. Biol. 2005, 19, 125–140. [Google Scholar] [CrossRef]
- Saint-Laurent, D.; Hähni, M.; St-Laurent, J.; Baril, F. Comparative Assessment of Soil Contamination by Lead and Heavy Metals in Riparian and Agricultural Areas (Southern Québec, Canada). Int. J. Environ. Res. Public Health 2010, 7, 3100–3114. [Google Scholar] [CrossRef]
- Cenci-Goga, B.T.; Sechi, P.; Karama, M.; Ciavarella, R.; Pipistrelli, M.V.; Goretti, E.; Elia, A.C.; Gardi, T.; Pallottini, M.; Rossi, R.; et al. Cross-sectional study to identify risk factors associated with the occurrence of antimicrobial resistance genes in honey bees Apis mellifera) in Umbria, Central Italy. Environ. Sci. Pollut. Res. 2020, 27, 9637–9645. [Google Scholar] [CrossRef]
- Jayakaran, A.D.; Ward, A.D. Geometry of Inset Channels and the Sediment Composition of Fluvian Benches in Agricultural Drainage Systems in Ohio. J. Soil Water Conserv. 2007, 62, 296–307. [Google Scholar]
- Murphy, J.; Riley, J.P. A Modified Single Solution Method for the Determination of Phosphate in Natural Waters. Anal. Chim. Acta 1962, 27, 31–36. [Google Scholar] [CrossRef]
- Robertson, G.P. Standard Soil Methods for Long-Term Ecological Research; Oxford University Press: Oxford, UK, 1999; p. 486. [Google Scholar]
- Meyer, M.T.; Bumgarner, J.E.; Varns, J.L.; Daughtridge, J.V.; Thurman, E.M.; Hostetler, K.A. Use of Radioimmunoassay as a Screen for Antibiotics in Confined Animal Feeding Operations and Confirmation by Liquid Chromatography/Mass Spectrometry. Sci. Total Environ. 2000, 248, 181–187. [Google Scholar] [CrossRef]
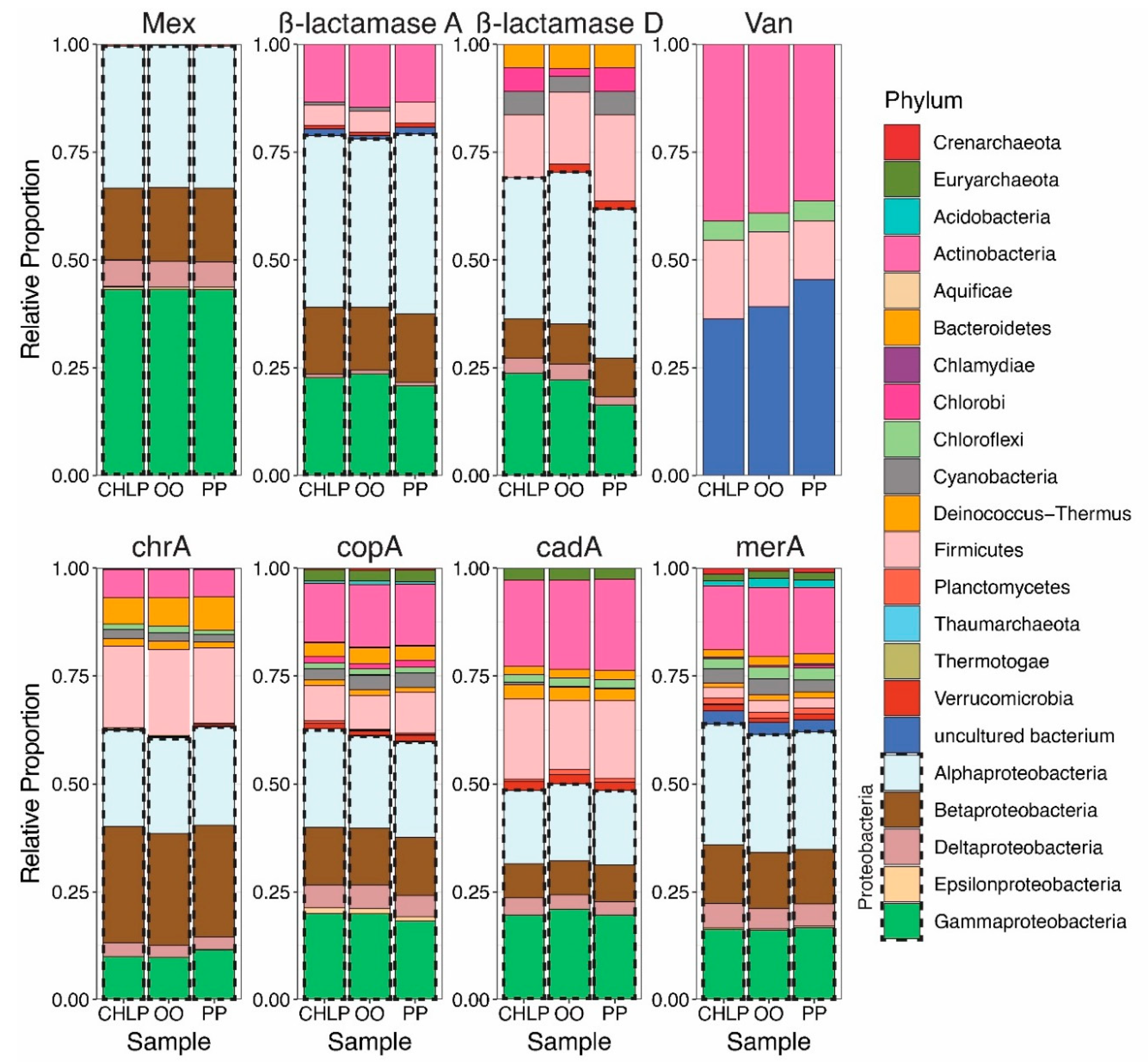
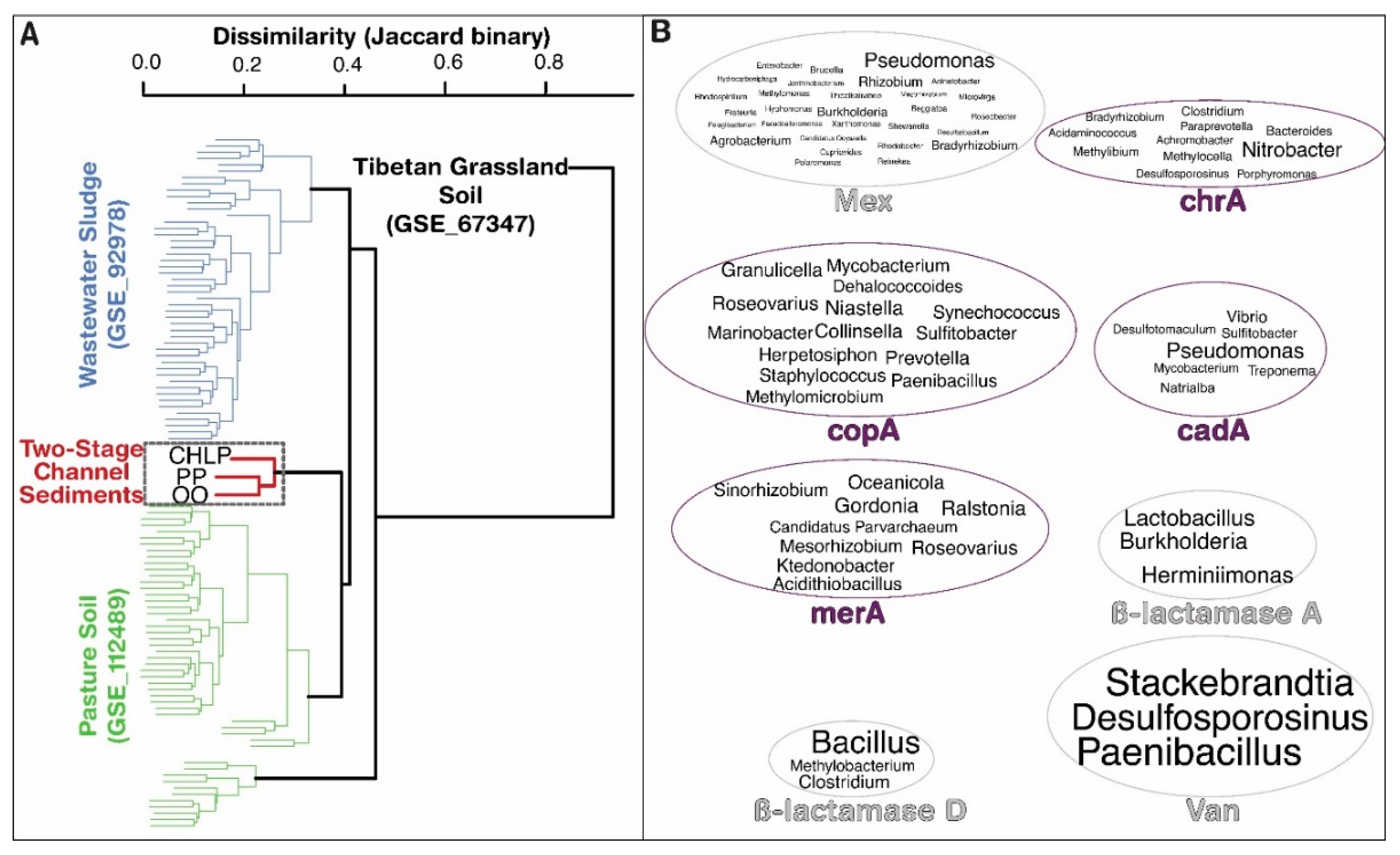
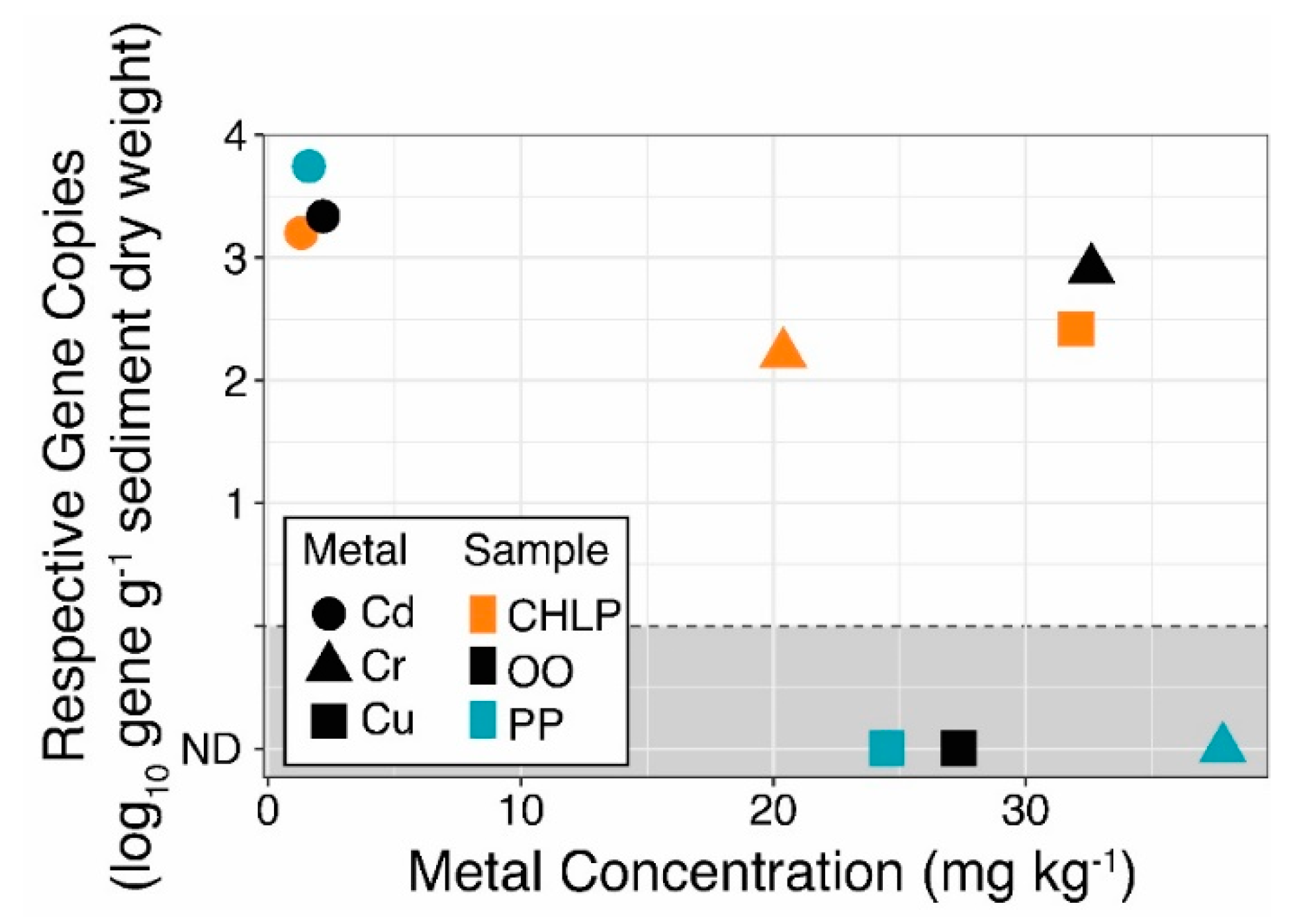
| Antibiotic | Site | ||||||
|---|---|---|---|---|---|---|---|
| CHLP | OO | PP | |||||
| Concnetration (µg kg−1 Dry Sediment) | |||||||
| Sulfanilamides | Sulfapyridine human | <0.3 | est. 0.4 | <0.4 | <0.4 | <0.3 | <0.3 |
| Sulfadiazine human, horse * | <0.02 | <0.02 | <0.02 | <0.02 | <0.02 | <0.02 | |
| Sulfamethoxazole human * | <0.3 | est. 0.5 | <0.4 | <0.4 | <0.4 | <0.4 | |
| Sulfamethazine swine, cattle * | <0.1 | est. 0.1 | <0.2 | <0.2 | <0.1 | <0.1 | |
| Sulfachloropyridazine swine, calf, dog * | <0.03 | <0.03 | <0.04 | <0.04 | <0.03 | <0.03 | |
| Sulfadimethoxine fish, poultry * | <0.1 | <0.1 | <0.3 | <0.3 | <0.2 | <0.2 | |
| Uncategorized | Carbadox swine * | <0.06 | <0.06 | <0.06 | <0.06 | <0.07 | <0.07 |
| Trimethoprim human, horse, dog * | <0.1 | <0.1 | <0.1 | 1.37 | <0.1 | <0.1 | |
| Lincomycin poultry, swine * | <0.01 | <0.01 | <0.01 | <0.01 | <0.8 | <0.8 | |
| Tetracyclines | Tetracycline human, dog, cattle * | <0.04 | <0.04 | <0.02 | <0.02 | <0.02 | <0.02 |
| Oxytetracycline fish, poultry, swine, cattle, sheep, bee, lobster * | <0.02 | <0.02 | <0.02 | <0.02 | <0.04 | <0.04 | |
| Chlortetracycline swine, poultry, cattle, sheep, duck * | <0.02 | <0.02 | <0.03 | <0.03 | <0.03 | <0.03 | |
| Fluoroquinolones | Norfloxacin human, poultry * | <3.1 | <3.1 | <10.3 | <10.3 | <4.1 | <4.1 |
| Ciprofloxacin human, poultry * | <1.3 | <1.3 | <4.5 | <4.5 | <1.7 | <1.7 | |
| Enrofloxacin swine, poultry, cattle, dog, cat | est. 0.8 | est. 1.0 | <0.8 | <0.8 | <0.7 | <0.7 | |
| Ofloxacin human, poultry * | est. 0.4 | est. 0.4 | <1.3 | <1.3 | <0.5 | est. 0.8 | |
| Macrolides | Erythromycin human, poultry, swine * | <0.5 | est. 1.1 | 9.19 | <0.7 | <1.1 | <1.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brooker, M.R.; Arnold, W.A.; Kerrigan, J.F.; LaPara, T.M.; Witter, J.D.; Mouser, P.J. Characterization of Antibiotic Resistance and Metal Homeostasis Genes in Midwest USA Agricultural Sediments. Water 2020, 12, 2476. https://doi.org/10.3390/w12092476
Brooker MR, Arnold WA, Kerrigan JF, LaPara TM, Witter JD, Mouser PJ. Characterization of Antibiotic Resistance and Metal Homeostasis Genes in Midwest USA Agricultural Sediments. Water. 2020; 12(9):2476. https://doi.org/10.3390/w12092476
Chicago/Turabian StyleBrooker, Michael R., William A. Arnold, Jill F. Kerrigan, Timothy M. LaPara, Jonathan D. Witter, and Paula J. Mouser. 2020. "Characterization of Antibiotic Resistance and Metal Homeostasis Genes in Midwest USA Agricultural Sediments" Water 12, no. 9: 2476. https://doi.org/10.3390/w12092476
APA StyleBrooker, M. R., Arnold, W. A., Kerrigan, J. F., LaPara, T. M., Witter, J. D., & Mouser, P. J. (2020). Characterization of Antibiotic Resistance and Metal Homeostasis Genes in Midwest USA Agricultural Sediments. Water, 12(9), 2476. https://doi.org/10.3390/w12092476





