Integrated Taxonomy for Halistemma Species from the Northwest Pacific Ocean
Abstract
1. Introduction
2. Materials and Methods
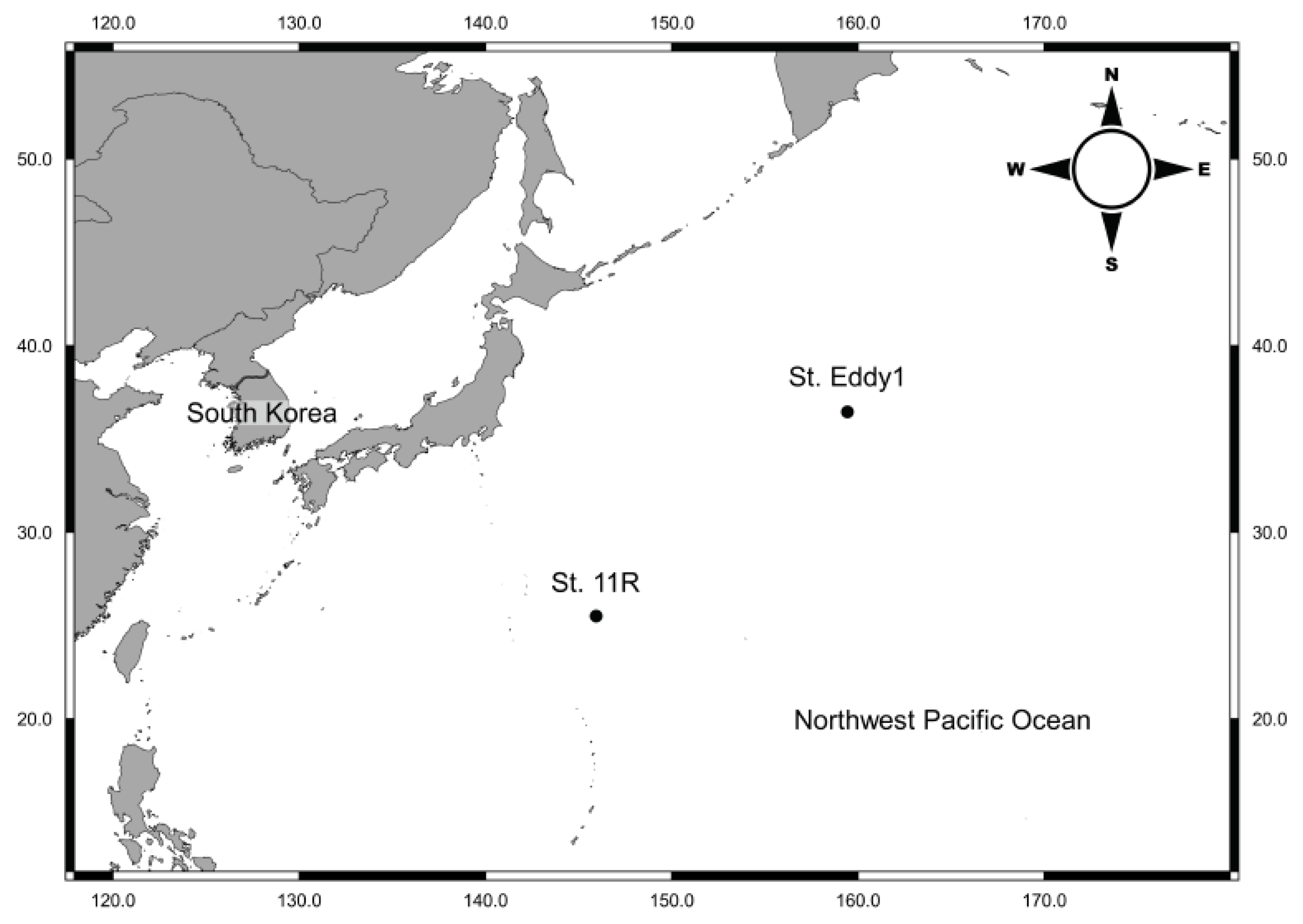
2.1. Sample Collections
2.2. Morphological Analysis
2.3. DNA Extraction, Amplification and Sequencing
2.4. Species Delimitation and Phylogenetic Analysis
3. Results
3.1. Systematics
- Phylum Cnidaria Hatschek, 1888
- Class Hydrozoa Owen, 1843
- Subclass Hydroidolina Collins, 2000
- Order Siphonophorae Eschscholtz, 1829
- Suborder Physonectae Haeckel, 1888
- Family Agalmatidae Brandt, 1834
- Genus Halistemma Huxley, 1859
- Halistemma isabu sp. nov.
3.1.1. Etymology
3.1.2. Locality
3.1.3. Material Examined
3.1.4. Measurements
3.1.5. Nectophore Descriptions
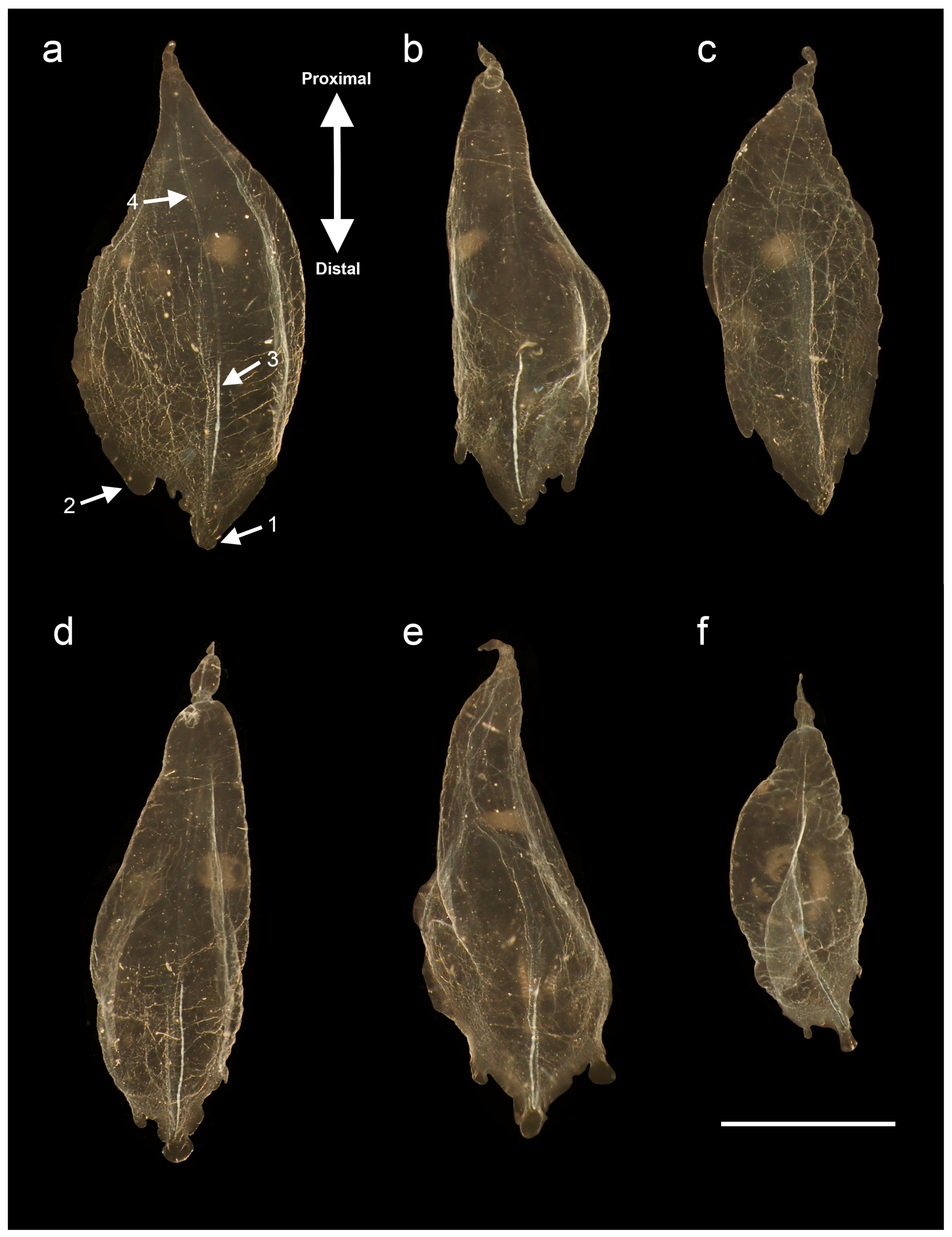
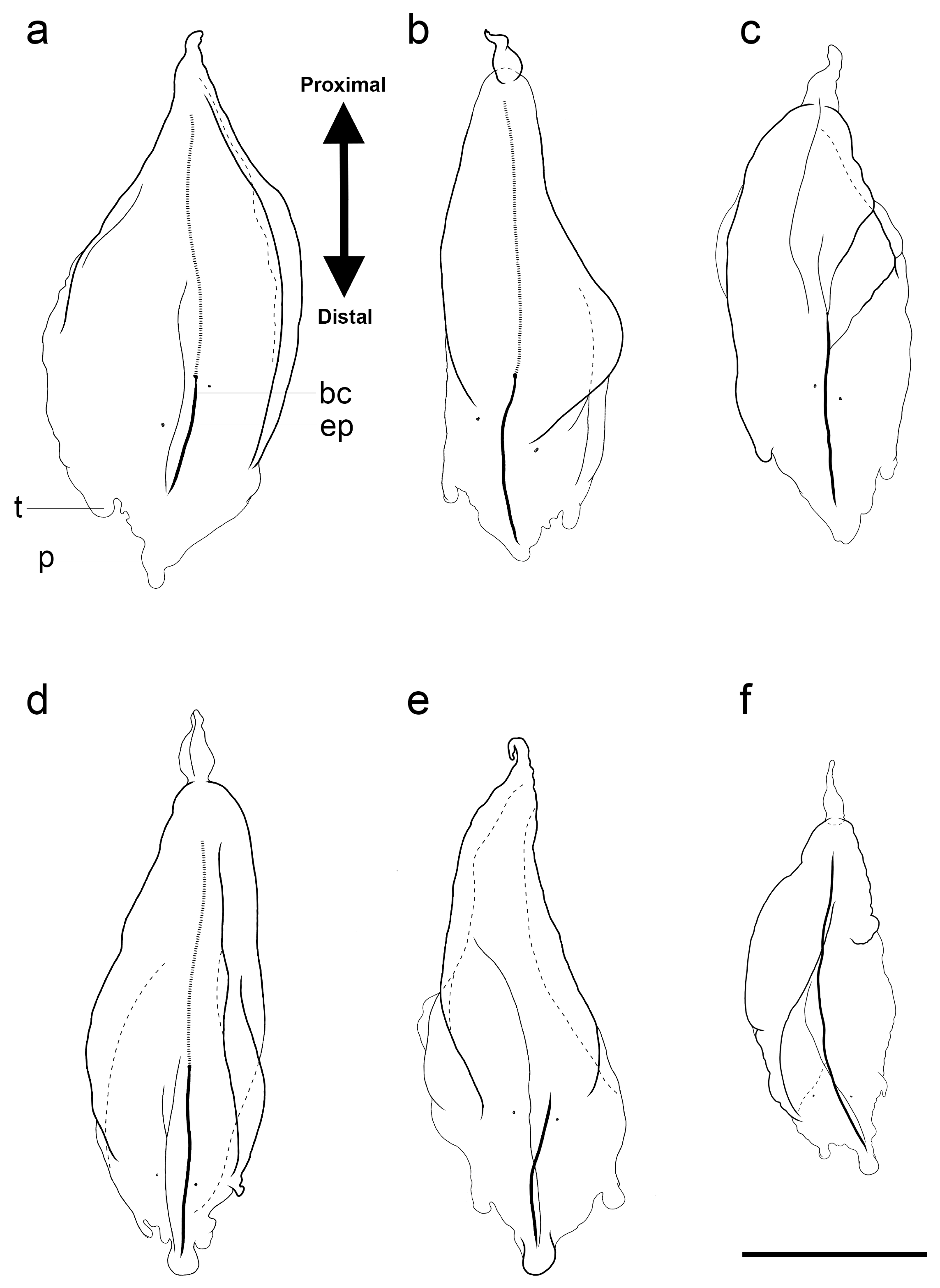
3.1.6. Bract Descriptions
3.2. Molecular Phylogenetic Analysis
3.3. Agalmatidae Sensu Stricto Genetic Diversity
4. Discussion
4.1. Agalmatid Family Phylogeny Based on Molecular Evidences
4.2. Morphological Comparisons of Halistemma Species
4.3. Geographic Distribution of Halistemma
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Totton, A.K.; Bargmann, H.E. British Museum (Natural History). In A Synopsis of the Siphonophora; British Museum (Natural History): London, UK, 1965; 232p. [Google Scholar]
- Munro, C.; Siebert, S.; Zapata, F.; Howison, M.; Damian-Serrano, A.; Church, S.H.; Goetz, F.E.; Pugh, P.R.; Haddock, S.H.; Dunn, C.W. Improved phylogenetic resolution within Siphonophora (Cnidaria) with implications for trait evolution. Mol. Phylogenetics Evol. 2018, 127, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Pugh, P. The vertical distribution of the siphonophores collected during the SOND cruise, 1965. J. Mar. Biol. Assoc. United Kingd. 1974, 54, 25–90. [Google Scholar] [CrossRef]
- Dunn, C.W.; Wagner, G.P. The evolution of colony-level development in the Siphonophora (Cnidaria: Hydrozoa). Dev. Genes Evol. 2006, 216, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Pugh, P. Siphonophorae. In South Atlantic zooplankton; Boltovskoy, D., Ed.; Backhuys Publishers: Leiden, The Netherlands, 1999; pp. 467–512. [Google Scholar]
- Mapstone, G.M. Global diversity and review of Siphonophorae (Cnidaria: Hydrozoa). PLoS ONE 2014, 9, e87737. [Google Scholar] [CrossRef] [PubMed]
- Mapstone, G. Systematics of siphonophores. In Evolution of Venomous Animals and Their Toxins; Springer: Dordrecht, The Netherlands, 2015; pp. 319–366. [Google Scholar]
- Dunn, C.W.; Pugh, P.R.; Haddock, S.H. Molecular phylogenetics of the Siphonophora (Cnidaria), with implications for the evolution of functional specialization. Syst. Biol. 2005, 54, 916–935. [Google Scholar] [CrossRef]
- Grossmann, M.M.; Lindsay, D.J. Diversity and distribution of the Siphonophora (Cnidaria) in Sagami Bay, Japan, and their association with tropical and subarctic water masses. J. Oceanogr. 2013, 69, 395–411. [Google Scholar] [CrossRef]
- Grossmann, M.M.; Lindsay, D.J.; Collins, A.G. The end of an enigmatic taxon: Eudoxia macra is the eudoxid stage of Lensia cossack (Siphonophora, Cnidaria). Syst. Biodivers. 2013, 11, 381–387. [Google Scholar] [CrossRef]
- Grossmann, M.M.; Collins, A.G.; Lindsay, D.J. Description of the eudoxid stages of Lensia havock and Lensia leloupi (Cnidaria: Siphonophora: Calycophorae), with a review of all known Lensia eudoxid bracts. Syst. Biodivers. 2014, 12, 163–180. [Google Scholar] [CrossRef]
- O’Hara, T.D.; Hugall, A.F.; MacIntosh, H.; Naughton, K.M.; Williams, A.; Moussalli, A. Dendrogramma is a siphonophore. Curr. Biol. 2016, 26, R457–R458. [Google Scholar] [CrossRef]
- Panasiuk, A.; Jażdżewska, A.; Słomska, A.; Irzycka, M.; Wawrzynek, J. Genetic identity of two physonect siphonophores from Southern Ocean waters–the enigmatic taxon Mica micula and Pyrostephos vanhoeffeni. J. Mar. Biol. Assoc. United Kingd. 2019, 99, 303–310. [Google Scholar] [CrossRef]
- Pugh, P. The taxonomic status of the genus Moseria (Siphonophora, Physonectae). Zootaxa 2006, 1343, 1–42. [Google Scholar] [CrossRef]
- Huxley, T.H. The Oceanic Hydrozoa; A Description of the Calycophoridae and Physophoridae Observed during the Voyage of HMS “Rattlesnake” in the Years 1846-50; Royal Society: London, UK, 1859. [Google Scholar]
- Pugh, P.; Baxter, E. A review of the physonect siphonophore genera Halistemma (Family Agalmatidae) and Stephanomia (Family Stephanomiidae). Zootaxa 2014, 3897, 1–111. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kirkpatrick, P.; Pugh, P. Siphonophores and Velellids: Keys and Notes for the Identification of the Species; Brill Archive: Leiden, The Netherlands, 1984; Volume 29. [Google Scholar]
- Mapstone, G.M. Siphonophora (Cnidaria, Hydrozoa) of Canadian Pacific Waters; NRC Research Press: Ottawa, ON, Canada, 2009. [Google Scholar]
- Totton, A.K. Siphonophora of the Indian Ocean together with systematic and biological notes on related specimens from other oceans. Disc Rep. 1954, 27, 1–162. [Google Scholar]
- Geller, J.; Meyer, C.; Parker, M.; Hawk, H. Redesign of PCR primers for mitochondrial cytochrome c oxidase subunit I for marine invertebrates and application in all-taxa biotic surveys. Mol. Ecol. Resour. 2013, 13, 851–861. [Google Scholar] [CrossRef]
- Schuchert, P. DNA barcoding of some Pandeidae species (Cnidaria, Hydrozoa, Anthoathecata). Rev. Suisse De Zool. 2018, 125, 101–127. [Google Scholar]
- Ender, A.; Schierwater, B. Placozoa are not derived cnidarians: Evidence from molecular morphology. Mol. Biol. Evol. 2003, 20, 130–134. [Google Scholar] [CrossRef]
- Zheng, L.; He, J.; Lin, Y.; Cao, W.; Zhang, W. 16S rRNA is a better choice than COI for DNA barcoding hydrozoans in the coastal waters of China. Acta Oceanol. Sin. 2014, 33, 55–76. [Google Scholar] [CrossRef]
- Medlin, L.; Elwood, H.J.; Stickel, S.; Sogin, M.L. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 1988, 71, 491–499. [Google Scholar] [CrossRef]
- Collins, A.G. Towards understanding the phylogenetic history of Hydrozoa: Hypothesis testing with 18S gene sequence data. Sci. Mar. 2000, 64, 5–22. [Google Scholar] [CrossRef]
- Strychar, K.B.; Hamilton, L.C.; Kenchington, E.L.; Scott, D.B. Genetic circumscription of deep-water coral species in Canada using 18S rRNA. In Cold-Water Corals and Ecosystems; Springer: Berlin, Germany, 2005; pp. 679–690. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Yamaoka, K.; Nakagawa, T.; Uno, T. Application of Akaike’s information criterion (AIC) in the evaluation of linear pharmacokinetic equations. J. Pharmacokinet. Biopharm. 1978, 6, 165–175. [Google Scholar] [CrossRef]
- Hurvich, C.M.; Tsai, C.-L. Regression and time series model selection in small samples. Biometrika 1989, 76, 297–307. [Google Scholar] [CrossRef]
- Simmons, M.P.; Ochoterena, H. Gaps as characters in sequence-based phylogenetic analyses. Syst. Biol. 2000, 49, 369–381. [Google Scholar] [CrossRef]
- Young, N.D.; Healy, J. GapCoder automates the use of indel characters in phylogenetic analysis. BMC Bioinform. 2003, 4, 6. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.; Ronquist, F. MrBayes: Bayesian inference of phylogeny. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Leloup, E.; Alberto, I. Siphonophores Calycophorides (Suite) et Physophorides Provenant des Campagnes du Prince Albert Ier de Monaco; Imprimerie de Monaco: Monaco, Monaco, 1936. [Google Scholar]
- Margulis, R.Y. Distribution of some siphonophore species of the suborder Physophorae in the Atlantic Ocean. Vestn. Mosk. Univ. 1969, 24, 17–38. [Google Scholar]
- Nogueira, C.R.; Oliveira, S.d.R., Jr. Siphonophora from the coast of Brazil (17ºS to 24ºS). Bol. Inst. Oceanogr. 1991, 39, 61–69. [Google Scholar] [CrossRef]
- Pagès, F.; Gili, J.M. Siphonophores (Cnidaria, Hydrozoa) of the Benguela Current (southeastern Atlantic). Sci. Mar. 1992, 56, 65–112. [Google Scholar]
- Kawamura, T. A report on Japanese siphonophores with special references to new and rare species. J. Shiga Pref. Jr. Coll. Ser. 1954, 2, 99–129. [Google Scholar]
- Yu, P.-W. Seasonal dynamics of siphonophores in the waters off southern and northern Taiwan, 2006. Master’s Thesis, National Sun Yat-Sen University, Kaohsiung City, Taiwan, 2013; pp. 1–137. [Google Scholar]
- Lo, W.-T.; Kang, P.-R.; Hsieh, H.-Y. Siphonophores from a transect off southern Taiwan between Kuroshio Current and South China Sea. Zool. Stud. 2012, 51, 1354–1366. [Google Scholar]
- Carré, D. Etude du développement d’Halistemma rubrum (Vogt 1852) siphonophore physonecte agalmidae. Cah. De Biol. Mar. 1971, 12, 77–93. [Google Scholar]
- Musayeva, E. Distribution of siphonophores in the eastern part of the Indian Ocean. Trans. Inst. Oceanol. 1976, 105, 171–197. [Google Scholar]
- Bouillon, J.; Medel, M.D.; Pagès, F.; Gili, J.M.; Boero, F.; Gravili, C. Fauna of the Mediterranean hydrozoa. Sci. Mar. 2004, 68, 5–438. [Google Scholar] [CrossRef]



| Specimens | Average (Min–Max) (mm) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nectophores | N | TL | TW | OW | NSH | NSW | LAWW | LAWH | RAWW | RAWH | TBW | TBH | LMW | LMH | RMW | RMH |
| 4 | 8.91 (7.99–9.87) | 9.27 (8.27–10.86) | 2.24 (1.57–2.73) | 6.23 (5.48–7.05) | 6.67 (5.64–7.45) | 2.77 (2.44–2.93) | 1.50 (0.74–1.80) | 2.93 (2.69–3.04) | 1.61 (0.95–2.01) | 3.90 (3.16–4.91) | 1.71 (1.54–1.97) | 0.65 (0.48–0.79) | 0.64 (0.49–0.84) | 0.70 (0.46–0.85) | 0.71 (0.59–0.82) | |
| Bracts | N | TL | TW | BALL | BCL | |||||||||||
| 17 | 15.39 (7.42–19.27) | 5.72 (3.64–8.14) | 5.22 (2.78–9.85) | 1.36 (0.49–3.33) | ||||||||||||
| Species | Size (Length × Width) (mm) | Upper Lateral Ridges Shape | Lateral Ridges | Vertical Lateral Ridges | Mouth Plate | Thrust Block Size | Nectosac Shape | Ridges between Upper Lateral and Lateral | Ascending Branch of Mantle Canal Length |
|---|---|---|---|---|---|---|---|---|---|
| H. isabu sp. nov. | ~9.9 × 10.9 | Obliquely inward (Y-shape with hook) | 2 pairs complete, 1 pair incomplete | 1 pair complete | O, deeply divided | >Axial wing | T-shape with stair | O, 1 pair incomplete | >descending branch |
| H. striata | ~23 × 25 | Obliquely inward (Y-shape with stair) | 1 pair complete | 3 pairs complete, 1 pair incomplete | O, deeply divided | ≤Axial wing | T-shape | X | >descending branch |
| H. foliacea | ~20 × 22 | Obliquely inward (Obscure Y-shape) | 1 pair incomplete | 2 pairs complete | O, slightly emarginated | <Axial wing | T-shape | X | =descending branch |
| H. rubrum * | ~7 × 7 | Vertically straight | 1 pair incomplete | 1 pair incomplete | X | <Axial wing | T-shape | X | =descending branch |
| H. maculatum | ~11 × 10 | Obliquely inward (V-shape) | 1 pair complete | 1 pair complete | O, deeply divided | ≥Axial wing | T-shape | X | =descending branch |
| H. cupulifera | ~9.5 × 9 | Obliquely inward (V-shape) | 1 pair complete | 1 pair complete | O, slightly emarginated | ≥Axial wing | T-shape | X | =descending branch |
| H. transliratum | ~13 × 13.4 | Obliquely inward (V-shape) | 1 pair complete | 1 pair complete | X | <Axial wing | T-shape | X | >descending branch |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, N.; Prudkovsky, A.A.; Lee, W. Integrated Taxonomy for Halistemma Species from the Northwest Pacific Ocean. Water 2020, 12, 3283. https://doi.org/10.3390/w12113283
Park N, Prudkovsky AA, Lee W. Integrated Taxonomy for Halistemma Species from the Northwest Pacific Ocean. Water. 2020; 12(11):3283. https://doi.org/10.3390/w12113283
Chicago/Turabian StylePark, Nayeon, Andrey A. Prudkovsky, and Wonchoel Lee. 2020. "Integrated Taxonomy for Halistemma Species from the Northwest Pacific Ocean" Water 12, no. 11: 3283. https://doi.org/10.3390/w12113283
APA StylePark, N., Prudkovsky, A. A., & Lee, W. (2020). Integrated Taxonomy for Halistemma Species from the Northwest Pacific Ocean. Water, 12(11), 3283. https://doi.org/10.3390/w12113283






