The Role of the Motility of Methylobacterium in Bacterial Interactions in Drinking Water
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
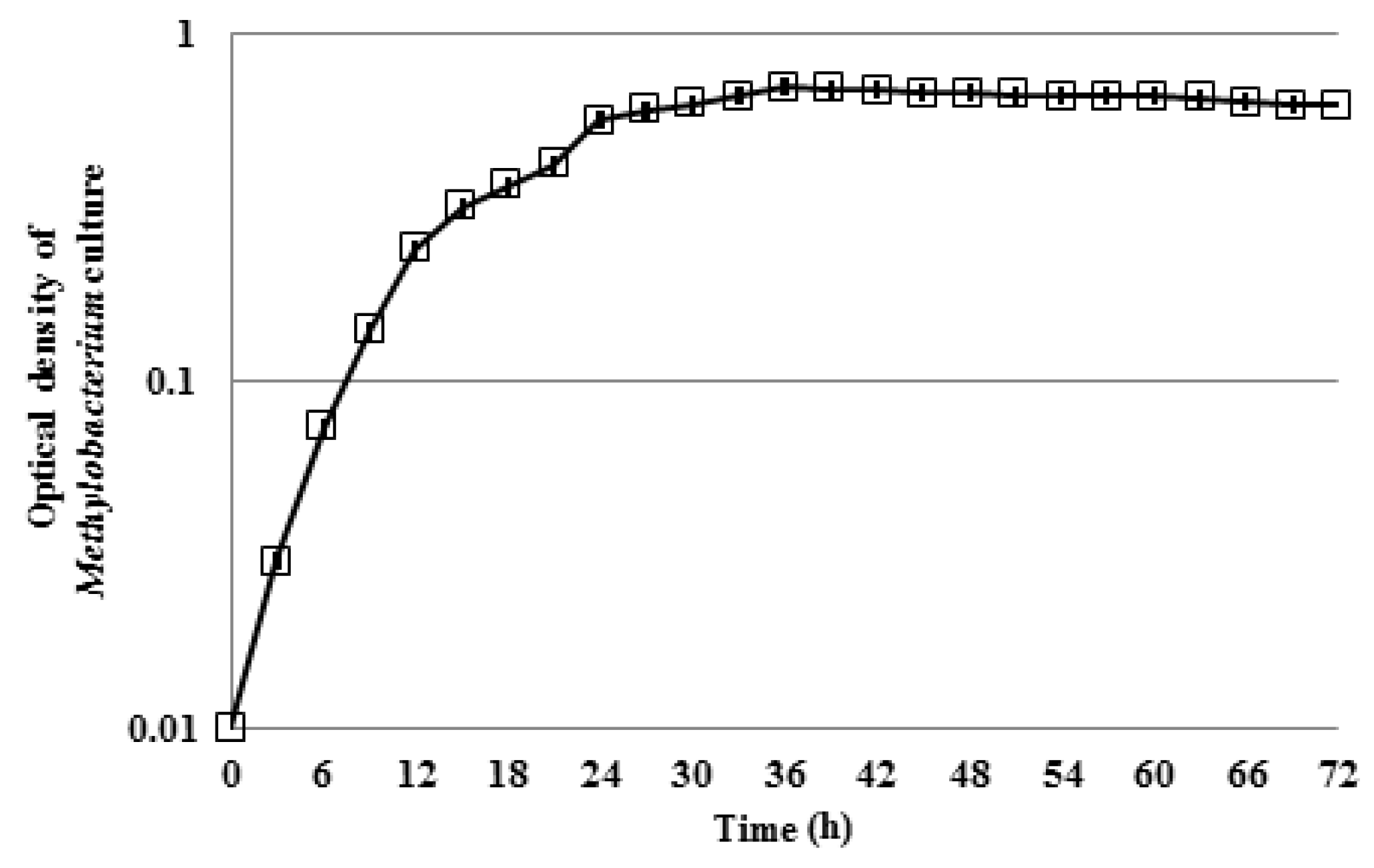
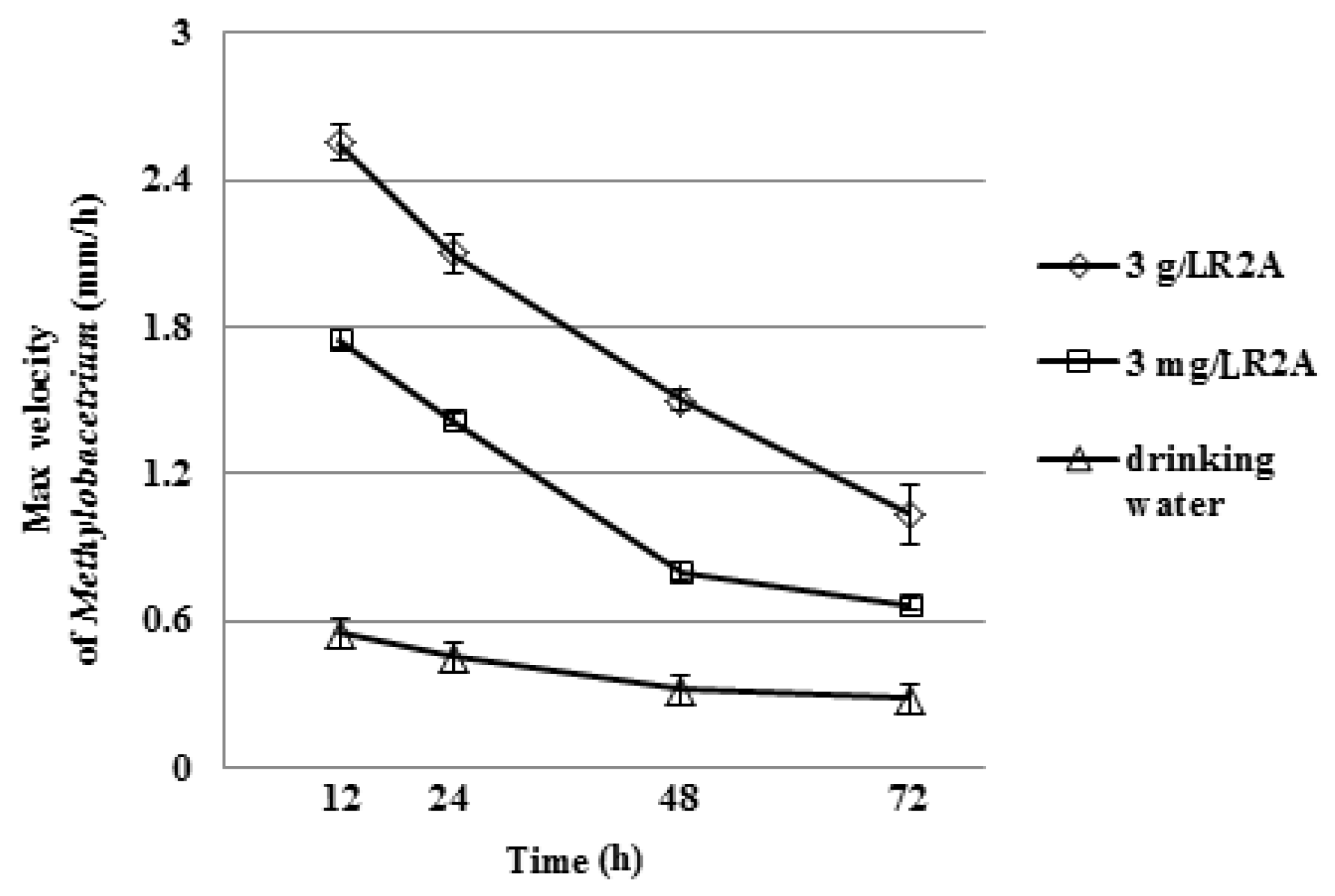
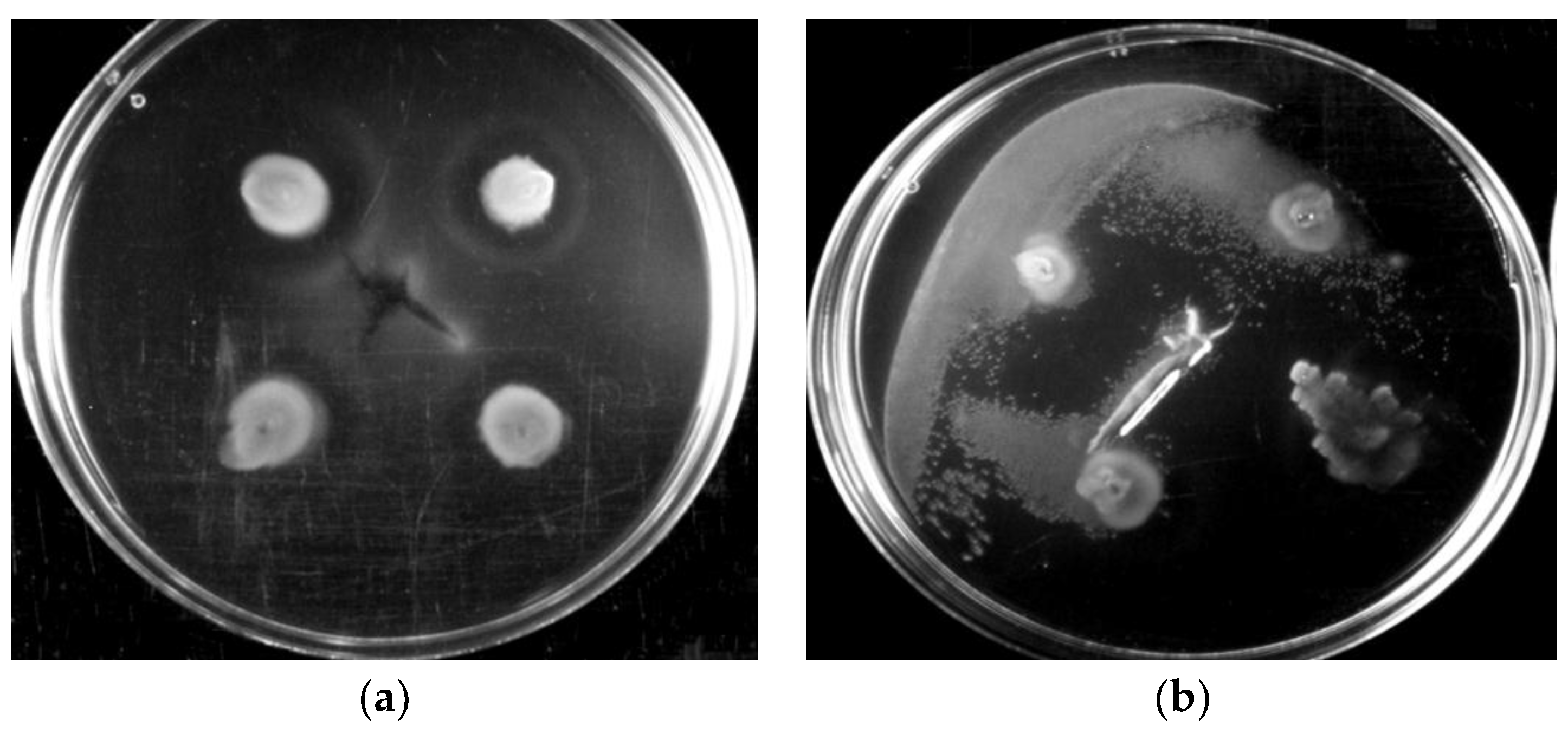
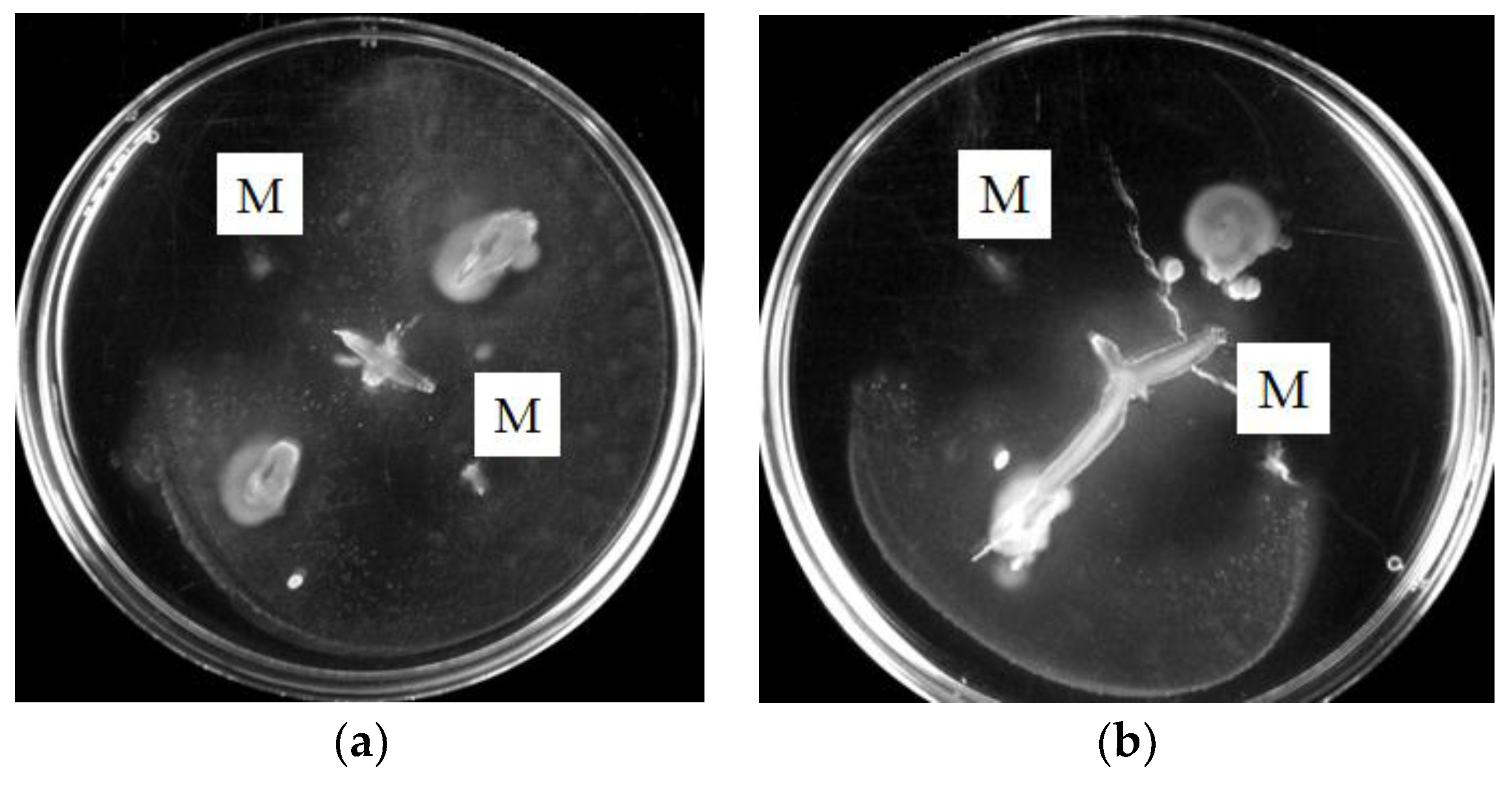
3.1. Motility of Pure Methylobacterium Colonies
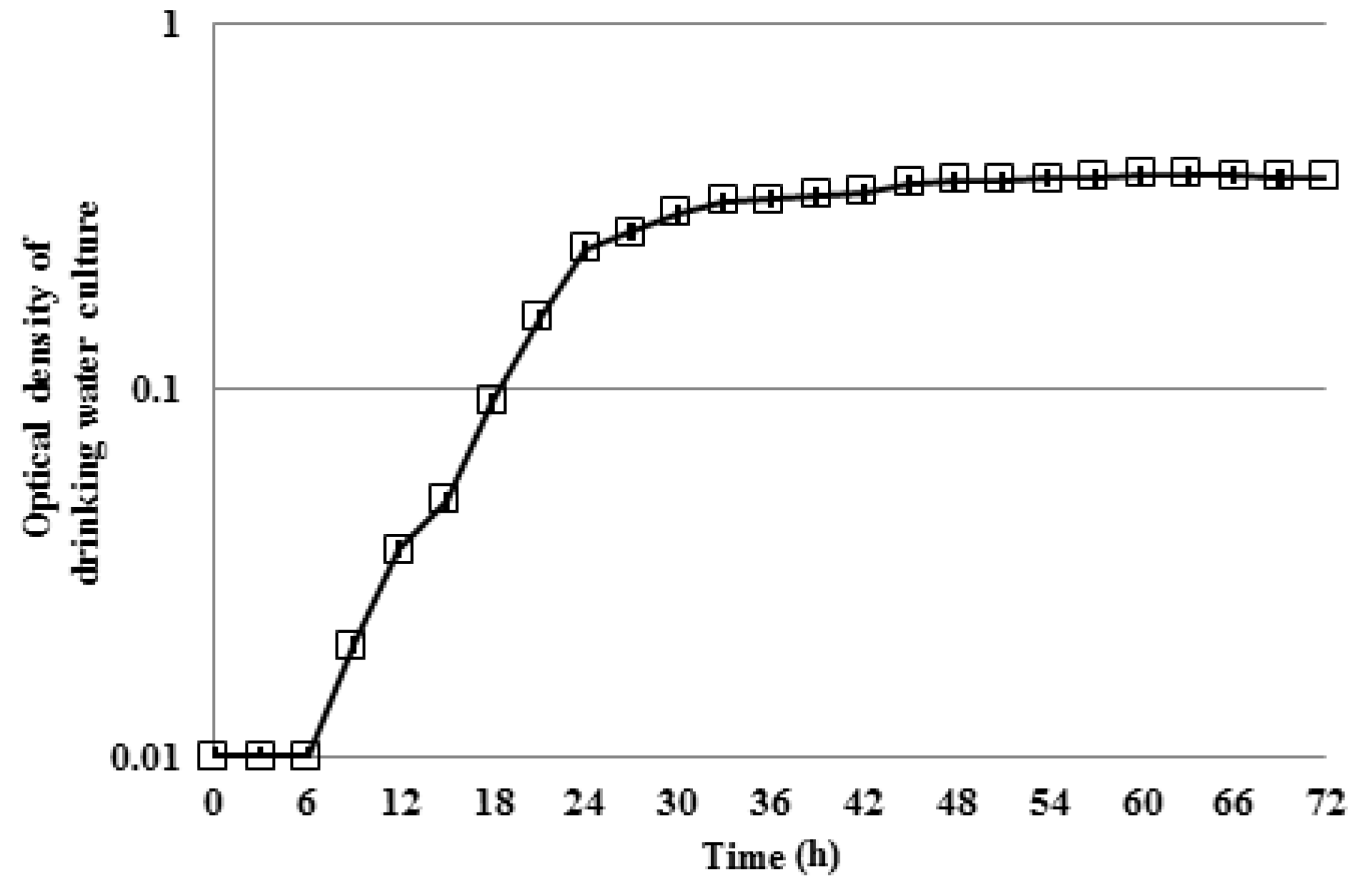
3.2. Interactions of Mixed Drinking Water and Pure Methylobacterium Colonies
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Garrett, T.R.; Bhakoo, M.; Zhang, Z. Bacterial adhesion and biofilms on surfaces. Prog. Nat. Sci. 2008, 18, 1049–1056. [Google Scholar] [CrossRef]
- Hori, K.; Matsumoto, S. Bacterial adhesion: From mechanism to control. Biochem. Eng. J. 2010, 48, 424–434. [Google Scholar] [CrossRef]
- Hall-Stoodley, L.; Stoodley, P. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 2005, 13, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Simões, L.C. Biofilms in Drinking Water, in Biofilms in Drinking Water: Formation and Control; Lambert Academic Publishing: Saarbrücken, Germany, 2012. [Google Scholar]
- Boe-Hansen, R. Microbial Growth in Drinking Water Distribution Systems. Environment & Resources; Technical University of Denmark: Copenhagen, Denmark, 2001. [Google Scholar]
- Kumarasamy, M.V.; Maharaj, P.M. The effect of biofilm growth on wall shear stress in drinking water PVC pipes. Pol. J. Environ. Stud. 2015, 24, 2479–2483. [Google Scholar] [CrossRef]
- Derlon, N.; Masse, A.; Escudie, R.; Bernet, N.; Paul, E. Stratification in the cohesion of biofilms grown under various environmental conditions. Water Res. 2008, 42, 2102–2110. [Google Scholar] [CrossRef] [PubMed]
- Paul, E.; Ochoa, J.C.; Pechaud, Y.; Liu, Y.; Line, A. Effect of shear stress and growth conditions on detachment and physical properties of biofilms. Water Res. 2012, 46, 5499–5508. [Google Scholar] [CrossRef] [PubMed]
- Fraser, G.M.; Claret, L.; Furness, R.; Gupta, S.; Hughes, C. Swarming-coupled expression of the Proteus mirabilis hpmBA haemolysin operon. Microbiology 2002, 148, 2191–2201. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S. Type IV pili and twitching motility. Annu. Rev. Microbiol. 2002, 56, 289–314. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, S.; Hopkins, M.J.; Macfarlane, G.T. Toxin synthesis and mucin breakdown are related to swarming phenomenon in Clostridium septicum. Infect. Immun. 2001, 69, 1120–1126. [Google Scholar] [CrossRef] [PubMed]
- Senesi, S.; Celandroni, F.; Salvetti, S.; Beecher, D.J.; Wong, A.C.L.; Ghelardi, E. Swarming motility in Bacillus cereus and characterization of a fliY mutant impaired in swarm cell differentiation. Microbiology 2002, 148, 1785–1794. [Google Scholar] [CrossRef] [PubMed]
- Mireles, J.R.; Toguchi, A.; Harshey, R.M. Salmonella enterica serovar typhimurium swarming mutants with altered biofilm-forming abilities: Surfactin inhibits biofilm formation. J. Bacteriol. 2001, 183, 5848–5854. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, G.A.; Kolter, R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998, 30, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Pratt, L.A.; Kolter, R. Genetic analysis of Escherichia coli biofilm formation: Roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 1998, 30, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Recht, J.; Kolter, R. Glycopeptidolipid acetylation affects sliding motility and biofilm formation in Mycobacterium smegmatis. J. Bacteriol. 2001, 183, 5718–5724. [Google Scholar] [CrossRef] [PubMed]
- Harshey, R.M. Bacterial motility on a surface: Many ways to a common goal. Ann. Rev. Microbiol. 2003, 57, 249–273. [Google Scholar] [CrossRef] [PubMed]
- Picioreanu, C.; Kreft, J.U.; Klausen, M.; Haagensen, J.A.J.; Tolker-Nielsen, T.; Molin, S. Microbial motility involvement in biofilm structure formation—A 3D modelling study. Water Sci. Technol. 2007, 55, 337. [Google Scholar] [CrossRef] [PubMed]
- Son, K.; Brumley, D.R.; Stocker, R. Live from under the lens: Exploring microbial motility with dynamic imaging and microfluidics. Nat. Rev. Microbiol. 2015, 13, 761–775. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, K.K. What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 2004, 236, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Riedel, K.; Hentzer, M.; Geisenberger, O.; Huber, B.; Steidle, A.; Wu, H.; Hoiby, N.; Givskov, M.; Molin, S.; Eberl, L. N-acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 2001, 147, 3249–3262. [Google Scholar] [CrossRef] [PubMed]
- Huber, B.; Riedel, K.; Hentzer, M.; Heydorn, A.; Gotschlich, A.; Givskov, M.; Molin, S.; Eberl, L. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 2001, 147, 2517–2528. [Google Scholar] [CrossRef] [PubMed]
- Poonguzhali, S.; Madhaiyan, M.; Sa, T. Production of acyl-homoserine lactone quorum-sensing signals is widespread in gram-negative Methylobacterium. J. Microbiol. Biotechnol. 2007, 17, 226–233. [Google Scholar] [PubMed]
- Mattick, J.S. Type IV pili and twitching motility. Ann. Rev. Microbiol. 2002, 56, 289–314. [Google Scholar] [CrossRef] [PubMed]
- Martinez, A.; Torello, S.; Kolter, R. Sliding motility in mycobacteria. J. Bacteriol. 1999, 181, 7331–7338. [Google Scholar] [PubMed]
- Matsuyama, T.; Harshey, R.M.; Matsushita, M. Self-similar colony morphogenesis by bacteria as the experimental model of fractal growth by a cell population. Fractals 1993, 1, 302–311. [Google Scholar] [CrossRef]
- Toguchi, A.; Siano, M.; Burkart, M.; Harshey, R.M. Genetics of swarming motility in Salmonella enterica serovar typhimurium: Critical role for lipopolysaccharide. J. Bacteriol. 2000, 182, 6308–6321. [Google Scholar] [CrossRef] [PubMed]
- Fraser, G.M.; Hughes, C. Swarming motility. Curr. Opin. Microbiol. 1999, 2, 630–635. [Google Scholar] [CrossRef]
- Matsuyama, T.; Bhasin, A.; Harshey, R.M. Mutational analysis of flagellum-independent surface spreading of Serratia marcescens 274 on a low-agar medium. J. Bacteriol. 1995, 177, 987–991. [Google Scholar] [CrossRef] [PubMed]
- Klausen, M.; Aaes-Jorgensen, A.; Molin, S.; Tolker-Nielsen, T. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 2003, 50, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Purevdorj, B.; Stoodley, P. Biofilm structure, behavior, and hydrodynamics. In Microbial Biofilms; ASM Press: Washington, DC, USA, 2004. [Google Scholar]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Craig, L.; Pique, M.E.; Tainer, J.A. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2004, 2, 363–378. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.H.; Kornberg, A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2000, 97, 4885–4890. [Google Scholar] [CrossRef] [PubMed]
- Soutourina, O.A.; Semenova, E.A.; Parfenova, V.V.; Danchin, A.; Bertin, P. Control of bacterial motility by environmental factors in polarly flagellated and peritrichous bacteria isolated from Lake Baikal. Appl. Environ. Microbiol. 2001, 67, 3852–3859. [Google Scholar] [CrossRef] [PubMed]
- Dennis, P.G.; Seymour, J.; Kumbun, K.; Tyson, G.W. Diverse populations of lake water bacteria exhibit chemotaxis towards inorganic nutrients. ISME J. 2013, 7, 1661–1664. [Google Scholar] [CrossRef] [PubMed]
- Stocker, R.; Seymour, J.R.; Samadani, A.; Hunt, D.E.; Polz, M.F. Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc. Natl. Acad. Sci. USA 2008, 105, 4209–4214. [Google Scholar] [CrossRef] [PubMed]
- Samad, T.; Billings, N.; Birjiniuk, A.; Crouzier, T.; Doyle, P.S.; Ribbeck, K. Swimming bacteria promote dispersal of non-motile staphylococcal species. ISME J. 2017, 11, 1933–1937. [Google Scholar] [CrossRef] [PubMed]
- Galajda, P.; Keymer, J.; Chaikin, P.; Austin, R. A wall of funnels concentrates swimming bacteria. J. Bacteriol. 2007, 189, 8704–8707. [Google Scholar] [CrossRef] [PubMed]
- Kearns, D.B. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 2010, 8, 634–644. [Google Scholar] [CrossRef] [PubMed]
- Schauer, S.; Kutschera, U. A novel growth-promoting microbe, Methylobacterium funariae sp. nov., isolated from the leaf surface of a common moss. Plant Signal. Behav. 2011, 6, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Doerges, L.; Kutschera, U. Assembly and loss of the polar flagellum in plant-associated methylobacteria. Naturwissenschaften 2014, 101, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Gallego, V.; Garcia, M.T.; Ventosa, A. Methylobacterium variabile sp. nov., a methylotrophic bacterium isolated from an aquatic environment. Int. J. Syst. Evol. Microbiol. 2005, 55, 1429–1433. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sahr, F.; Xue, T.; Sun, B. Methylobacterium salsuginis sp. nov., isolated from seawater. Int. J. Syst. Evol. Microbiol. 2007, 57, 1699–1703. [Google Scholar] [CrossRef] [PubMed]
- Veyisoglu, A.; Camas, M.; Tatar, D.; Guven, K.; Sazaki, A.; Sahin, N. Methylobacterium tarhaniae sp. nov., isolated from arid soil. Int. J. Syst. Evol. Microbiol. 2013, 63, 2823–2828. [Google Scholar] [CrossRef] [PubMed]
- Weon, H.Y.; Kim, B.Y.; Joa, J.H.; Son, J.A.; Song, M.H.; Kwon, S.W.; Go, S.J.; Yoon, S.H. Methylobacterium iners sp. nov. and Methylobacterium aerolatum sp. nov., isolated from air samples in Korea. Int. J. Syst. Evol. Microbiol. 2008, 58, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Gunawan, C.; Barraud, N.; Rice, S.A.; Harry, E.J.; Amal, R. Understanding, monitoring, and controlling biofilm growth in drinking water distribution systems. Environ. Sci. Technol. 2016, 50, 8954–8976. [Google Scholar] [CrossRef] [PubMed]
- Tsagkari, E.; Sloan, W.T. Turbulence accelerates the growth of drinking water biofilms. Bioprocess Biosyst. Eng. 2018, 41, 757–770. [Google Scholar] [CrossRef] [PubMed]
- Tsagkari, E.; Keating, C.; Couto, J.M.; Sloan, W.T. A keystone Methylobacterium strain in biofilm formation in drinking water. Water 2017, 9, 778. [Google Scholar] [CrossRef]
- Hauwaerts, D.; Alexandre, G.; Das, S.K.; Vanderleyden, J.; Zhulin, I.B. A major chemotaxis gene cluster in Azospirillum brasilense and relationships between chemotaxis operons in alpha-proteobacteria. FEMS Microbiol. Lett. 2002, 208, 61–67. [Google Scholar] [PubMed]
- Rasmussen, L.; White, E.L.; Pathak, A.; Ayala, J.C.; Wang, H.X.; Wu, J.H.; Benitez, J.A.; Silva, A.J. A high-throughput screening assay for inhibitors of bacterial motility identifies a novel inhibitor of the Na+-driven flagellar motor and virulence gene expression in Vibrio cholerae. Antimicrob. Agents Chemother. 2011, 55, 4134–4143. [Google Scholar] [CrossRef] [PubMed]
- Simões, L.C.; Simões, M.; Vieira, M.J. Biofilm interactions between distinct bacterial genera isolated from drinking water. Appl. Environ. Microbiol. 2007, 73, 6192–6200. [Google Scholar] [CrossRef] [PubMed]
- Reasoner, D.J.; Geldreich, E.E. A New Medium for the Enumeration and Subculture of Bacteria from Potable Water. Appl. Environ. Microbiol. 1985, 49, 1–7. [Google Scholar] [PubMed]
- Wolfe, A.J.; Berg, H.C. Migration of bacteria in semisolid agar. Proc. Natl. Acad. Sci. USA 1989, 86, 6973–6977. [Google Scholar] [CrossRef] [PubMed]
- Ben-Jacob, E.; Schohet, O.; Tenenbaum, A.; Cohen, I.; Czirók, A.; Vicsek, T. Generic modelling of cooperative growth patterns in bacterial colonies. Nature 1994, 368, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Douterelo, I.; Sharpe, R.L.; Boxall, J.B. Influence of hydraulic regimes on bacterial community structure and composition in an experimental drinking water distribution system. Water Res. 2013, 47, 503–516. [Google Scholar] [CrossRef] [PubMed]


| First Set of Experiments | Colonies | Medium | Agar (%) | Temperature (°C) |
|---|---|---|---|---|
| 1 | 4 pure Methylobacterium | 3 g/L R2A | 0.2, 0.3, 1 | 28, 16 |
| 2 | 4 pure Methylobacterium | 3 g/L R2A and 3 mg/L R2A | 0.2 | 28 |
| 3 | 4 pure Methylobacterium | Drinking water | 0.2 | 28 |
| Second Set of Experiments | Colonies | Medium | Agar (%) | Temperature (°C) |
|---|---|---|---|---|
| 1 | 4 drinking water colonies versus 4 drinking water colonies with 1% Methylobacterium | Drinking water | 0.2 | 28 |
| 2 | 2 drinking water colonies + 2 Methylobacterium colonies versus 2 drinking water colonies with 1% Methylobacterium + 2 Methylobacterium colonies | Drinking water | 0.2 | 28 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsagkari, E.; Sloan, W.T. The Role of the Motility of Methylobacterium in Bacterial Interactions in Drinking Water. Water 2018, 10, 1386. https://doi.org/10.3390/w10101386
Tsagkari E, Sloan WT. The Role of the Motility of Methylobacterium in Bacterial Interactions in Drinking Water. Water. 2018; 10(10):1386. https://doi.org/10.3390/w10101386
Chicago/Turabian StyleTsagkari, Erifyli, and William T. Sloan. 2018. "The Role of the Motility of Methylobacterium in Bacterial Interactions in Drinking Water" Water 10, no. 10: 1386. https://doi.org/10.3390/w10101386
APA StyleTsagkari, E., & Sloan, W. T. (2018). The Role of the Motility of Methylobacterium in Bacterial Interactions in Drinking Water. Water, 10(10), 1386. https://doi.org/10.3390/w10101386





