PRDM Histone Methyltransferase mRNA Levels Increase in Response to Curative Hormone Treatment for Cryptorchidism-Dependent Male Infertility
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Biopsy Sample Collection
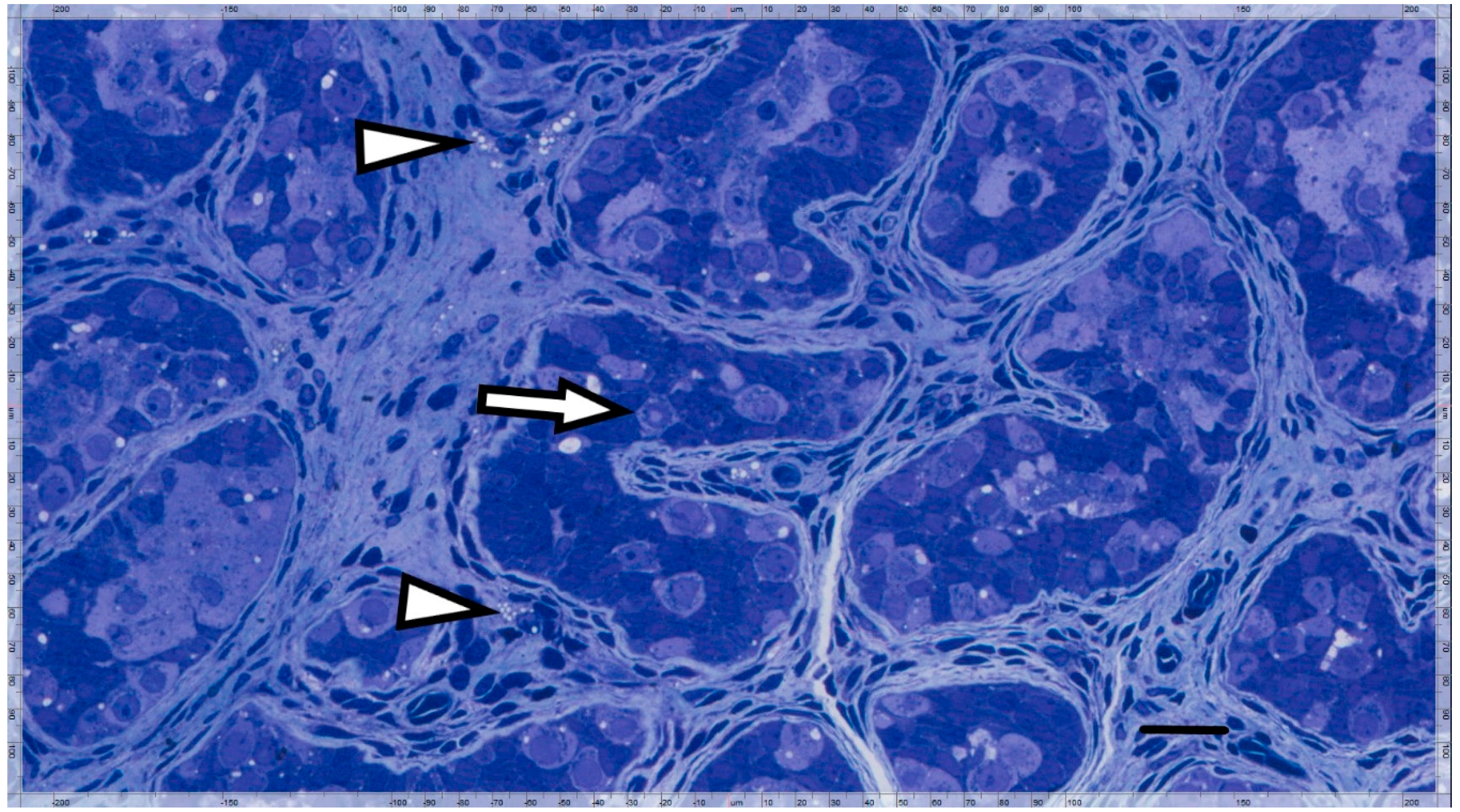
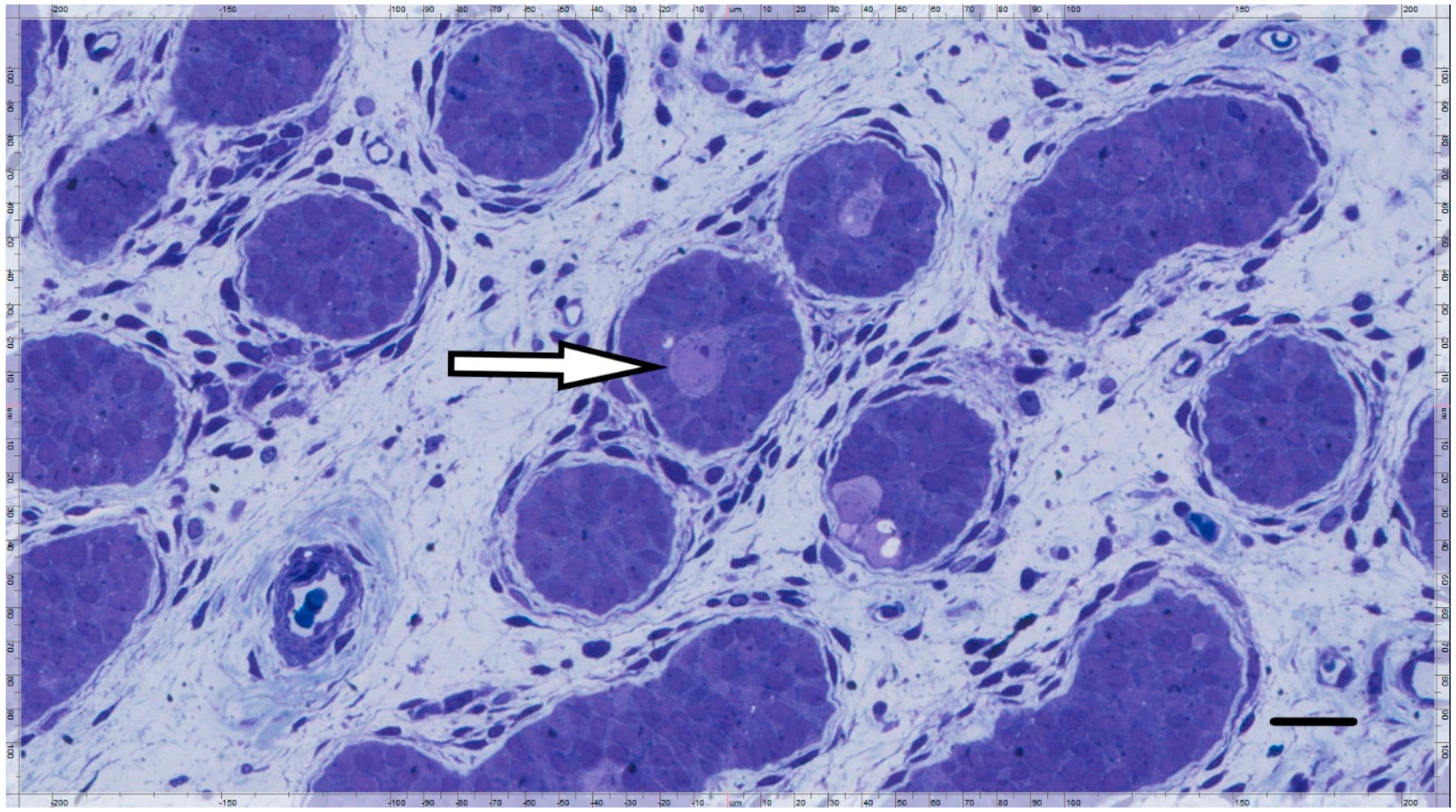
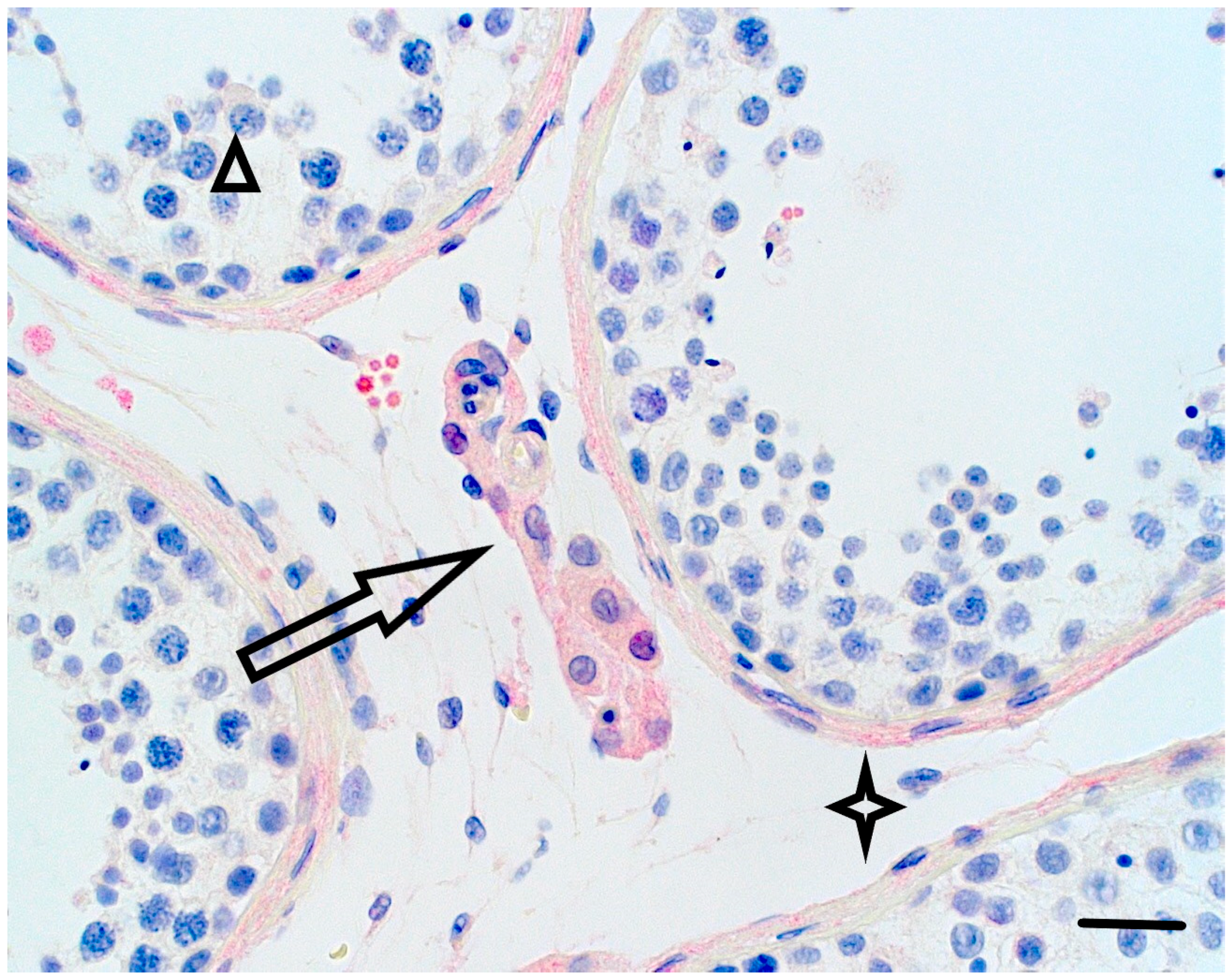
2.2. Immunohistochemical Analyses
2.3. RNA Preparation, Sequencing, Data Analyses, and RNA Expression Levels
2.4. RNA-Sequencing Data and Differential Gene Expression Analysis
2.5. Ethics Statement
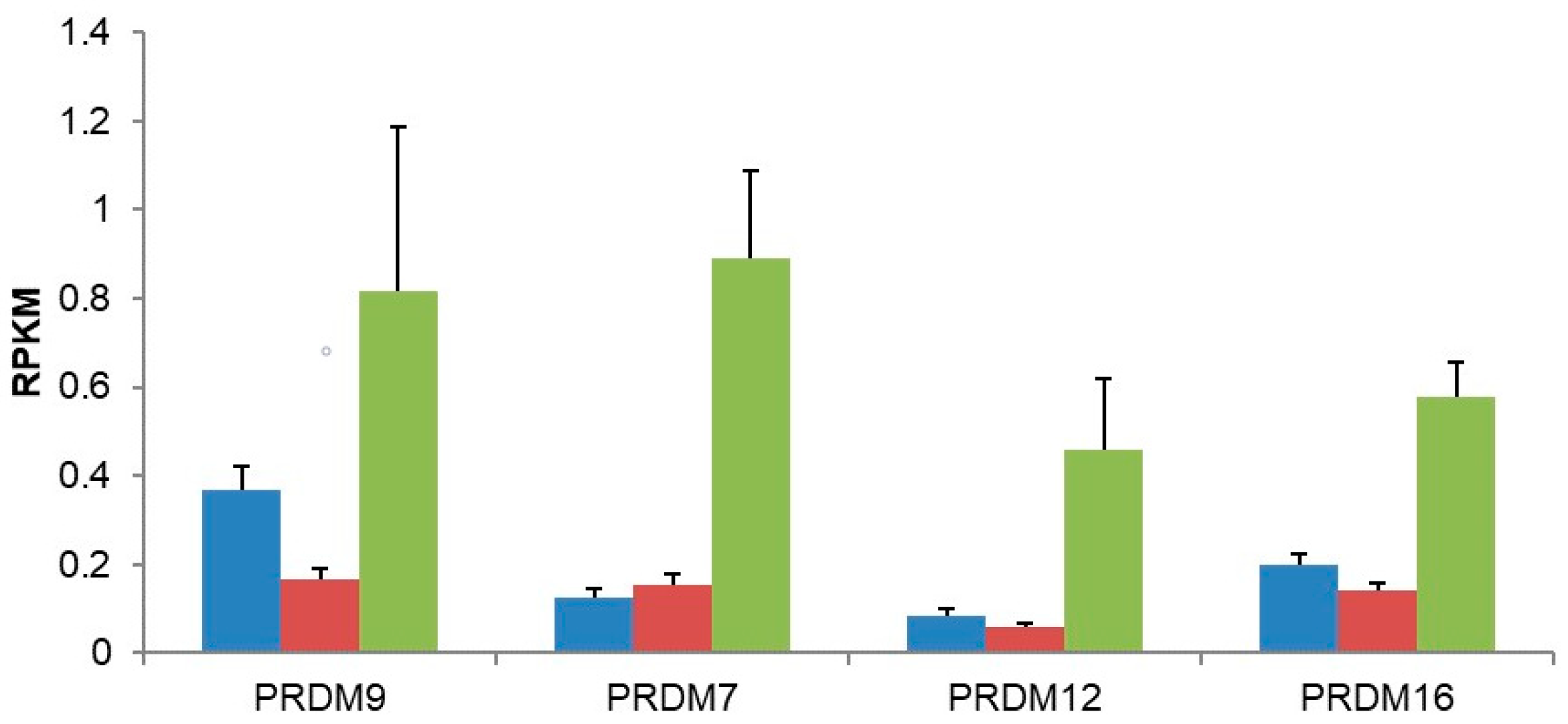
3. Results
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Amann, R.P.; Veeramachaneni, D.N. Cryptorchidism in common eutherian mammals. Reproduction 2007, 133, 541–561. [Google Scholar] [CrossRef] [PubMed]
- Gurney, J.K.; McGlynn, K.A.; Stanley, J.; Merriman, T.; Signal, V.; Shaw, C.; Edwards, R.; Richiardi, L.; Hutson, J.; Sarfati, D. Risk factors for cryptorchidism. Nat. Rev. Urol. 2017, 14, 534–548. [Google Scholar] [CrossRef] [PubMed]
- Docampo, M.J.; Hadziselimovic, F. Molecular pathology of cryptorchidism-induced infertility. Sex. Dev. 2015, 9, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Cortes, D.; Holt, R.; de Knegt, V.E. Hormonal aspects of the pathogenesis and treatment of cryptorchidism. Eur. J. Pediatr. Surg. 2016, 26, 409–417. [Google Scholar] [PubMed]
- Hadziselimovic, F.; Herzog, B. The importance of both an early orchidopexy and germ cell maturation for fertility. Lancet 2001, 358, 1156–1157. [Google Scholar] [CrossRef]
- Hadziselimovic, F.; Hocht, B.; Herzog, B.; Buser, M.W. Infertility in cryptorchidism is linked to the stage of germ cell development at orchidopexy. Horm. Res. 2007, 68, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Zivkovic, D.; Bica, D.G.; Hadziselimovic, F. Effects of hormonal treatment on the contralateral descended testis in unilateral cryptorchidism. J. Pediatr. Urol. 2006, 2, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Gegenschatz-Schmid, K.; Verkauskas, G.; Demougin, P.; Bilius, V.; Dasevicius, D.; Stadler, M.B.; Hadziselimovic, F. DMRTC2, PAX7, BRACHYURY/T and TERT are implicated in male germ cell development following curative hormone treatment for cryptorchidism-induced infertility. Genes 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Hadziselimovic, F.; Gegenschatz-Schmid, K.; Verkauskas, G.; Docampo-Garcia, M.J.; Demougin, P.; Bilius, V.; Malcius, D.; Dasevicius, D.; Stadtler, M.B. Gene expression changes underlying idiopathic central hypogonadism in cryptorchidism with defective mini-puberty. Sex. Dev. 2016, 10, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Hadziselimovic, F.; Hadziselimovic, N.O.; Demougin, P.; Krey, G.; Hoecht, B.; Oakeley, E.J. EGR4 is a master gene responsible for fertility in cryptorchidism. Sex. Dev. 2009, 3, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Hadziselimovic, F.; Hadziselimovic, N.O.; Demougin, P.; Krey, G.; Oakeley, E. Piwi-pathway alteration induces LINE-1 transposon derepression and infertility development in cryptorchidism. Sex. Dev. 2015, 9, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Hadziselimovic, F.; Hadziselimovic, N.O.; Demougin, P.; Krey, G.; Oakeley, E.J. Deficient expression of genes involved in the endogenous defense system against transposons in cryptorchid boys with impaired mini-puberty. Sex. Dev. 2011, 5, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Hadziselimovic, F.; Hadziselimovic, N.O.; Demougin, P.; Oakeley, E.J. Testicular gene expression in cryptorchid boys at risk of azoospermia. Sex. Dev. 2011, 5, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Hadziselimovic, F.; Hadziselimovic, N.O.; Demougin, P.; Oakeley, E.J. Decreased expression of genes associated with memory and X-linked mental retardation in boys with non-syndromic cryptorchidism and high infertility risk. Mol. Syndromol. 2014, 5, 76–80. [Google Scholar] [CrossRef] [PubMed]
- Krausz, C.; Escamilla, A.R.; Chianese, C. Genetics of male infertility: From research to clinic. Reproduction 2015, 150, R159–R174. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, T.G.; Aston, K.I.; James, E.R.; Carrell, D.T. Sperm epigenetics in the study of male fertility, offspring health, and potential clinical applications. Syst. Biol. Reprod. Med. 2017, 63, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Stewart, K.R.; Veselovska, L.; Kelsey, G. Establishment and functions of DNA methylation in the germline. Epigenomics 2016, 8, 1399–1413. [Google Scholar] [CrossRef] [PubMed]
- Carrell, D.T.; Aston, K.I.; Oliva, R.; Emery, B.R.; De Jonge, C.J. The “omics” of human male infertility: Integrating big data in a systems biology approach. Cell. Tissue Res. 2016, 363, 295–312. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yan, W.; Duan, E. Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat. Rev. Genet. 2016, 17, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Mzoughi, S.; Tan, Y.X.; Low, D.; Guccione, E. The role of PRDMs in cancer: One family, two sides. Curr. Opin. Genet. Dev. 2016, 36, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Hohenauer, T.; Moore, A.W. The Prdm family: Expanding roles in stem cells and development. Development 2012, 139, 2267–2282. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Zhang, H.; Tang, W.W.C.; Irie, N.; Withey, S.; Klisch, D.; Sybirna, A.; Dietmann, S.; Contreras, D.A.; Webb, R.; et al. Principles of early human development and germ cell program from conserved model systems. Nature 2017, 546, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Shirane, K.; Kurimoto, K.; Yabuta, Y.; Yamaji, M.; Satoh, J.; Ito, S.; Watanabe, A.; Hayashi, K.; Saitou, M.; Sasaki, H. Global landscape and regulatory principles of DNA methylation reprogramming for germ cell specification by mouse pluripotent stem cells. Dev. Cell. 2016, 39, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, M.E.; Flenniken, A.M.; Ji, X.; Teboul, L.; Wong, M.D.; White, J.K.; Meehan, T.F.; Weninger, W.J.; Westerberg, H.; Adissu, H.; et al. High-throuput discovery of novel developmental phenotypes. Nature 2016, 537, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Smagulova, F.; Brick, K.; Pu, Y.; Camerini-Otero, R.D.; Petukhova, G.V. The evolutionary turnover of recombination hot spots contributes to speciation in mice. Genes Dev. 2016, 30, 266–280. [Google Scholar] [CrossRef] [PubMed]
- Parvanov, E.D.; Tian, H.; Billings, T.; Saxl, R.L.; Spruce, C.; Aithal, R.; Krejci, L.; Paigen, K.; Petkov, P.M. PRDM9 interactions with other proteins provide a link between recombination hotspots and the chromosomal axis in meiosis. Mol. Biol. Cell 2017, 28, 488–499. [Google Scholar] [CrossRef] [PubMed]
- Smagulova, F.; Gregoretti, I.V.; Brick, K.; Khil, P.; Camerini-Otero, R.D.; Petukhova, G.V. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 2011, 472, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Hadziselimovic, F.; Gegenschatz-Schmid, K.; Verkauskas, G.; Demougin, P.; Bilius, V.; Dasevicius, D.; Stadler, M.B. GnRHa treatment of cryptorchid boys affects genes involved in hormonal control of the HPG axis and fertility. Sex. Dev. 2017, 11, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Vincel, B.; Verkauskas, G.; Bilius, V.; Dasevicius, D.; Malcius, D.; Jones, B.; Hadziselimovic, F. Gonadotropin-releasing hormone agonist corrects defective mini- puberty in boys with cryptorchidism: A prospective randomized study. BioMed Res. Int. 2018. [Google Scholar] [CrossRef]
- Gaudet, P.; Michel, P.A.; Zahn-Zabal, M.; Britan, A.; Cusin, I.; Domagalski, M.; Duek, P.D.; Gateau, A.; Gleizes, A.; Hinard, V.; et al. The neXtProt knowledgebase on human proteins: 2017 update. Nucleic Acids Res. 2017, 45, D177–D182. [Google Scholar] [CrossRef] [PubMed]
- Mona, B.; Uruena, A.; Kollipara, R.K.; Ma, Z.; Borromeo, M.D.; Chang, J.C.; Johnson, J.E. Repression by PRDM13 is critical for generating precision in neuronal identity. Elife 2017, 29, E25787. [Google Scholar] [CrossRef] [PubMed]
- Meehan, T.F.; Conte, N.; West, D.B.; Jacobsen, J.O.; Mason, J.; Warren, J.; Chen, C.K.; Tudose, I.; Relac, M.; Matthews, P.; et al. Disease model discovery from 3328 gene knockouts by the international mouse phenotyping consortium. Nat. Genet. 2017, 49, 1231–1238. [Google Scholar] [CrossRef] [PubMed]
- Ohinata, Y.; Ohta, H.; Shigeta, M.; Yamanaka, K.; Wakayama, T.; Saitou, M. A signaling principle for the specification of the germ cell lineage in mice. Cell 2009, 137, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Nakaki, F.; Hayashi, K.; Ohta, H.; Kurimoto, K.; Yabuta, Y.; Saitou, M. Induction of mouse germ-cell fate by transcription factors in vitro. Nature 2013, 501, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Ohinata, Y.; Payer, B.; O’Carroll, D.; Ancelin, K.; Ono, Y.; Sano, M.; Barton, S.C.; Obukhanych, T.; Nussenzweig, M.; Tarakhovsky, A.; et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 2005, 436, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Vincent, S.D.; Dunn, N.R.; Sciammas, R.; Shapiro-Shalef, M.; Davis, M.M.; Calame, K.; Bikoff, E.K.; Robertson, E.J. The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development 2005, 132, 1315–1325. [Google Scholar] [CrossRef] [PubMed]
- Kurimoto, K.; Yamaji, M.; Seki, Y.; Saitou, M. Specification of the germ cell lineage in mice: A process orchestrated by the PR-domain proteins, Blimp1 and Prdm14. Cell Cycle 2008, 7, 3514–3518. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, M.; Seki, Y.; Kurimoto, K.; Yabuta, Y.; Yuasa, M.; Shigeta, M.; Yamanaka, K.; Ohinata, Y.; Saitou, M. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat. Genet. 2008, 40, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Saitou, M.; Kagiwada, S.; Kurimoto, K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development 2012, 139, 15–31. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.M.; Shinkai, Y. Prdm12 is induced by retinoic acid and exhibits anti-proliferative properties through the cell cycle modulation of P19 embryonic carcinoma cells. Cell. Struct. Funct. 2013, 38, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Auer-Grumbach, M.; Matsukawa, S.; Zitzelsberger, M.; Themistocleous, A.C.; Strom, T.M.; Samara, C.; Moore, A.W.; Cho, L.T.; Young, G.T.; et al. Transcriptional regulator PRDM12 is essential for human pain perception. Nat. Genet. 2015, 47, 803–808. [Google Scholar] [CrossRef] [PubMed]
- Nagy, V.; Cole, T.; Van Campenhout, C.; Khoung, T.M.; Leung, C.; Vermeiren, S.; Novatchkova, M.; Wenzel, D.; Cikes, D.; Polyansky, A.A.; et al. The evolutionarily conserved transcription factor PRDM12 controls sensory neuron development and pain perception. Cell Cycle 2015, 14, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, S.; Yamazaki, C.; Masumoto, K.H.; Nagano, M.; Naito, M.; Soga, T.; Hiyama, H.; Matsumoto, M.; Takasaki, J.; Kamohara, M.; et al. Abnormal development of the olfactory bulb and reproductive system in mice lacking prokineticin receptor PKR2. Proc. Natl. Acad. Sci. USA 2006, 103, 4140–4145. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.Y.; Cheng, M.Y. Prokineticin 2 and circadian clock output. FEBS J. 2005, 272, 5703–5709. [Google Scholar] [CrossRef] [PubMed]
- Satoh, A.; Brace, C.S.; Rensing, N.; Imai, S. Deficiency of Prdm13, a dorsomedial hypothalamus-enriched gene, mimics age-associated changes in sleep quality and adiposity. Aging Cell. 2015, 14, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Blazer, L.L.; Lima-Fernandes, E.; Gibson, E.; Eram, M.S.; Loppnau, P.; Arrowsmith, C.H.; Schapira, M.; Vedadi, M. PR domain-containing protein 7 (PRDM7) is a histone 3 lysine 4 trimethyltransferase. J. Biol. Chem. 2016, 291, 13509–13519. [Google Scholar] [CrossRef] [PubMed]
- Shimada, I.S.; Acar, M.; Burgess, R.J.; Zhao, Z.; Morrison, S.J. Prdm16 is required for the maintenance of neural stem cells in the postnatal forebrain and their differentiation into ependymal cells. Genes Dev. 2017, 31, 1134–1146. [Google Scholar] [CrossRef] [PubMed]
- Inoue, M.; Iwai, R.; Tabata, H.; Konno, D.; Komabayashi-Suzuki, M.; Watanabe, C.; Iwanari, H.; Mochizuki, Y.; Hamakubo, T.; Matsuzaki, F.; et al. Prdm16 is crucial for progression of the multipolar phase during neural differentiation of the developing neocortex. Development 2017, 144, 385–399. [Google Scholar] [CrossRef] [PubMed]
- Powers, N.R.; Parvanov, E.D.; Baker, C.L.; Walker, M.; Petkov, P.M.; Paigen, K. The meiotic recombination activator PRDM9 trimethylates both H3K36 and H3K4 at recombination hotspots in vivo. PLoS Genet. 2016, 12, e1006146. [Google Scholar] [CrossRef] [PubMed]
- Altemose, N.; Noor, N.; Bitoun, E.; Tumian, A.; Imbeault, M.; Chapman, J.R.; Aricescu, A.R.; Myers, S.R. A map of human PRDM9 binding provides evidence for novel behaviors of PRDM9 and other zinc-finger proteins in meiosis. Elife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Qin, Y.; Wang, R.; Huang, Z.; Zhang, Y.; Zhou, R.; Song, L.; Ling, X.; Hu, Z.; Miao, D.; et al. Copy number gain of VCX, X-linked multi-copy gene, leads to cell proliferation and apoptosis during spermatogenesis. Oncotarget 2016, 7, 78532–78540. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.W.; Zhang, J.C.; Zhang, X.D.; Miao, S.Y.; Zong, S.D.; Sheng, Q.; Wang, L.F. Expression and localization of VCX/Y proteins and their possible involvement in regulation of ribosome assembly during spermatogenesis. Cell Res. 2003, 13, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Sleutels, F.; Soochit, W.; Bartkuhn, M.; Heath, H.; Dienstbach, S.; Bergmaier, P.; Franke, V.; Rosa-Garrido, M.; van de Nobelen, S.; Caesar, L.; et al. The male germ cell gene regulator CTCFL is functionally different from CTCF and binds CTCF-like consensus sites in a nucleosome composition-dependent manner. Epigenet. Chromatin 2012, 5, 8. [Google Scholar] [CrossRef] [PubMed]
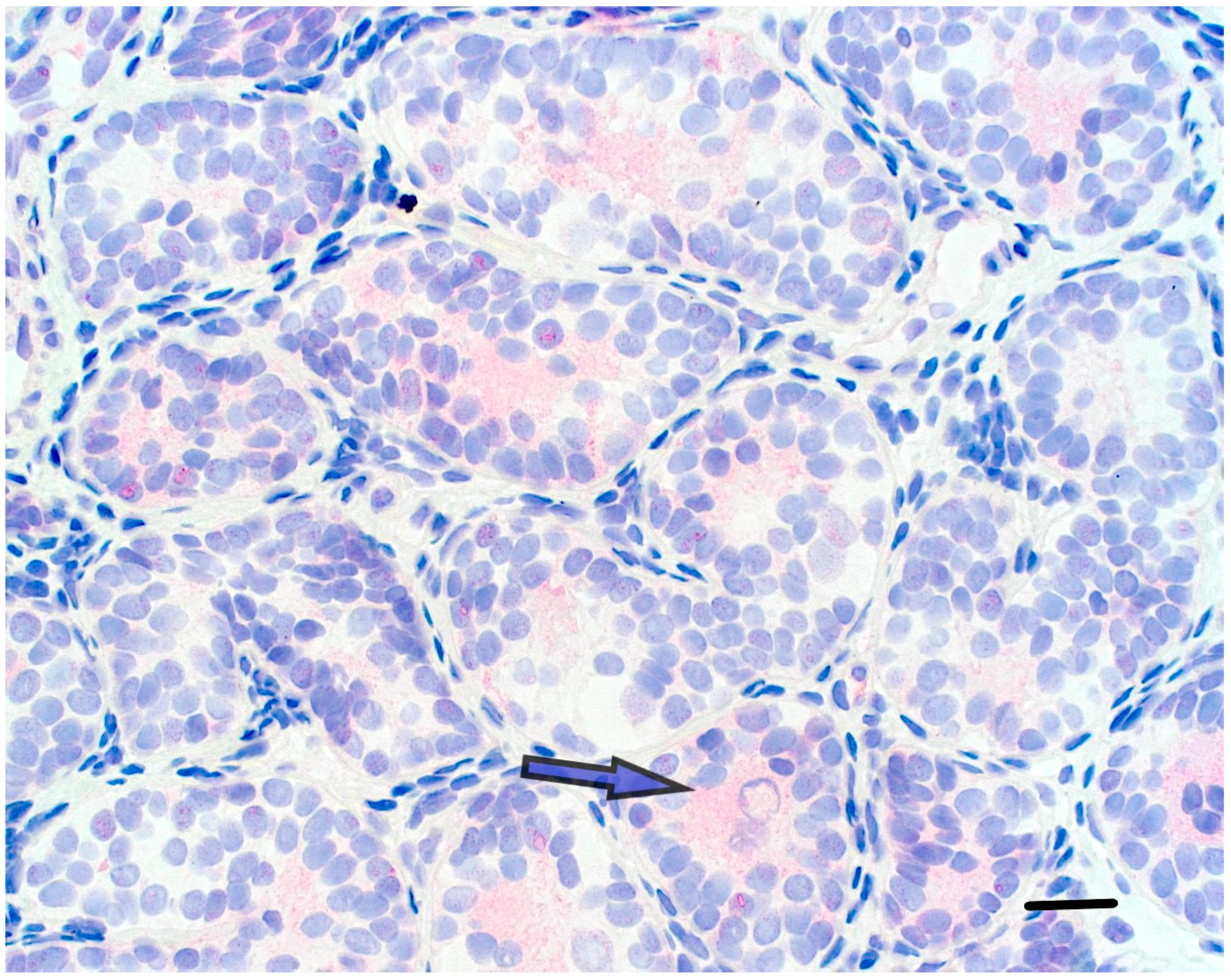
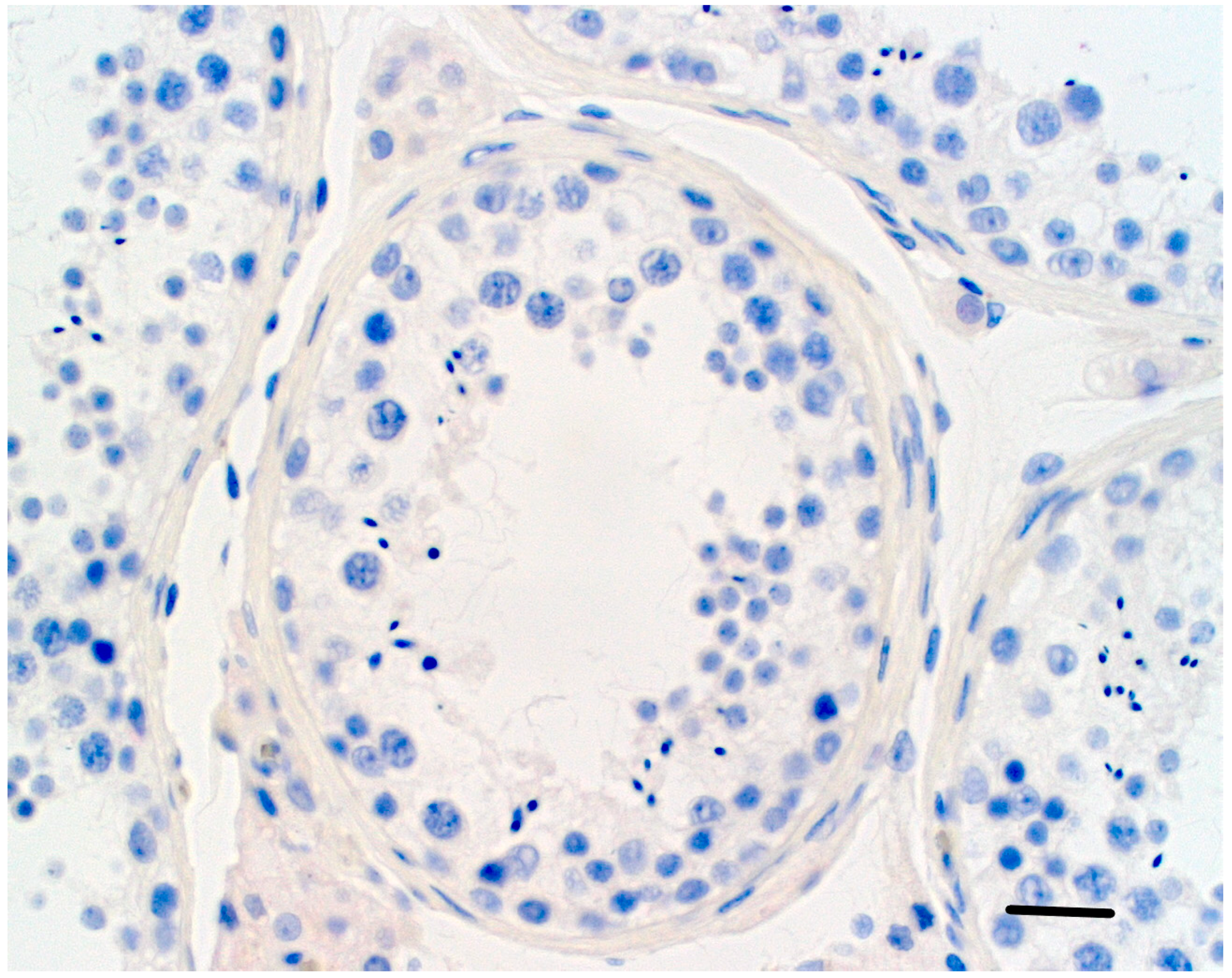
| ENTREZID | SYMBOL | IogFC HIR/LIR | FDR HIR/LIR | Pvalue HIR/LIR |
|---|---|---|---|---|
| 639 | PRDM1 | −1.11 | 0.003 | 0.000217 |
| 7799 | PRDM2 | 0.29 | 0.059 | 0.012406 |
| 2122 | MECOM | −0.36 | 0.12 | 0.028340 |
| 11108 | PRDM4 | 0.07 | 0.46 | 0.289401 |
| 11107 | PRDM5 | −0.23 | 0.244 | 0.107015 |
| 93166 | PRDM6 | −2.15 | 0.0002 | 1.1 × 10−6 |
| 56978 | PRDM8 | −0.13 | 0.807 | 0.693591 |
| 56979 | PRDM9 | −1.17 | 0.007 | 0.000639 |
| 56980 | PRDM10 | −0.12 | 0.255 | 0.114300 |
| 56981 | PRDM11 | 0.25 | 0.109 | 0.031354 |
| 59336 | PRDM13 | −1.58 | 0.024 | 0.003636 |
| 63978 | PRDM14 | −2.55 | 0.001 | 5.2 × 10−5 |
| 63977 | PRDM15 | −0.25 | 0.103 | 0.103668 |
| 63976 | PRDM16 | −0.42 | 0.225 | 0.094767 |
| ENTREZID | SYMBOL | IogFC HIR/LIR | FDR HIR/LIR | Pvalue HIR/LIR |
|---|---|---|---|---|
| 11108 | PRDM4 | −0.53 | 0.0105 | 0.007595 |
| 11107 | PRDM5 | −0.53 | 0.0248 | 0.010239 |
| 11105 | PRDM7 | 2.18 | 0.0003 | 2.1 × 10−5 |
| 56979 | PRDM9 | 1.68 | 0.0014 | 0.000219 |
| 59335 | PRDM12 | 2.11 | 0.0113 | 0.031354 |
| 63976 | PRDM16 | 1.15 | 0.0044 | 0.001111 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hadziselimovic, F.; Cathomas, G.; Verkauskas, G.; Dasevicius, D.; Stadler, M.B. PRDM Histone Methyltransferase mRNA Levels Increase in Response to Curative Hormone Treatment for Cryptorchidism-Dependent Male Infertility. Genes 2018, 9, 391. https://doi.org/10.3390/genes9080391
Hadziselimovic F, Cathomas G, Verkauskas G, Dasevicius D, Stadler MB. PRDM Histone Methyltransferase mRNA Levels Increase in Response to Curative Hormone Treatment for Cryptorchidism-Dependent Male Infertility. Genes. 2018; 9(8):391. https://doi.org/10.3390/genes9080391
Chicago/Turabian StyleHadziselimovic, Faruk, Gieri Cathomas, Gilvydas Verkauskas, Darius Dasevicius, and Michael B. Stadler. 2018. "PRDM Histone Methyltransferase mRNA Levels Increase in Response to Curative Hormone Treatment for Cryptorchidism-Dependent Male Infertility" Genes 9, no. 8: 391. https://doi.org/10.3390/genes9080391
APA StyleHadziselimovic, F., Cathomas, G., Verkauskas, G., Dasevicius, D., & Stadler, M. B. (2018). PRDM Histone Methyltransferase mRNA Levels Increase in Response to Curative Hormone Treatment for Cryptorchidism-Dependent Male Infertility. Genes, 9(8), 391. https://doi.org/10.3390/genes9080391




