Genome Mining of Non-Conventional Yeasts: Search and Analysis of MAL Clusters and Proteins
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Yeasts and the Genomes
2.2. Extraction of DNA and Protein Sequences and Analysis of Genomic Neighborhood of AG Genes to Detect MAL Clusters
2.3. Alignment of Gene and Protein Sequences for Identity Evaluation, Construction of Phylogenetic Trees, and Defining Signature Amino Acids
2.4. Heterologous Expression of AGs Encoded in the Genome of Scheffersomyces stipitis and Substrate Specificity Assay of the Enzymes
2.5. Assay of Scheffersomyces stipitis Growth Ability on Sugars and Evaluation of Hydrolysis of α-Glucosidic Sugars by S. stipitis Cell Extracts
3. Results
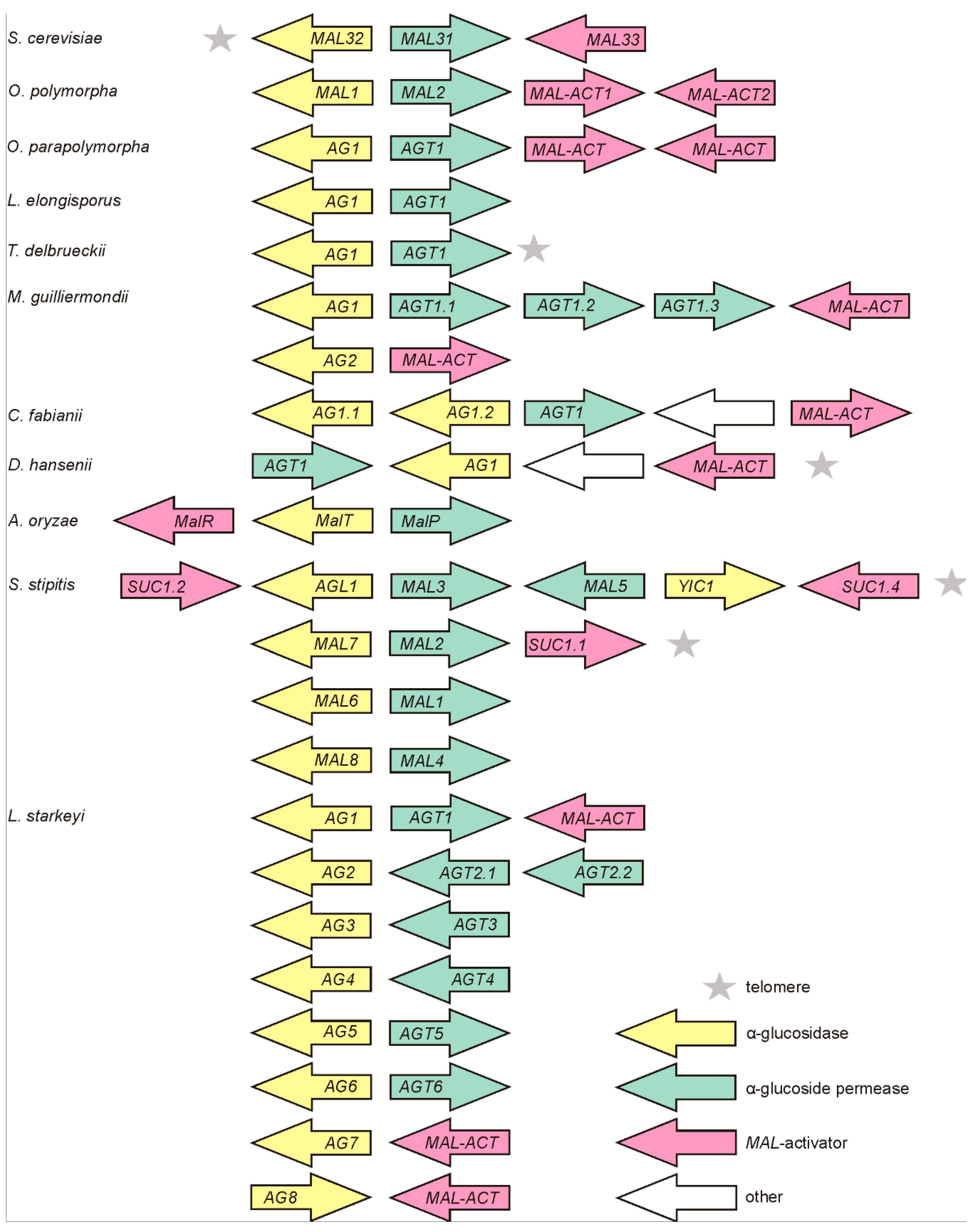
3.1. MAL Genes are Clustered in Ogataea polymorpha (Op) and O. parapolymorpha (Opp)
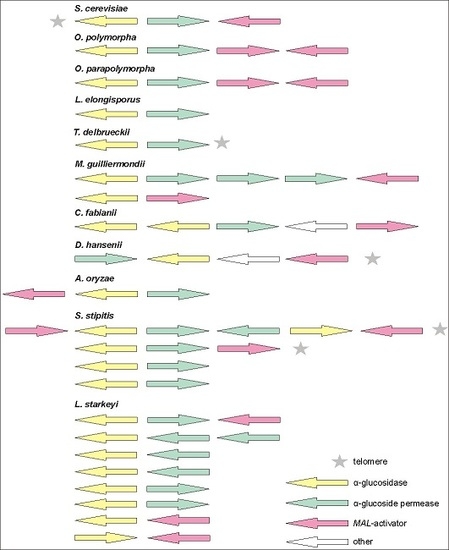
3.2. MAL Clusters Are Also Present in the Genomes of Other Non-Conventional Yeasts
3.3. MAL-Activator Genes Are Often Genomically Clustered with AG and AGT Genes
3.4. Phylograms of AGTs and AGs Encoded by the MAL Clusters Largely Agree with Phylograms of Yeast Species
3.5. Analysis of Yeast AGs for Signature Amino Acids: Prediction of Substrate Specificity
3.6. Substrate Specificity Evaluation of Scheffersomyces stipitis AGs: Verifying the Prediction
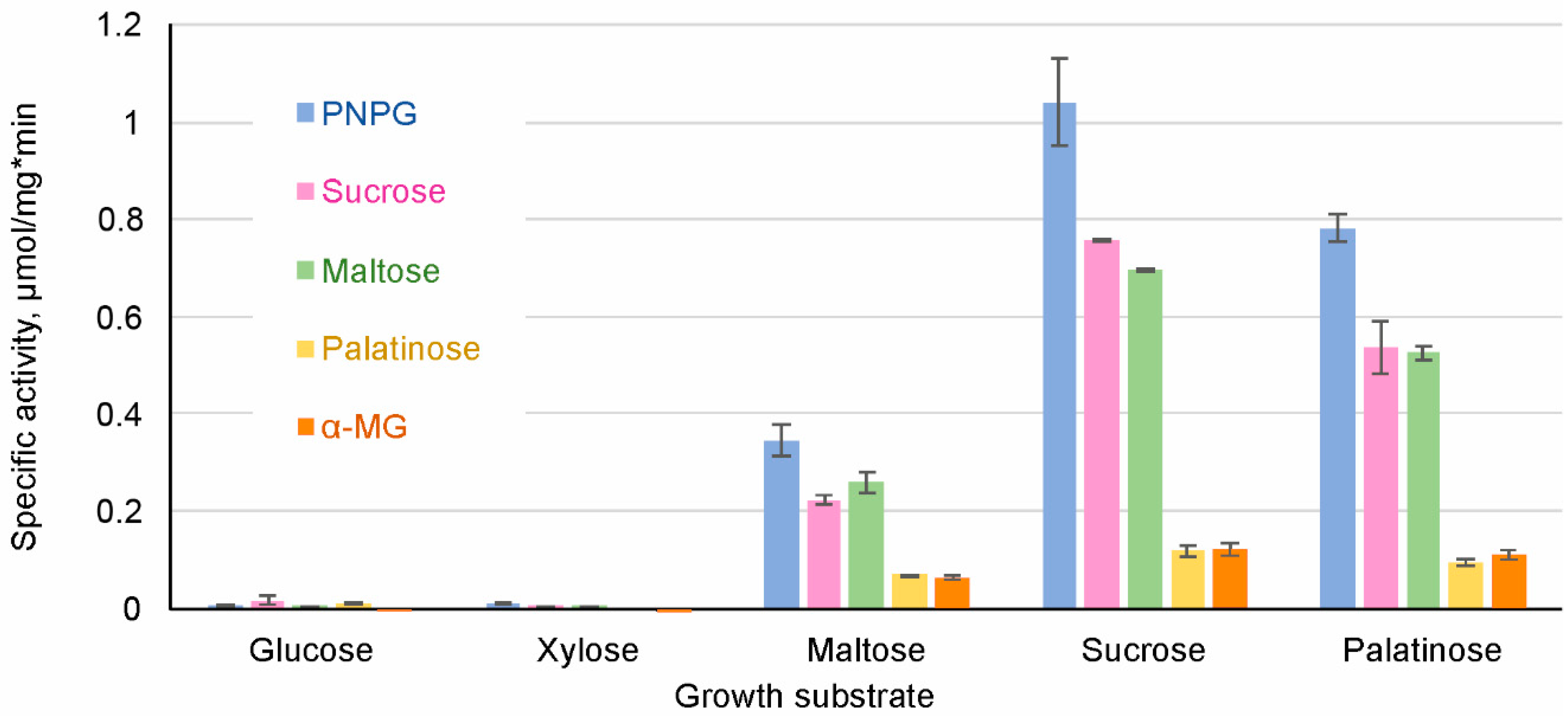
3.6.1. S. stipitis Assimilates Both Maltose-Like and Isomaltose-Like Sugars
3.6.2. Cell Extracts of S. stipitis Hydrolyzed both Maltose-Like and Isomaltose-Like Sugars
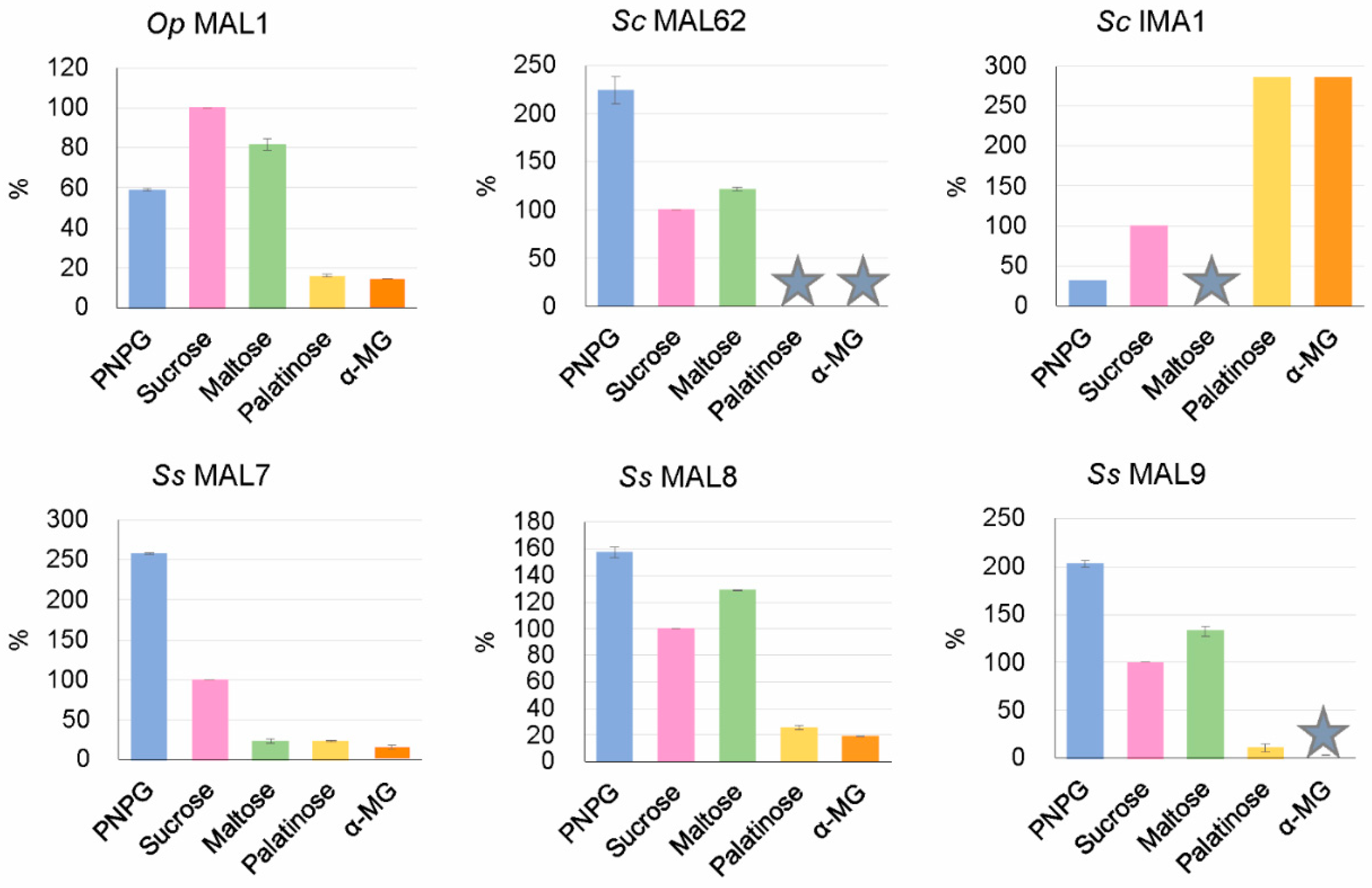
3.6.3. MAL7, MAL8, and MAL9 of S. stipitis Proven to be Maltase-Isomaltases
4. Discussion
4.1. The Natural Habitat of Non-Conventional Yeasts Possessing MAL Genes Contains α-Glucosidic Sugars
4.2. A Bi-Directional Promoter between the AGT and AG Genes Contributes to Balance the Transport and Further Metabolism of Disaccharides
4.3. How have the MAL Clusters Emerged and Evolved?
4.4. Evolution of AGs: Repeated Changes in Substrate Specificity
5. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stewart, G.G. Saccharomyces species in the production of beer. Beverages 2016, 2, 34. [Google Scholar] [CrossRef]
- Yurimoto, H. Molecular basis of methanol-inducible gene expression and its application in the methylotrophic yeast Candida boidinii. Biosci. Biotechnol. Biochem. 2009, 73, 793–800. [Google Scholar] [CrossRef] [PubMed]
- Weninger, A.; Hatzl, A.-M.; Schmid, C.; Vogl, T.; Glieder, A. Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris. J. Biotechnol. 2016, 235, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Kickenweiz, T.; Glieder, A.; Wu, J.C. Construction of a cellulose-metabolizing Komagataella phaffii (Pichia pastoris) by co-expressing glucanases and β-glucosidase. Appl. Microbiol. Biotechnol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Alamäe, T.; Liiv, L. Glucose repression of maltase and methanol-oxidizing enzymes in the methylotrophic yeast Hansenula polymorpha: Isolation and study of regulatory mutants. Folia Microbiol. 1998, 43, 443–452. [Google Scholar] [CrossRef]
- Liiv, L.; Pärn, P.; Alamäe, T. Cloning of maltase gene from a methylotrophic yeast, Hansenula polymorpha. Gene 2001, 265, 77–85. [Google Scholar] [CrossRef]
- Kramarenko, T.; Karp, H.; Järviste, A.; Alamäe, T. Sugar repression in the methylotrophic yeast Hansenula polymorpha studied by using hexokinase-negative, glucokinase-negative and double kinase-negative mutants. Folia Microbiol. 2000, 45, 521–529. [Google Scholar] [CrossRef]
- Alamäe, T.; Pärn, P.; Viigand, K.; Karp, H. Regulation of the Hansenula polymorpha maltase gene promoter in H. polymorpha and Saccharomyces cerevisiae. FEMS Yeast Res. 2003, 4, 165–173. [Google Scholar] [CrossRef]
- Karp, H.; Järviste, A.; Kriegel, T.M.; Alamäe, T. Cloning and biochemical characterization of hexokinase from the methylotrophic yeast Hansenula polymorpha. Curr. Genet. 2003, 44, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Viigand, K.; Tammus, K.; Alamäe, T. Clustering of MAL genes in Hansenula polymorpha: Cloning of the maltose permease gene and expression from the divergent intergenic region between the maltose permease and maltase genes. FEMS Yeast Res. 2005, 5, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Suppi, S.; Michelson, T.; Viigand, K.; Alamäe, T. Repression vs. activation of MOX, FMD, MPP1 and MAL1 promoters by sugars in Hansenula polymorpha: The outcome depends on cell’s ability to phosphorylate sugar. FEMS Yeast Res. 2013, 13, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Viigand, K.; Visnapuu, T.; Mardo, K.; Aasamets, A.; Alamäe, T. Maltase protein of Ogataea (Hansenula) polymorpha is a counterpart to the resurrected ancestor protein ancMALS of yeast maltases and isomaltases. Yeast 2016, 33, 415–432. [Google Scholar] [CrossRef] [PubMed]
- Voordeckers, K.; Brown, C.A.; Vanneste, K.; van der Zande, E.; Voet, A.; Maere, S.; Verstrepen, K.J. Reconstruction of ancestral metabolic enzymes reveals molecular mechanisms underlying evolutionary innovation through gene duplication. PLoS Biol. 2012, 10, e1001446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naumoff, D.G.; Naumov, G.I. Discovery of a novel family of α-glucosidase IMA genes in yeast Saccharomyces cerevisiae. Dokl. Biochem. Biophys. 2010, 432, 114–116. [Google Scholar] [CrossRef] [PubMed]
- Teste, M.-A.; Francois, J.M.; Parrou, J.-L. Characterization of a new multigene family encoding isomaltases in the yeast Saccharomyces cerevisiae, the IMA family. J. Biol. Chem. 2010, 285, 26815–26824. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Mateo, R.Q.; Kolecka, A.; Theelen, B.; Robert, V.; Boekhout, T. Advances in yeast systematics and phylogeny and their use as predictors of biotechnologically important metabolic pathways. FEMS Yeast Res. 2015, 15, fov050. [Google Scholar] [CrossRef] [PubMed]
- Ávila, J.; González, C.; Brito, N.; Machín, M.F.; Pérez, D.; Siverio, J.M. A second Zn(II)2Cys6 transcriptional factor encoded by the YNA2 gene is indispensable for the transcriptional activation of the genes involved in nitrate assimilation in the yeast Hansenula polymorpha. Yeast 2002, 19, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.; Wolfe, K.H. Birth of a metabolic gene cluster in yeast by adaptive gene relocation. Nat. Genet. 2005, 37, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Slot, J.C.; Rokas, A. Multiple GAL pathway gene clusters evolved independently and by different mechanisms in fungi. Proc. Natl. Acad. Sci. USA 2010, 107, 10136–10141. [Google Scholar] [CrossRef] [PubMed]
- Kunze, G.; Gaillardin, C.; Czernicka, M.; Durrens, P.; Martin, T.; Böer, E.; Gabaldón, T.; Cruz, J.A.; Talla, E.; Marck, C.; et al. The complete genome of Blastobotrys (Arxula) adeninivorans LS3—A yeast of biotechnological interest. Biotechnol. Biofuels 2014, 7, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeffries, T.W.; Van Vleet, J.R.H. Pichia stipitis genomics, transcriptomics, and gene clusters. Fems Yeast Res. 2009, 9, 793–807. [Google Scholar] [CrossRef] [PubMed]
- Naumov, G.I.; Naumova, E.S.; Michels, C.A. Genetic variation of the repeated Mal loci in natural populations of Saccharomyces cerevisiae and Saccharomyces paradoxus. Genetics 1994, 136, 803–812. [Google Scholar] [PubMed]
- Brown, C.A.; Murray, A.W.; Verstrepen, K.J. Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr. Biol. CB 2010, 20, 895–903. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, F.; Vidgren, V.; Ruohonen, L.; Gibson, B. Maltose and maltotriose utilisation by group I strains of the hybrid lager yeast Saccharomyces pastorianus. FEMS Yeast Res. 2016, 16, fow053. [Google Scholar] [CrossRef] [PubMed]
- Jeffries, T.W.; Grigoriev, I.V.; Grimwood, J.; Laplaza, J.M.; Aerts, A.; Salamov, A.; Schmutz, J.; Lindquist, E.; Dehal, P.; Shapiro, H.; et al. Genome sequence of the lignocellulose-bioconverting and xylose-fermenting yeast Pichia stipitis. Nat. Biotechnol. 2007, 25, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Ravin, N.V.; Eldarov, M.A.; Kadnikov, V.V.; Beletsky, A.V.; Schneider, J.; Mardanova, E.S.; Smekalova, E.M.; Zvereva, M.I.; Dontsova, O.A.; Mardanov, A.V.; et al. Genome sequence and analysis of methylotrophic yeast Hansenula polymorpha DL1. BMC Genom. 2013, 14, 837. [Google Scholar] [CrossRef] [PubMed]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.; Otillar, R.; Riley, R.; Salamov, A.; Zhao, X.; Korzeniewski, F.; et al. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014, 42, D699–D704. [Google Scholar] [CrossRef] [PubMed]
- Ramezani-Rad, M.; Hollenberg, C.P.; Lauber, J.; Wedler, H.; Griess, E.; Wagner, C.; Albermann, K.; Hani, J.; Piontek, M.; Dahlems, U.; et al. The Hansenula polymorpha (strain CBS4732) genome sequencing and analysis. FEMS Yeast Res. 2003, 4, 207–215. [Google Scholar] [CrossRef]
- Lahtchev, K.L.; Semenova, V.D.; Tolstorukov, I.I.; van der Klei, I.; Veenhuis, M. Isolation and properties of genetically defined strains of the methylotrophic yeast Hansenula polymorpha CBS4732. Arch. Microbiol. 2002, 177, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Riley, R.; Haridas, S.; Wolfe, K.H.; Lopes, M.R.; Hittinger, C.T.; Göker, M.; Salamov, A.A.; Wisecaver, J.H.; Long, T.M.; Calvey, C.H.; et al. Comparative genomics of biotechnologically important yeasts. Proc. Natl. Acad. Sci. USA 2016, 113, 9882–9887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoyanov, A.; Lahtchev, K. The methylotrophic yeast Ogataea (Hansenula) polymorpha as a model organism for studying reproductive isolation. Genet. Plant Physiol. 2016, 6, 14–26. [Google Scholar]
- Butler, G.; Rasmussen, M.D.; Lin, M.F.; Santos, M.A.; Sakthikumar, S.; Munro, C.A.; Rheinbay, E.; Grabherr, M.; Forche, A.; Reedy, J.L.; et al. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 2009, 459, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Sherman, D.J.; Martin, T.; Nikolski, M.; Cayla, C.; Souciet, J.-L.; Durrens, P. Génolevures: Protein families and synteny among complete hemiascomycetous yeast proteomes and genomes. Nucleic Acids Res. 2009, 37, D550–D554. [Google Scholar] [CrossRef] [PubMed]
- Dujon, B.; Sherman, D.; Fischer, G.; Durrens, P.; Casaregola, S.; Lafontaine, I.; de Montigny, J.; Marck, C.; Neuvéglise, C.; Talla, E.; et al. Genome evolution in yeasts. Nature 2004, 430, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Papon, N.; Savini, V.; Lanoue, A.; Simkin, A.J.; Crèche, J.; Giglioli-Guivarc’h, N.; Clastre, M.; Courdavault, V.; Sibirny, A.A. Candida guilliermondii: Biotechnological applications, perspectives for biological control, emerging clinical importance and recent advances in genetics. Curr. Genet. 2013, 59, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Wood, V.; Gwilliam, R.; Rajandream, M.-A.; Lyne, M.; Lyne, R.; Stewart, A.; Sgouros, J.; Peat, N.; Hayles, J.; Baker, S.; et al. The genome sequence of Schizosaccharomyces pombe. Nature 2002, 415, 871–880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freel, K.C.; Sarilar, V.; Neuvéglise, C.; Devillers, H.; Friedrich, A.; Schacherer, J. Genome sequence of the yeast Cyberlindnera fabianii (Hansenula fabianii). Genome Announc. 2014, 2, e00638-14. [Google Scholar] [CrossRef] [PubMed]
- Gordon, J.L.; Armisén, D.; Proux-Wéra, E.; ÓhÉigeartaigh, S.S.; Byrne, K.P.; Wolfe, K.H. Evolutionary erosion of yeast sex chromosomes by mating-type switching accidents. Proc. Natl. Acad. Sci. USA 2011, 108, 20024–20029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goffeau, A.; Barrell, B.G.; Bussey, H.; Davis, R.W.; Dujon, B.; Feldmann, H.; Galibert, F.; Hoheisel, J.D.; Jacq, C.; Johnston, M.; et al. Life with 6000 Genes. Science 1996, 274, 546–567. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0. Molecular Biology and Evolution. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Dayhoff, M.O.; Schwartz, R.M.; Orcutt, B.C. A model of evolutionary change in proteins. In Atlas of Protein Sequence and Structure; National Biomedical Research Foundation: Washington, DC, USA, 1978; Volume 5, pp. 345–352. [Google Scholar]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charron, M.J.; Read, E.; Haut, S.R.; Michels, C.A. Molecular evolution of the telomere-associated MAL loci of Saccharomyces. Genetics 1989, 122, 307–316. [Google Scholar] [PubMed]
- Curiel, J.A.; de las Rivas, B.; Mancheño, J.M.; Muñoz, R. The pURI family of expression vectors: A versatile set of ligation independent cloning plasmids for producing recombinant His-fusion proteins. Protein Expr. Purif. 2011, 76, 44–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ernits, K.; Viigand, K.; Visnapuu, T.; Põšnograjeva, K.; Alamäe, T. Thermostability measurement of an α-glucosidase using a classical activity-based assay and a novel Thermofluor method. Bio-Protocol 2017, 7. [Google Scholar] [CrossRef]
- Michels, C.A.; Needleman, R.B. The dispersed, repeated family of MAL loci in Saccharomyces spp. J. Bacteriol. 1984, 157, 949–952. [Google Scholar] [PubMed]
- Agaphonov, M.; Romanova, N.; Choi, E.-S.; Ter-Avanesyan, M. A novel kanamycin/G418 resistance marker for direct selection of transformants in Escherichia coli and different yeast species. Yeast 2010, 27, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, S.; Takizawa, M.; Suyama, H.; Shintani, T.; Gomi, K. Characterization and expression analysis of a maltose-utilizing (MAL) cluster in Aspergillus oryzae. Fungal Genet. Biol. 2010, 47, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Fairhead, C.; Dujon, B. Structure of Kluyveromyces lactis subtelomeres: Duplications and gene content. FEMS Yeast Res. 2006, 6, 428–441. [Google Scholar] [CrossRef] [PubMed]
- Leifso, K.R.; Williams, D.; Hintz, W.E. Heterologous expression of cyan and yellow fluorescent proteins from the Kluyveromyces lactis KlMAL21–KlMAL22 bi-directional promoter. Biotechnol. Lett. 2007, 29, 1233–1241. [Google Scholar] [CrossRef] [PubMed]
- Change, Y.S.; Dubin, R.A.; Perkins, E.; Forrest, D.; Michels, C.A.; Needleman, R.B. MAL63 codes for a positive regulator of maltose fermentation in Saccharomyces cerevisiae. Curr. Genet. 1988, 14, 201–209. [Google Scholar] [CrossRef]
- Kelly, R.; Kwon-Chung, K.J. A zinc finger protein from Candida albicans is involved in sucrose utilization. J. Bacteriol. 1992, 174, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Pougach, K.; Voet, A.; Kondrashov, F.A.; Voordeckers, K.; Christiaens, J.F.; Baying, B.; Benes, V.; Sakai, R.; Aerts, J.; Zhu, B.; et al. Duplication of a promiscuous transcription factor drives the emergence of a new regulatory network. Nat. Commun. 2014, 5, 4868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Day, R.E.; Higgins, V.J.; Rogers, P.J.; Dawes, I.W. Characterization of the putative maltose transporters encoded by YDL247w and YJR160c. Yeast 2002, 19, 1015–1027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Day, R.E.; Rogers, P.J.; Dawes, I.W.; Higgins, V.J. Molecular analysis of maltotriose transport and utilization by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2002, 68, 5326–5335. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Michels, C.A. The MAL63 gene of Saccharomyces encodes a cysteine-zinc finger protein. Curr. Genet. 1988, 14, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.-X.; Zhou, X.; Kominek, J.; Kurtzman, C.P.; Hittinger, C.T.; Rokas, A. Reconstructing the backbone of the Saccharomycotina yeast phylogeny using genome-scale data. G3 Genes Genomes Genet. 2016, 6, 3927–3939. [Google Scholar] [CrossRef] [PubMed]
- Dujon, B.A.; Louis, E.J. Genome diversity and evolution in the budding yeasts (Saccharomycotina). Genetics 2017, 206, 717–750. [Google Scholar] [CrossRef] [PubMed]
- Reinders, A.; Ward, J.M. Functional characterization of the α-glucoside transporter Sut1p from Schizosaccharomyces pombe, the first fungal homologue of plant sucrose transporters. Mol. Microbiol. 2001, 39, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, Y.; Honda, H.; Taniguchi-Morimura, J.; Iwasaki, S. The codon CUG is read as serine in an asporogenic yeast Candida cylindracea. Nature 1989, 341, 164–166. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Nakayama, A.; Yamamoto, Y.; Tabata, S. Val216 decides the substrate specificity of α-glucosidase in Saccharomyces cerevisiae: Substrate specificity of α-glucosidase. Eur. J. Biochem. 2004, 271, 3414–3420. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Miyake, H.; Kusunoki, M.; Osaki, S. Crystal structures of isomaltase from Saccharomyces cerevisiae and in complex with its competitive inhibitor maltose: Crystal structure of isomaltase. FEBS J. 2010, 277, 4205–4214. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Miyake, H.; Kusunoki, M.; Osaki, S. Steric hindrance by 2 amino acid residues determines the substrate specificity of isomaltase from Saccharomyces cerevisiae. J. Biosci. Bioeng. 2011, 112, 545–550. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Petitjean, M.; Teste, M.-A.; Kooli, W.; Tranier, S.; François, J.M.; Parrou, J.-L. Similarities and differences in the biochemical and enzymological properties of the four isomaltases from Saccharomyces cerevisiae. FEBS Open Bio 2014, 4, 200–212. [Google Scholar] [CrossRef] [PubMed]
- Chi, Z.; Ni, X.; Yao, S. Cloning and overexpression of a maltase gene from Schizosaccharomyces pombe in Escherichia coli and characterization of the recombinant maltase. Mycol. Res. 2008, 112, 983–989. [Google Scholar] [CrossRef] [PubMed]
- Tsujimoto, Y.; Tanaka, H.; Takemura, R.; Yokogawa, T.; Shimonaka, A.; Matsui, H.; Kashiwabara, S.-I.; Watanabe, K.; Suzuki, Y. Molecular determinants of substrate recognition in thermostable alpha-glucosidases belonging to glycoside hydrolase family 13. J. Biochem. 2007, 142, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Eisawa, H.; Ogawa, S.; Yamazaki, N.; Maekawa, K.; Yamaguchi, T.; Sato, S.; Shiota, K.; Yoshida, T. Characterization of three fungal isomaltases belonging to glycoside hydrolase family 13 that do not show transglycosylation activity. J. Appl. Glycosci. 2017, 64, 9–13. [Google Scholar] [CrossRef] [Green Version]
- Sainz-Polo, M.A.; Ramírez-Escudero, M.; Lafraya, A.; González, B.; Marín-Navarro, J.; Polaina, J.; Sanz-Aparicio, J. Three-dimensional structure of Saccharomyces Invertase: Role of a non-catalytic domain in oligomerization and substrate specificity. J. Biol. Chem. 2013, 288, 9755–9766. [Google Scholar] [CrossRef] [PubMed]
- Geber, A.; Williamson, P.R.; Rex, J.H.; Sweeney, E.C.; Bennett, J.E. Cloning and characterization of a Candida albicans maltase gene involved in sucrose utilization. J. Bacteriol. 1992, 174, 6992–6996. [Google Scholar] [CrossRef] [PubMed]
- Bell, P.J.L.; Bissinger, P.H.; Evans, R.J.; Dawes, I.W. A two-reporter gene system for the analysis of bi-directional transcription from the divergent MAL6T-MAL6S promoter in Saccharomyces cerevisiae. Curr. Genet. 1995, 28, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Keppler, F.; Hamilton, J.T.G.; Braß, M.; Röckmann, T. Methane emissions from terrestrial plants under aerobic conditions. Nature 2006, 439, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Abbas, C.A.; Sibirny, A.A. Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers. Microbiol. Mol. Biol. Rev. MMBR 2011, 75, 321–360. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Randhawa, A.; Ganesan, K.; Raghava, G.P.S.; Mondal, A.K. Draft genome sequence of salt-tolerant yeast Debaryomyces hansenii var. hansenii MTCC 234. Eukaryot. Cell 2012, 11, 961–962. [Google Scholar] [CrossRef] [PubMed]
- Angerbauer, C.; Siebenhofer, M.; Mittelbach, M.; Guebitz, G.M. Conversion of sewage sludge into lipids by Lipomyces starkeyi for biodiesel production. Bioresour. Technol. 2008, 99, 3051–3056. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, C.S.; Wood, V.; Fantes, P.A. An ancient yeast for young geneticists: A primer on the Schizosaccharomyces pombe model system. Genetics 2015, 201, 403–423. [Google Scholar] [CrossRef] [PubMed]
- Levine, J.; Tanouye, L.; Michels, C.A. The UASMAL is a bidirectional promotor element required for the expression of both the MAL61 and MAL62 genes of the Saccharomyces MAL6 locus. Curr. Genet. 1992, 22, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Bali, M.; Medintz, I.; Michels, C.A. Intracellular maltose is sufficient to induce MAL gene expression in Saccharomyces cerevisiae. Eukaryot. Cell 2002, 1, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Viigand, K.; Alamäe, T. Further study of the Hansenula polymorpha MAL locus: Characterization of the α-glucoside permease encoded by the HpMAL2 gene. FEMS Yeast Res. 2007, 7, 1134–1144. [Google Scholar] [CrossRef] [PubMed]
- Henderson, R.; Poolman, B. Proton-solute coupling mechanism of the maltose transporter from Saccharomyces cerevisiae. Sci. Rep. 2017, 7, 14375. [Google Scholar] [CrossRef] [PubMed]
- Jansen, M.L.A.; De Winde, J.H.; Pronk, J.T. Hxt-carrier-mediated glucose efflux upon exposure of Saccharomyces cerevisiae to excess maltose. Appl. Environ. Microbiol. 2002, 68, 4259–4265. [Google Scholar] [CrossRef] [PubMed]
- Stajich, J.E.; Dietrich, F.S.; Roy, S.W. Comparative genomic analysis of fungal genomes reveals intron-rich ancestors. Genome Biol. 2007, 8, R223. [Google Scholar] [CrossRef] [PubMed]
- Da Lage, J.-L.; Binder, M.; Hua-Van, A.; Janeček, Š.; Casane, D. Gene make-up: Rapid and massive intron gains after horizontal transfer of a bacterial α-amylase gene to Basidiomycetes. BMC Evol. Biol. 2013, 13, 40. [Google Scholar] [CrossRef] [PubMed]
- Gabriško, M. Evolutionary history of eukaryotic α-glucosidases from the α-amylase family. J. Mol. Evol. 2013, 76, 129–145. [Google Scholar] [CrossRef] [PubMed]
- Visnapuu, T.; Mäe, A.; Alamäe, T. Hansenula polymorpha maltase gene promoter with sigma 70-like elements is feasible for Escherichia coli-based biotechnological applications: Expression of three genomic levansucrase genes of Pseudomonas syringae pv. tomato. Process Biochem. 2008, 43, 414–422. [Google Scholar] [CrossRef]
- Kelly, C.T.; Moriarty, M.E.; Fogarty, W.M. Thermostable extracellular α-amylase and α-glucosidase of Lipomyces starkeyi. Appl. Microbiol. Biotechnol. 1985, 22, 352–358. [Google Scholar] [CrossRef]

| Yeast | Culture Collection Numbers | Genome Accession from | Reference |
|---|---|---|---|
| Ogataea polymorpha leu1.1 | NCYC 495; ATCC MYA-335; CBS 1976, NRRL Y-1789 | MycoCosm | [30] |
| Ogataea parapolymorpha DL-1 | ATCC 26012; CBS 12304; NRRL Y-7560 | MycoCosm | [26] |
| Lipomyces starkeyi | NRRL Y-11557; ATCC 58680; CBS 1807 | MycoCosm | [30] |
| Debaryomyces hansenii | CBS 767; ATCC 36239 | MycoCosm | [33,34] |
| Meyerozyma (Pichia) guillermondii | CBS 566; ATCC 6260 | MycoCosm | [32,35] |
| Scheffersomyces (Pichia) stipitis | CBS 6054 | MycoCosm | [25] |
| Lodderomyces elongisporus | NRRL YB-4239; CBS 2605; ATCC 11503 | MycoCosm | [32] |
| Blastobotrys (Arxula) adeninivorans LS3 | CBS 8244 | MycoCosm | [20,33] |
| Schizosaccharomyces pombe | ATCC 24843, CBS 10395 | MycoCosm | [36] |
| Cyberlindnera fabianii | YJS4271 | European Nucleotide Archive (ENA) | [37] |
| Torulaspora delbrueckii | CBS 1146 | MycoCosm | [38] |
| Saccharomyces cerevisiae S288C | CBS 8803; ATCC 204508 | MycoCosm | [39] |
| α-Glucosidase | Signature Amino Acids (Numbering as in Sc IMA1) | Function (Prediction) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 158 | 216 | 217 | 218 | 219 | 278 | 279 | 307 | 411 | ||
| Op MAL1/Opp AG1 | F | T | A | G | L | V | G | D | N | maltase-isomaltase |
| Le AG1 | H | T | A | G | M | V | G | D | N | maltase-isomaltase |
| ancMALS | F | T | A | G | L | V | G | D | E | maltase-isomaltase |
| Sc MAL12/Sc MAL32/Sc MAL62 | F | T | A | G | L | V | A | E | D | maltase |
| Sc IMA1/Sc IMA2 | Y | V | G | S | L | M | Q | D | E | isomaltase |
| Sc IMA3/4 | Y | V | G | S | L | M | R | D | E | isomaltase |
| Sc IMA5 | F | V | G | S | M | V | G | S | E | isomaltase |
| Td AG1 | Y | V | G | S | L | M | Q | D | E | isomaltase |
| Cf AG1.2 | H | T | A | G | L | V | G | D | N | maltase-isomaltase |
| Cf AG1.1 | M | V | C | S | L | V | G | S | Q | isomaltase |
| Ss MAL6 | Y | T | A | G | L | V | G | N | N | maltase-isomaltase |
| Ss MAL7 | F | T | A | G | L | V | G | T | N | maltase-isomaltase |
| Ss MAL8 | Y | T | A | G | L | V | G | E | N | maltase-isomaltase |
| Ss MAL9 | Y | T | A | G | M | V | G | E | N | maltase-isomaltase |
| Ss AGL1 | Y | T | A | G | L | V | G | W | N | maltase-isomaltase |
| Mg AG2 | Y | T | A | G | M | V | G | D | N | maltase-isomaltase |
| Mg AG1 | C | V | A | A | L | V | G | E | E | isomaltase |
| Ls AG1 | Y | T | V | N | K | L | S | H | E | maltase |
| Ls AG6 | N | T | V | N | R | L | P | G | R | maltase |
| Ba AG2 | Y | T | V | Q | I | G | S | R | N | maltase |
| Ao MalT | I | T | V | N | M | L | P | D | D | maltase |
| Ls AG2 | L | A | I | N | F | M | A | D | E | maltase |
| Ls AG4 | H | A | I | N | F | M | G | T | E | maltase |
| Ls AG5 | A | A | I | N | F | M | A | D | E | maltase |
| Sp Mal1 | Y | A | I | N | M | M | P | D | E | maltase |
| Bs α-1,4-glucosidase | I | A | I | S | H | A | N | G | A | maltase |
| Ls AG3 | C | V | I | N | F | M | P | D | E | isomaltase |
| Ls AG7 | E | V | I | N | Y | M | G | Q | E | isomaltase |
| Ls AG8 | - | V | I | N | F | M | P | D | E | isomaltase |
| Dh AG1 | A | V | I | N | F | M | P | D | E | isomaltase |
| Ba AG1 | Y | V | I | N | L | M | P | Q | E | isomaltase |
| Bt oligo-1,6-glucosidase | V | V | I | N | M | T | P | D | E | isomaltase |
| An AgdC | F | V | I | N | F | M | P | D | D | isomaltase |
| Fo Foagl1 | F | V | I | N | F | M | P | D | D | isomaltase |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Viigand, K.; Põšnograjeva, K.; Visnapuu, T.; Alamäe, T. Genome Mining of Non-Conventional Yeasts: Search and Analysis of MAL Clusters and Proteins. Genes 2018, 9, 354. https://doi.org/10.3390/genes9070354
Viigand K, Põšnograjeva K, Visnapuu T, Alamäe T. Genome Mining of Non-Conventional Yeasts: Search and Analysis of MAL Clusters and Proteins. Genes. 2018; 9(7):354. https://doi.org/10.3390/genes9070354
Chicago/Turabian StyleViigand, Katrin, Kristina Põšnograjeva, Triinu Visnapuu, and Tiina Alamäe. 2018. "Genome Mining of Non-Conventional Yeasts: Search and Analysis of MAL Clusters and Proteins" Genes 9, no. 7: 354. https://doi.org/10.3390/genes9070354
APA StyleViigand, K., Põšnograjeva, K., Visnapuu, T., & Alamäe, T. (2018). Genome Mining of Non-Conventional Yeasts: Search and Analysis of MAL Clusters and Proteins. Genes, 9(7), 354. https://doi.org/10.3390/genes9070354