AtPAP2, a Unique Member of the PAP Family, Functions in the Plasma Membrane
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatics Analysis of AtPAP2
2.2. Construction of Plasmids
2.3. Transient Expression and Microscopy
3. Results and Discussion
3.1. Bioinformatics Analysis of Purple Acid Phosphatases in Arabidopsis
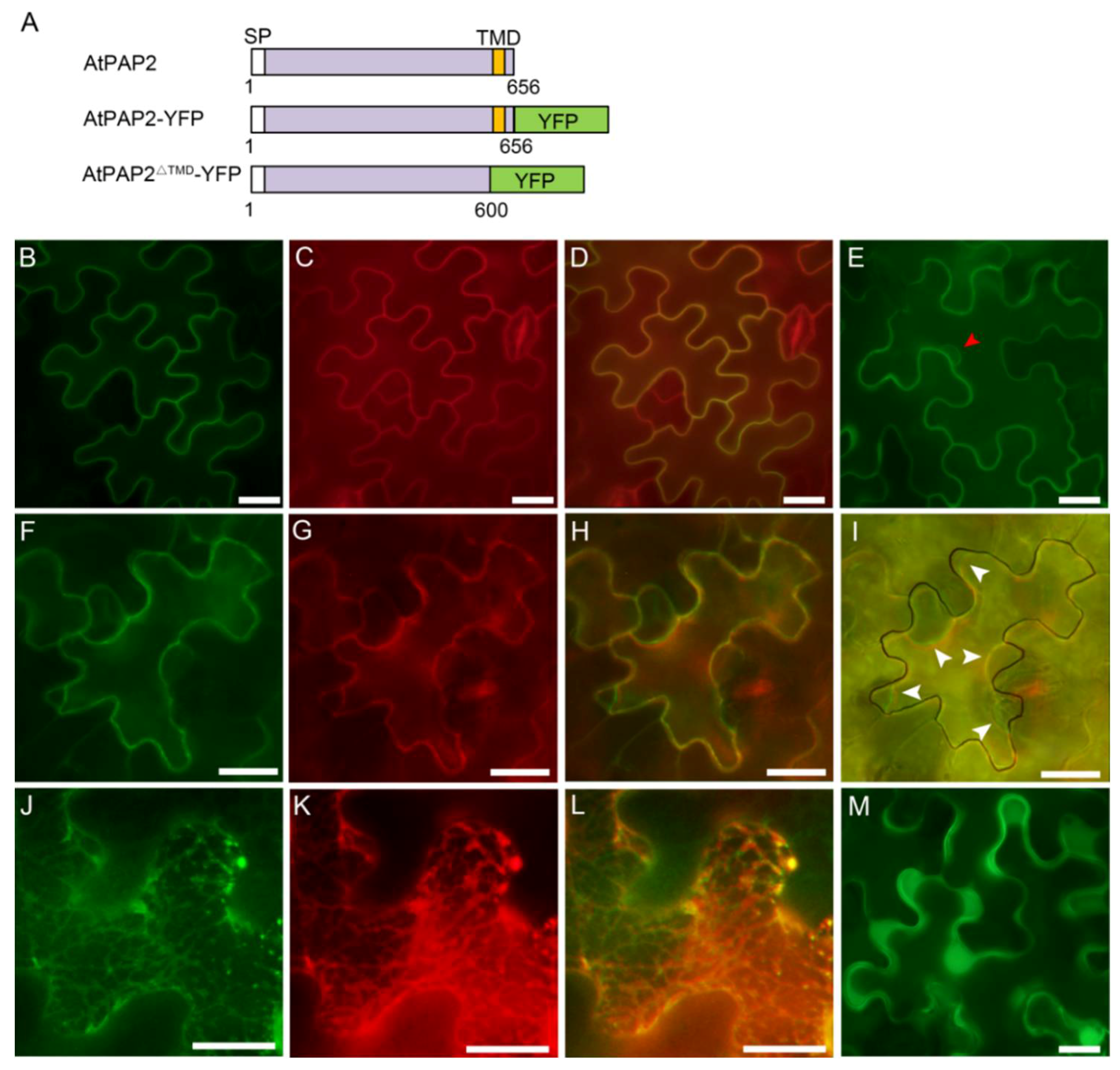
3.2. AtPAP2 Is Localized to the Plasma Membrane
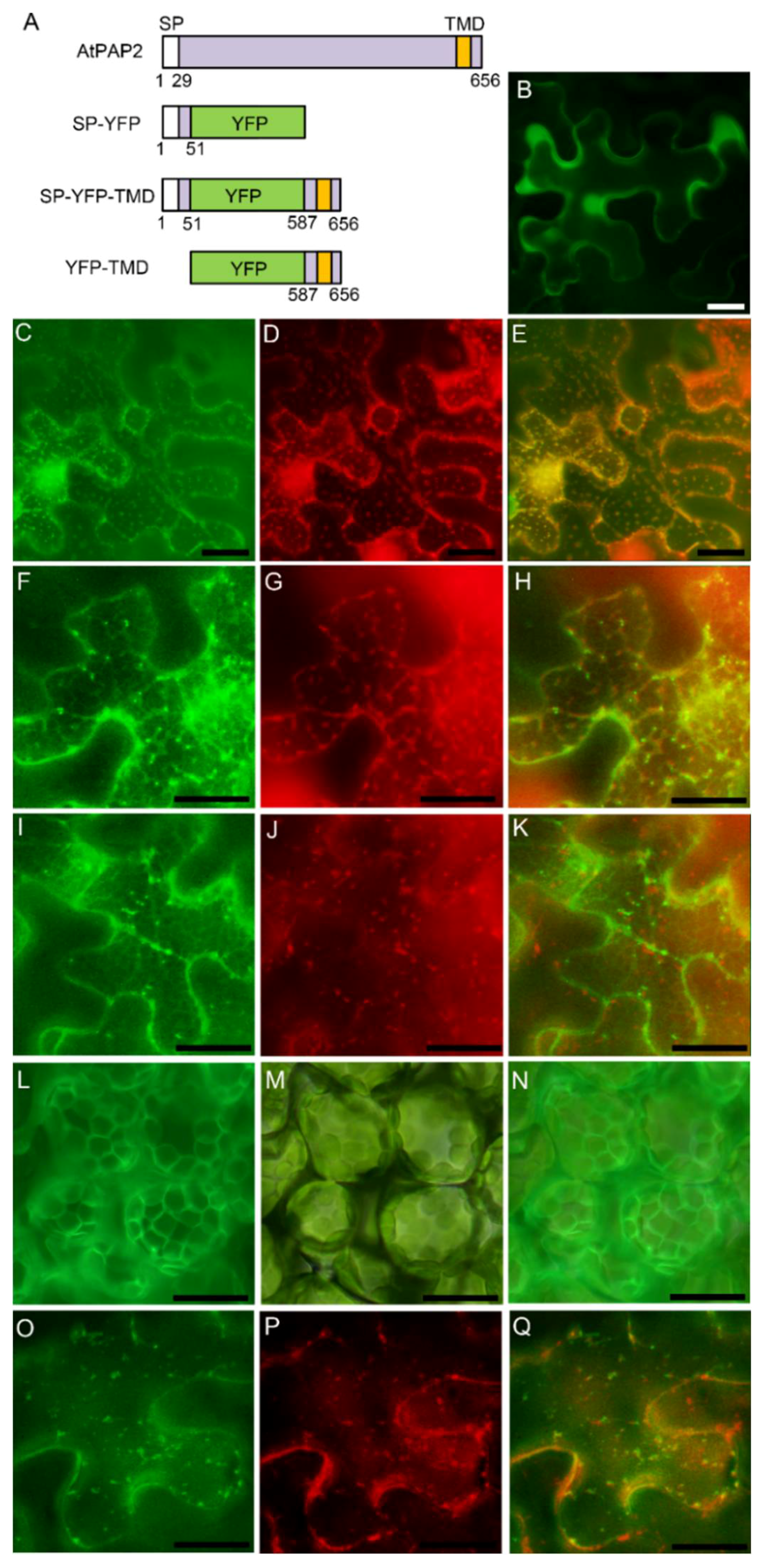
3.3. AtPAP2 is Targeted to the Plasma Membrane through Secretory Pathway
3.4. The Evolutionary Origin of AtPAP2 and AtPAP9
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hesterberg, D. Macroscale chemical properties and X-ray absorption spectroscopy of soil phosphorus. In Developments in Soil Science; Singh, B., Gräfe, M., Eds.; Elsevier: New York, NY, USA, 2010; Volume 34, pp. 313–356. [Google Scholar]
- Shen, J.B.; Yuan, L.X.; Zhang, J.L.; Li, H.G.; Bai, Z.H.; Chen, X.P.; Zhang, W.F.; Zhang, F.S. Phosphorus dynamics: from soil to plant. Plant Physiol. 2011, 156, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.T.; Hurley, B.A.; Plaxton, W.C. Feeding hungry plants: The role of purple acid phosphatases in phosphate nutrition. Plant Sci. 2010, 179, 14–27. [Google Scholar] [CrossRef]
- Wang, L.; Li, Z.; Qian, W.; Guo, W.; Gao, X.; Huang, L.; Wang, H.; Zhu, H.; Wu, J.W.; Wang, D.; et al. The Arabidopsis purple acid phosphatase AtPAP10 is predominantly associated with the root surface and plays an important role in plant tolerance to phosphate limitation. Plant Physiol. 2011, 157, 1283–1299. [Google Scholar] [CrossRef] [PubMed]
- Zamani, K.; Lohrasebi, T.; Sabet, M.S.; Malboobi, M.A.; Mousavi, A. Expression pattern and subcellular localization of Arabidopsis purple acid phosphatase AtPAP9. Gene Expr. Patterns 2014, 14, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Antonyuk, S.V.; Olczak, M.; Olczak, T.; Ciuraszkiewicz, J.; Strange, R.W. The structure of a purple acid phosphatase involved in plant growth and pathogen defence exhibits a novel immunoglobulin-like fold. IUCrJ 2014, 1, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.S.; Qi, H.; Sun, Y.Z.; Xiao, S.; Lim, B.L. Transgenic Arabidopsis thaliana containing increased levels of ATP and sucrose is more susceptible to Pseudomonas syringae. PLoS ONE 2017, 12, e0171040. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, S.; Stone, S.L.; Benkel, B.; Zhang, J.; Berrue, F.; Prithiviraj, B. Optimal level of purple acid phosphatase5 is required for maintaining complete resistance to Pseudomonas syringae. Front. Plant Sci. 2015, 6, 568. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, S.; Stone, S.L.; Benkel, B.; Prithiviraj, B. Purple Acid Phosphatase5 is required for maintaining basal resistance against Pseudomonas syringae in Arabidopsis. BMC Plant Biol. 2013, 13, 107. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhu, H.; Liu, K.; Liu, X.; Leggewie, G.; Udvardi, M.; Wang, D. Purple acid phosphatases of Arabidopsis thaliana. Comparative analysis and differential regulation by phosphate deprivation. J. Biol. Chem. 2002, 277, 27772–27781. [Google Scholar] [CrossRef] [PubMed]
- Schenk, G.; Guddat, L.W.; Ge, Y.; Carrington, L.E.; Hume, D.A.; Hamilton, S.; de Jersey, J. Identification of mammalian-like purple acid phosphatases in a wide range of plants. Gene 2000, 250, 117–125. [Google Scholar] [CrossRef]
- Gonzalez-Munoz, E.; Avendano-Vazquez, A.O.; Montes, R.A.; de Folter, S.; Andres-Hernandez, L.; Abreu-Goodger, C.; Sawers, R.J. The maize (Zea mays ssp. (mays var. B73) genome encodes 33 members of the purple acid phosphatase family. Front. Plant Sci. 2015, 6, 341. [Google Scholar] [PubMed]
- Bozzo, G.G.; Raghothama, K.G.; Plaxton, W.C. Purification and characterization of two secreted purple acid phosphatase isozymes from phosphate-starved tomato (Lycopersicon esculentum) cell cultures. Eur. J. Biochem. 2002, 269, 6278–6286. [Google Scholar] [CrossRef] [PubMed]
- Schenk, G.; Ge, Y.; Carrington, L.E.; Wynne, C.J.; Searle, I.R.; Carroll, B.J.; Hamilton, S.; de Jersey, J. Binuclear metal centers in plant purple acid phosphatases: Fe-Mn in sweet potato and Fe-Zn in soybean. Arch. Biochem. Biophys. 1999, 370, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, P.; Regierer, B.; Kossmann, J.; Frossard, E.; Amrhein, N.; Bucher, M. Differential expression of three purple acid phosphatases from potato. Plant Biol. 2004, 6, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Qian, W.; Lu, X.; Li, D.; Liu, X.; Liu, K.; Wang, D. Expression patterns of purple acid phosphatase genes in Arabidopsis organs and functional analysis of AtPAP23 predominantly transcribed in flower. Plant Mol. Biol. 2005, 59, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lu, S.; Zhang, Y.; Li, Z.; Du, X.; Liu, D. Comparative genetic analysis of Arabidopsis purple acid phosphatases AtPAP10, AtPAP12, and AtPAP26 provides new insights into their roles in plant adaptation to phosphate deprivation. J. Integr. Plant Biol. 2014, 56, 299–314. [Google Scholar] [CrossRef] [PubMed]
- Robinson, W.D.; Park, J.; Tran, H.T.; Del Vecchio, H.A.; Ying, S.; Zins, J.L.; Patel, K.; McKnight, T.D.; Plaxton, W.C. The secreted purple acid phosphatase isozymes AtPAP12 and AtPAP26 play a pivotal role in extracellular phosphate-scavenging by Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 6531–6542. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.T.; Qian, W.; Hurley, B.A.; She, Y.M.; Wang, D.; Plaxton, W.C. Biochemical and molecular characterization of AtPAP12 and AtPAP26: The predominant purple acid phosphatase isozymes secreted by phosphate-starved Arabidopsis thaliana. Plant Cell Environ. 2010, 33, 1789–1803. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Liu, D. Arabidopsis phosphatase under-producer mutants pup1 and pup3 contain mutations in the AtPAP10 and AtPAP26 genes. Plant Signal. Behav. 2015, 10, e1035851. [Google Scholar] [PubMed]
- Robinson, W.D.; Carson, I.; Ying, S.; Ellis, K.; Plaxton, W.C. Eliminating the purple acid phosphatase AtPAP26 in Arabidopsis thaliana delays leaf senescence and impairs phosphorus remobilization. New Phytol. 2012, 196, 1024–1029. [Google Scholar] [CrossRef] [PubMed]
- Kuang, R.; Chan, K.H.; Yeung, E.; Lim, B.L. Molecular and biochemical characterization of AtPAP15, a purple acid phosphatase with phytase activity, in Arabidopsis. Plant Physiol. 2009, 151, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Del Vecchio, H.A.; Ying, S.; Park, J.; Knowles, V.L.; Kanno, S.; Tanoi, K.; She, Y.M.; Plaxton, W.C. The cell wall-targeted purple acid phosphatase AtPAP25 is critical for acclimation of Arabidopsis thaliana to nutritional phosphorus deprivation. Plant J. 2014, 80, 569–581. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Suen, P.K.; Zhang, Y.J.; Liang, C.; Carrie, C.; Whelan, J.; Ward, J.L.; Hawkins, N.D.; Jiang, L.W.; Lim, B.L. A dual-targeted purple acid phosphatase in Arabidopsis thaliana moderates carbon metabolism and its overexpression leads to faster plant growth and higher seed yield. New Phytol. 2012, 194, 206–219. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Carrie, C.; Law, S.; Murcha, M.W.; Zhang, R.; Law, Y.S.; Suen, P.K.; Whelan, J.; Lim, B.L. AtPAP2 is a tail-anchored protein in the outer membrane of chloroplasts and mitochondria. Plant Signal. Behav. 2012, 7, 927–932. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, L.; Yung, K.F.; Leung, D.Y.; Sun, F.; Lim, B.L. Over-expression of AtPAP2 in Camelina sativa leads to faster plant growth and higher seed yield. Biotechnol. Biofuels 2012, 5, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, F.; Fettke, J.; Schottler, M.A.; Ramsden, L.; Fernie, A.R.; Lim, B.L. Heterologous expression of AtPAP2 in transgenic potato influences carbon metabolism and tuber development. FEBS Lett. 2014, 588, 3726–3731. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Kerppola, T.K. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 2006, 1, 1278–1286. [Google Scholar] [CrossRef] [PubMed]
- Batoko, H.; Zheng, H.Q.; Hawes, C.; Moore, I. A Rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 2000, 12, 2201. [Google Scholar] [CrossRef] [PubMed]
- Li, H.J.; Xue, Y.; Jia, D.J.; Wang, T.; Hi, D.Q.; Liu, J.; Cui, F.; Xie, Q.; Ye, D.; Yang, W.C. POD1 regulates pollen tube guidance in response to micropylar female signaling and acts in early embryo patterning in Arabidopsis. Plant Cell 2011, 23, 3288–3302. [Google Scholar] [CrossRef] [PubMed]
- Daschner, K.; Thalheim, C.; Guha, C.; Brennicke, A.; Binder, S. In plants a putative isovaleryl-CoA-dehydrogenase is located in mitochondria. Plant Mol. Biol. 1999, 39, 1275–1282. [Google Scholar] [PubMed]
- Nikolovski, N.; Shliaha, P.V.; Gatto, L.; Dupree, P.; Lilley, K.S. Label-free protein quantification for plant Golgi protein localization and abundance. Plant Physiol. 2014, 166, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Zhang, Y.; Cheng, S.; Osorio, S.; Sun, Y.; Fernie, A.R.; Cheung, C.Y.; Lim, B.L. Impacts of high ATP supply from chloroplasts and mitochondria on the leaf metabolism of Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 922. [Google Scholar] [CrossRef] [PubMed]
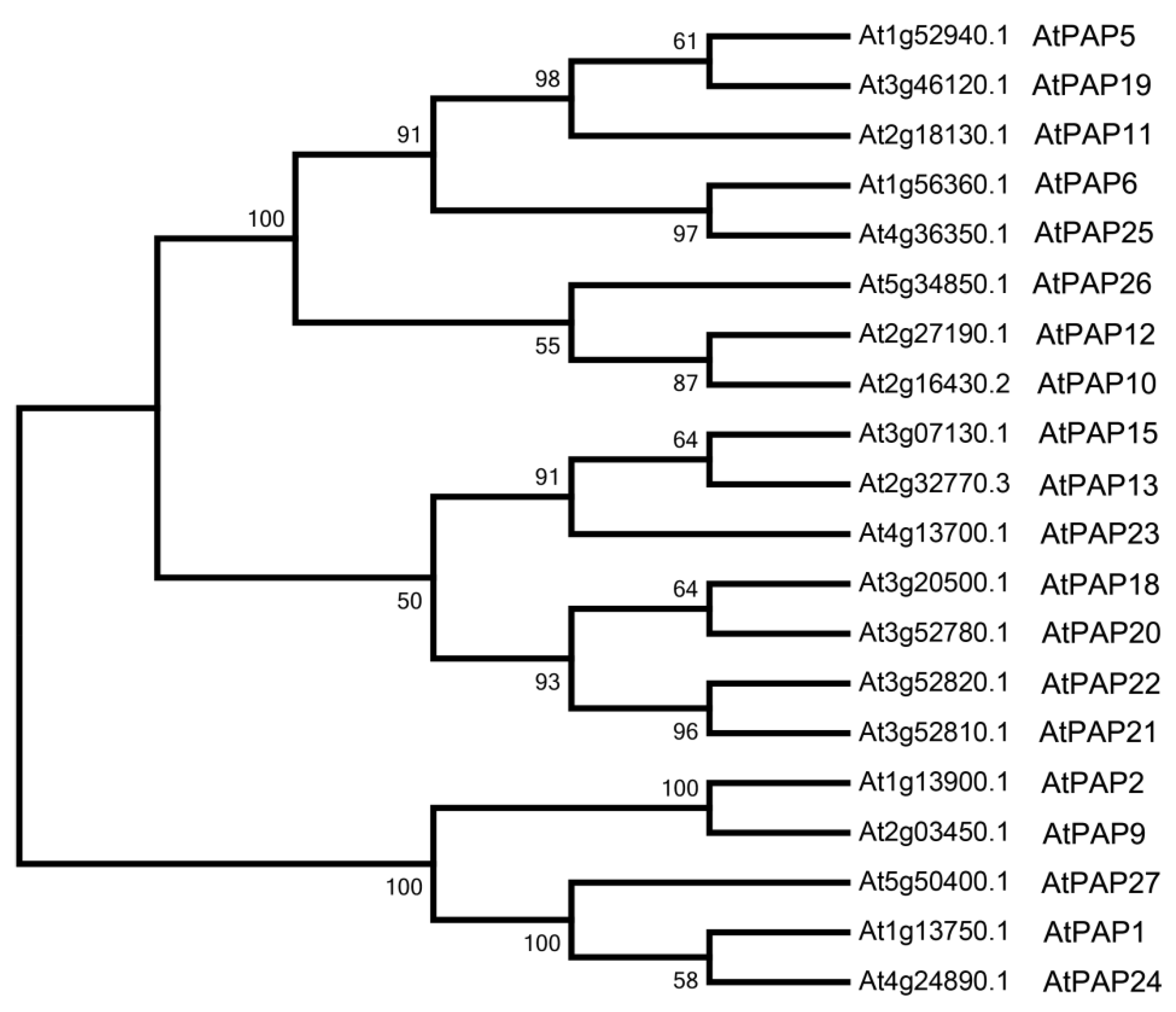

| Gene Name | AGI 1 Code | Protein Length | No. and Position of Transmembrane Domain (TMD) | Subcellular Localization 2 |
|---|---|---|---|---|
| AtPAP1 | At1g13750.1 | 613 aa 3 | - 4 | Secretory Pathway |
| AtPAP2 | At1g13900.1 | 656 aa | 1: 615–635 aa | Secretory Pathway |
| AtPAP5 | At1g52940.1 | 396 aa | - | Other |
| AtPAP6 | At1g56360.1 | 466 aa | - | Secretory Pathway |
| AtPAP9 | At2g03450.1 | 651 aa | 1: 606–626 aa | Secretory Pathway |
| AtPAP10 | At2g16430.1 | 348 aa | - | Mitochondrion |
| AtPAP10 | At2g16430.2 | 468 aa | - | Secretory Pathway |
| AtPAP11 | At2g18130.1 | 441 aa | - | Secretory Pathway |
| AtPAP12 | At2g27190.1 | 469 aa | - | Secretory Pathway |
| AtPAP13 | At2g32770.1 | 516 aa | - | Secretory Pathway |
| AtPAP13 | At2g32770.2 | 428 aa | - | Other |
| AtPAP13 | At2g32770.3 | 545 aa | - | Secretory Pathway |
| AtPAP15 | At3g07130.1 | 532 aa | - | Secretory Pathway |
| AtPAP18 | At3g20500.1 | 437 aa | - | Secretory Pathway |
| AtPAP19 | At3g46120.1 | 388 aa | - | Secretory Pathway |
| AtPAP20 | At3g52780.1 | 427 aa | - | Secretory Pathway |
| AtPAP20 | At3g52780.2 | 361 aa | - | Secretory Pathway |
| AtPAP21 | At3g52810.1 | 437 aa | - | Secretory Pathway |
| AtPAP22 | At3g52820.1 | 434 aa | - | Secretory Pathway |
| AtPAP23 | At4g13700.1 | 458 aa | - | Secretory Pathway |
| AtPAP24 | At4g24890.1 | 615 aa | - | Secretory Pathway |
| AtPAP25 | At4g36350.1 | 466 aa | - | Secretory Pathway |
| AtPAP26 | At5g34850.1 | 475 aa | - | Secretory Pathway |
| AtPAP27 | At5g50400.1 | 611 aa | - | Secretory Pathway |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Q.; Li, J.; Cheng, W.; Guo, H.; Liu, X.; Gao, H. AtPAP2, a Unique Member of the PAP Family, Functions in the Plasma Membrane. Genes 2018, 9, 257. https://doi.org/10.3390/genes9050257
Sun Q, Li J, Cheng W, Guo H, Liu X, Gao H. AtPAP2, a Unique Member of the PAP Family, Functions in the Plasma Membrane. Genes. 2018; 9(5):257. https://doi.org/10.3390/genes9050257
Chicago/Turabian StyleSun, Qingqing, Jinyu Li, Wenzhen Cheng, Huihong Guo, Xiaomin Liu, and Hongbo Gao. 2018. "AtPAP2, a Unique Member of the PAP Family, Functions in the Plasma Membrane" Genes 9, no. 5: 257. https://doi.org/10.3390/genes9050257
APA StyleSun, Q., Li, J., Cheng, W., Guo, H., Liu, X., & Gao, H. (2018). AtPAP2, a Unique Member of the PAP Family, Functions in the Plasma Membrane. Genes, 9(5), 257. https://doi.org/10.3390/genes9050257





