Low Maternal Microbiota Sharing across Gut, Breast Milk and Vagina, as Revealed by 16S rRNA Gene and Reduced Metagenomic Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohort
2.2. Generation of the Common 16S rRNA Dataset
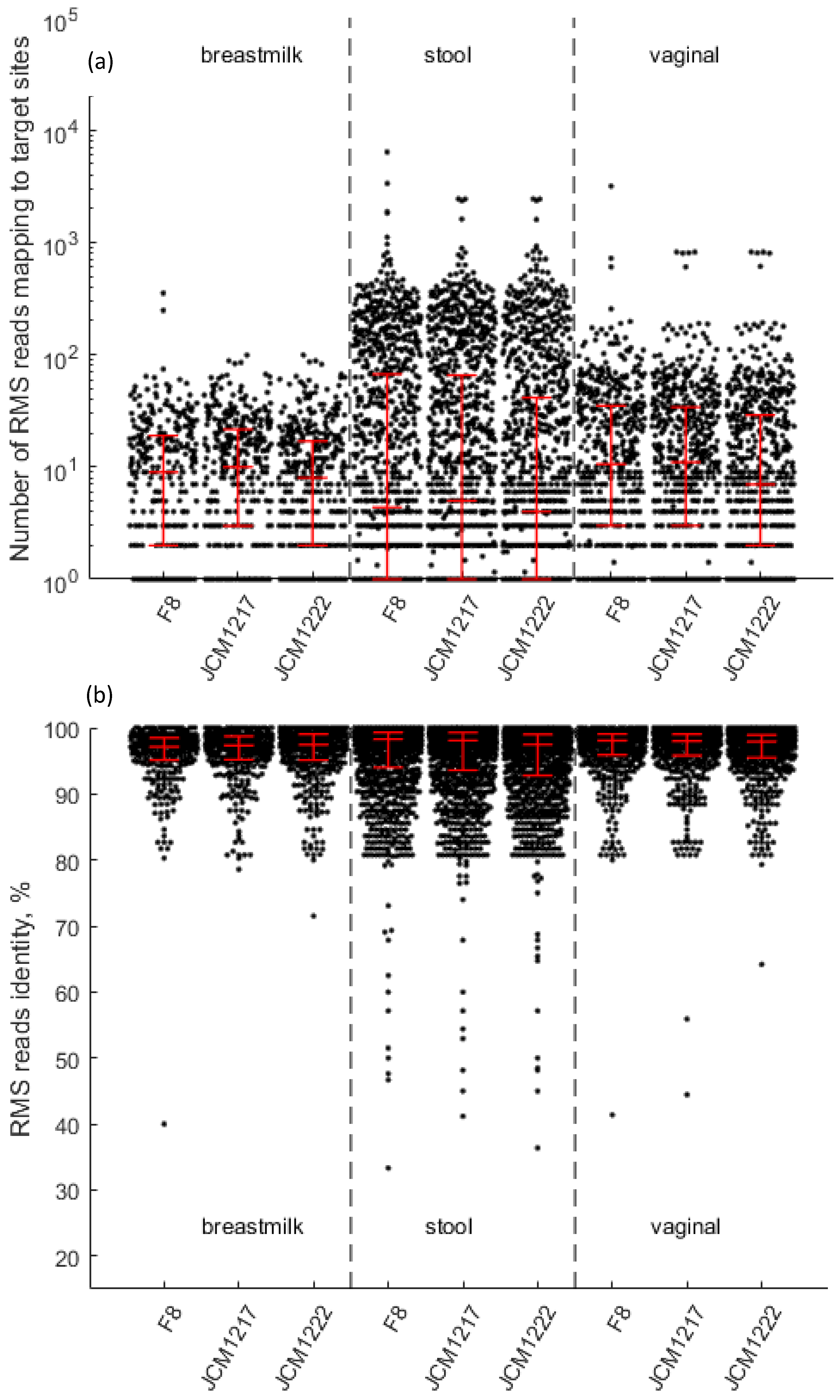
2.3. Generation of the RMS Dataset
2.4. Generation of the RMS Database Based on the Human Microbiome Project Representative Isolates (HMP-RMS Database)
2.5. Statistical Analyses
2.6. Data Availability
3. Results
3.1. Sharing of Bacteria between Breast Milk, Vaginal and Stool Microbiota, as Determined by 16S rRNA Gene Sequencing
3.2. RMS Analysis of Samples from Mothers with Shared BVS-OTUs
3.3. Microbiota Sharing between the Study Cohort and the HMP Database
3.4. Correlation between RMS and 16S rRNA Analysis
3.5. Assessment of the DNA Kitome
3.6. Search for Probiotic Bacteria in RMS Dataset
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Appendix A
Verification of the RMS Sequencing Approach
References
- Maynard, C.L.; Elcon, C.O.; Hatton, R.D.; Weaver, C.T. Reciprocal interactions of the intestinal microbiota and immune system. Nature 2012, 489, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Wilson, I.D.; Nicholson, J.K. The role of gut microbiota in drug response. Curr. Pharm. Des. 2009, 15, 1519–1523. [Google Scholar] [CrossRef] [PubMed]
- Foster, J.A.; McVey Neufeld, K.A. Gut-brain axis: How the microbiome influences anxiety and depression. Trends Neurosci. 2013, 36, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Devaraj, S.; Hemarajata, P.; Versalovic, J. The human gut microbiome and body metabolism: Implications for obesity and diabetes. Clin. Chem. 2013, 59, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Tremaroli, V.; Backhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Richardson, L.A. Evolving as a holobiont. PLoS Biol. 2017, 15, e2002168. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, S.F.; Sapp, J.; Tauber, A.I. A symbiotic view of life: We have never been individuals. Q. Rev. Biol. 2012, 87, 325–341. [Google Scholar] [CrossRef] [PubMed]
- Douglas, A.E.; Werren, J.H. Holes in the hologenome: Why host-microbe symbioses are not holobionts. Mbio 2016, 7, e02099. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, H.E.; Abrahamsson, T.R.; Jenmalm, M.C.; Harris, K.; Quince, C.; Jernberg, C.; Björksténet, B.; Engstrand, L.; Andersson, A.F. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut 2014, 63, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, E.; Fernández, L.; Marín, M.L.; Martín, R.; Odriozola, J.M.; Nueno-Palop, C.; Narbad, A.; Olivares, M.; Xaus, J.; Rodríguez, J.M. Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr. Microbiol. 2005, 51, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Perez-Munoz, M.E.; Arrieta, M.C.; Ramer-Tait, A.E.; Walter, J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: Implications for research on the pioneer infant microbiome. Microbiome 2017, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- Martin, V.; Maldonado-Barragán, A.; Moles, L.; Rodriguez-Baños, M.; Campo, R.D.; Fernández, L.; Rodríguez, J.M.; Jiménez, E. Sharing of bacterial strains between breast milk and infant feces. J. Hum. Lact. 2012, 28, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Jost, T.; Lacroix, C.; Braegger, C.P.; Rochat, F.; Chassard, C. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ. Microbiol. 2014, 16, 2891–2904. [Google Scholar] [CrossRef] [PubMed]
- Perez, P.F.; Doré, J.; Leclerc, M.; Levenez, F.; Benyacoub, J.; Serrant, P.; Segura-Roggero, I.; Schiffrin, E.J.; Donnet-Hughes, A. Bacterial imprinting of the neonatal immune system: Lessons from maternal cells? Pediatrics 2007, 119, e724–e732. [Google Scholar] [CrossRef] [PubMed]
- Benitez-Paez, A.; Portune, K.J.; Sanz, Y. Species-level resolution of 16S rRNA gene amplicons sequenced through the MinION portable nanopore sequencer. Gigascience 2016, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Knight, R.; Siljander, H.; Knip, M.; Xavier, R.J.; Gevers, D. ConStrains identifies microbial strains in metagenomic datasets. Nat. Biotechnol. 2015, 33, 1045–1052. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Z.; Bushman, F.D.; Li, H.Z. A Model-based approach for species abundance quantification based on shotgun metagenomic data. Stat. Biosci. 2017, 9, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.Y.; Worden, P.; Monahan, L.G.; DeMaere, M.Z.; Burke, C.M.; Djordjevic, S.P.; Charles, I.G.; Darling, A.E. Evaluation of ddRAD seq for reduced representation metagenome sequencing. PeerJ 2017, 5, e3837. [Google Scholar] [CrossRef] [PubMed]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; van de Lee, T.; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef] [PubMed]
- Ravi, A.; Avershina, E.; Angell, I.L.; Ludvigsen, J.; Manohar, P.; Padmanaban, S.; Nachimuthu, R.; Snipen, L.; Rudi, K. Comparison of reduced metagenome and 16S rRNA gene sequencing for determination of genetic diversity and mother-child overlap of the gut associated microbiota. J. Microbiol. Methods 2018. [Google Scholar] [CrossRef] [PubMed]
- Dotterud, C.K.; Storrø, O.; Johnsen, R.; Oien, T. Probiotics in pregnant women to prevent allergic disease: A randomized, double-blind trial. Br. J. Dermatol. 2010, 163, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Simpson, M.R.; Avershina, E.; Storrø, O.; Johnsen, R.; Rudi, K.; Øien, T. Breastfeeding-associated microbiota in human milk following supplementation with Lactobacillus rhamnosus GG, Lactobacillus acidophilus La-5, and Bifidobacterium animalis ssp. lactis Bb-12. J. Dairy Sci. 2018, 101, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Avershina, E.; Slangsvold, S.; Simpson, M.R.; Storrø, O.; Johnsen, R.; Øien, T.; Rudi, K. Diversity of vaginal microbiota increases by the time of labor onset. Sci. Rep. 2017, 7, 17558. [Google Scholar] [CrossRef] [PubMed]
- Avershina, E.; Lundgård, K.; Sekelja, M.; Dotterud, C.; Storrø, O.; Øien, T.; Johnsen, R.; Rudi, K. Transition from infant- to adult-like gut microbiota. Environ. Microbiol. 2016, 18, 2226–2236. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, B. BBMap Short Read Aligner; University of California: Berkeley, CA, USA, 2015. [Google Scholar]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Price, J.; Mahurkar, A.; Rahnavard, G.; Crabtree, J.; Orvis, J.; Hall, A.B.; Brady, A.; Creasy, H.H.; McCracken, C.; Giglio, M.G.; et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 2017, 550, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Song, S.J.; Lauber, C.; Costello, E.K.; Lozupone, C.A.; Humphrey, G.; Berg-Lyons, D.; Caporaso, J.G.; Knights, D.; Clemente, J.C.; Nakielny, S.; et al. Cohabiting family members share microbiota with one another and with their dogs. Elife 2013, 2, e00458. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, E.; Fernández, L.; Maldonado, A.; Martín, R.; Olivares, M.; Xaus, J.; Rodríguez, J.M. Oral administration of Lactobacillus strains isolated from breast milk as an alternative for the treatment of infectious mastitis during lactation. Appl. Environ. Microbiol. 2008, 74, 4650–4655. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, R.; Martín, V.; Maldonado, A.; Jiménez, E.; Fernández, L.; Rodríguez, J.M. Treatment of infectious mastitis during lactation: Antibiotics versus oral administration of Lactobacilli isolated from breast milk. Clin. Infect. Dis. 2010, 50, 1551–1558. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.M. The origin of human milk bacteria: Is there a bacterial entero-mammary pathway during late pregnancy and lactation? Adv. Nutr. 2014, 5, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Kozak, K.; Charbonneau, D.; Sanozky-Dawes, R.; Klaenhammer, T. Characterization of bacterial isolates from the microbiota of mothers’ breast milk and their infants. Gut Microbes 2015, 6, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Freitas, A.C.; Hill, J.E. Quantification, isolation and characterization of Bifidobacterium from the vaginal microbiomes of reproductive aged women. Anaerobe 2017, 47, 145–156. [Google Scholar] [CrossRef] [PubMed]
- DeMarco, A.L.; Rabe, L.K.; Austin, M.N.; Stoner, K.A.; Avolia, H.A.; Meyn, L.A.; Hillier, S.L. Survival of vaginal microorganisms in three commercially available transport systems. Anaerobe 2017, 45, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Lawley, B.; Munro, K.; Hughes, A.; Hodgkinson, A.J.; Prosser, C.G.; Lowry, D.; Zhou, S.J.; Makrides, M.; Gibson, R.A.; Lay, C.; et al. Differentiation of Bifidobacterium longum subspecies longum and infantis by quantitative PCR using functional gene targets. PeerJ 2017, 5, e3375. [Google Scholar] [CrossRef] [PubMed]
- Avershina, E.; Storrø, O.; Øien, T.; Johnsen, R.; Wilson, R.; Egeland, T.; Rudi, K. Bifidobacterial succession and correlation networks in a large unselected cohort of mothers and their children. Appl. Environ. Microbiol. 2013, 79, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.; Heilig, H.G.; Zoetendal, E.G.; Jiménez, E.; Fernández, L.; Smidt, H.; Rodríguez, J.M. Cultivation-independent assessment of the bacterial diversity of breast milk among healthy women. Res. Microbiol. 2007, 158, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Saez-Lopez, E.; Guiral, E.; Fernández-Orth, D.; Villanueva, S.; Goncé, A.; López, M.; Teixidó, I.; Pericot, A.; Figueras, F.; Palacio, M.; et al. Vaginal versus obstetric infection Escherichia coli isolates among pregnant women: Antimicrobial resistance and genetic virulence profile. PLoS ONE 2016, 11, e0146531. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avershina, E.; Angell, I.L.; Simpson, M.; Storrø, O.; Øien, T.; Johnsen, R.; Rudi, K. Low Maternal Microbiota Sharing across Gut, Breast Milk and Vagina, as Revealed by 16S rRNA Gene and Reduced Metagenomic Sequencing. Genes 2018, 9, 231. https://doi.org/10.3390/genes9050231
Avershina E, Angell IL, Simpson M, Storrø O, Øien T, Johnsen R, Rudi K. Low Maternal Microbiota Sharing across Gut, Breast Milk and Vagina, as Revealed by 16S rRNA Gene and Reduced Metagenomic Sequencing. Genes. 2018; 9(5):231. https://doi.org/10.3390/genes9050231
Chicago/Turabian StyleAvershina, Ekaterina, Inga Leena Angell, Melanie Simpson, Ola Storrø, Torbjørn Øien, Roar Johnsen, and Knut Rudi. 2018. "Low Maternal Microbiota Sharing across Gut, Breast Milk and Vagina, as Revealed by 16S rRNA Gene and Reduced Metagenomic Sequencing" Genes 9, no. 5: 231. https://doi.org/10.3390/genes9050231
APA StyleAvershina, E., Angell, I. L., Simpson, M., Storrø, O., Øien, T., Johnsen, R., & Rudi, K. (2018). Low Maternal Microbiota Sharing across Gut, Breast Milk and Vagina, as Revealed by 16S rRNA Gene and Reduced Metagenomic Sequencing. Genes, 9(5), 231. https://doi.org/10.3390/genes9050231





