Genome-Wide Analysis of the PYL Gene Family and Identification of PYL Genes That Respond to Abiotic Stress in Brassica napus
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Stress Treatments
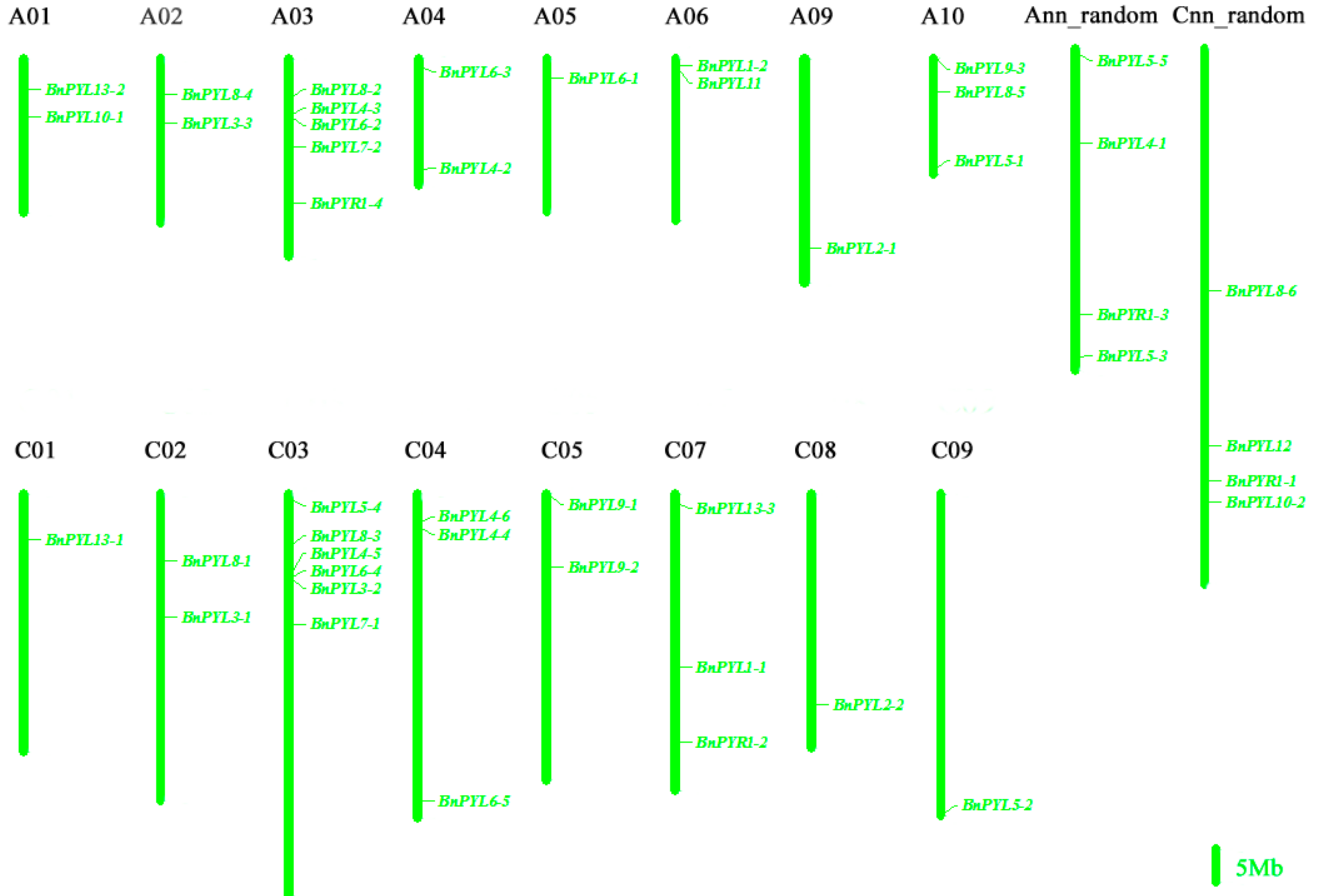
2.2. Genome-Wide Identification and Chromosomal Location of PYL Gene Family in B. napus
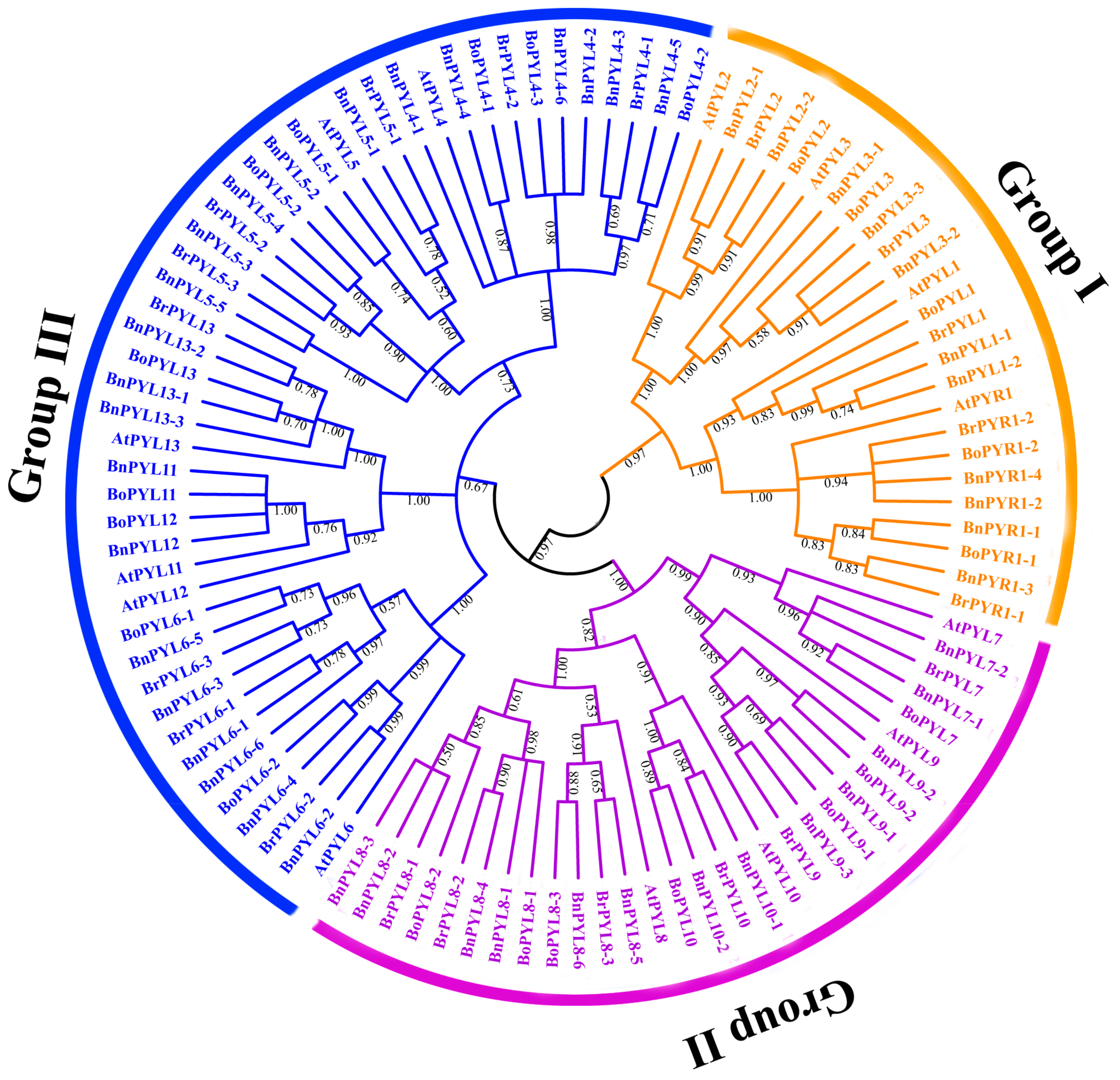
2.3. Phylogenetic Tree Analysis of PYL Gene Family in B. napus, B. rapa, B. oleracea and A. thaliana
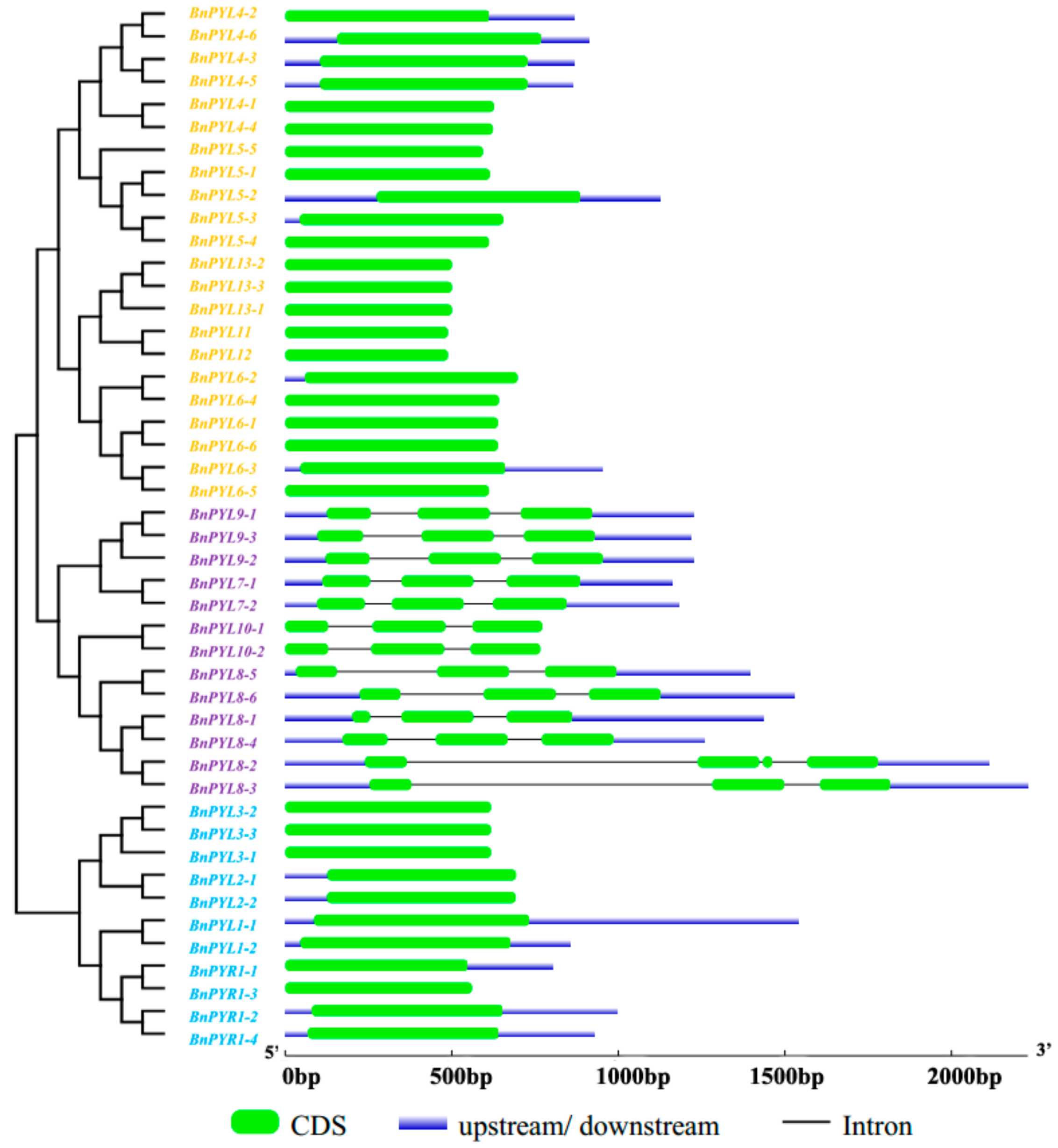
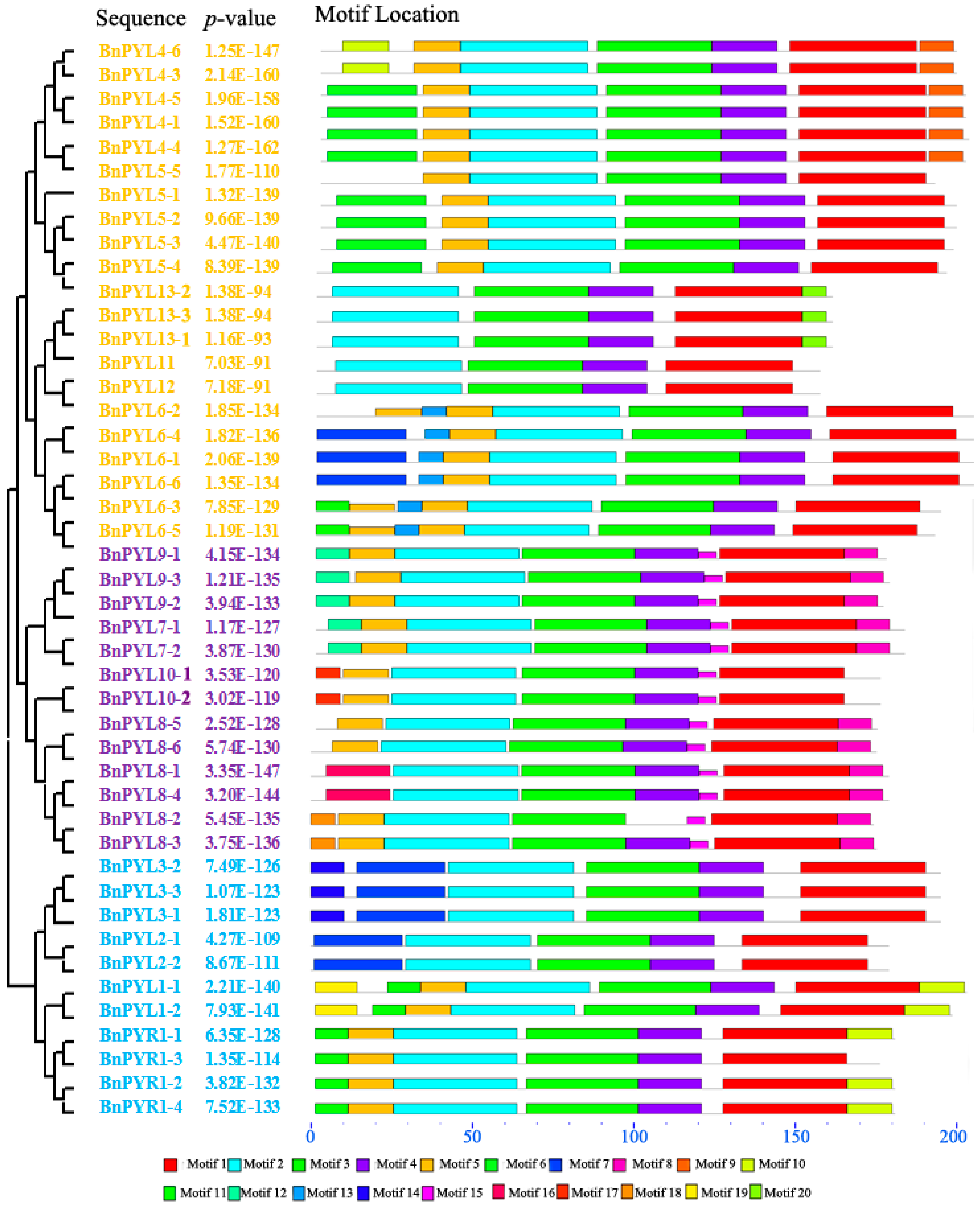
2.4. Analysis of Gene Exon-Intron Structures and Protein Conserved Motifs
2.5. Analyzing cis-Elements in the BnPYL Promoters
2.6. Prediction of miRNAs Targeting BnPYL Genes
2.7. Analysis of Gene Expression Profiles and Gene Ontology Enrichment
2.8. RNA Extraction and Real-Time RT-PCR
3. Results
3.1. Characterization of BnPYL Gene Family
3.2. Analysis of Phylogenetic Relationships and Gene Structures of BnPYLs
3.3. Analysis of BnPYL Conserved Motifs
3.4. Cis-Elements in BnPYL Promoters
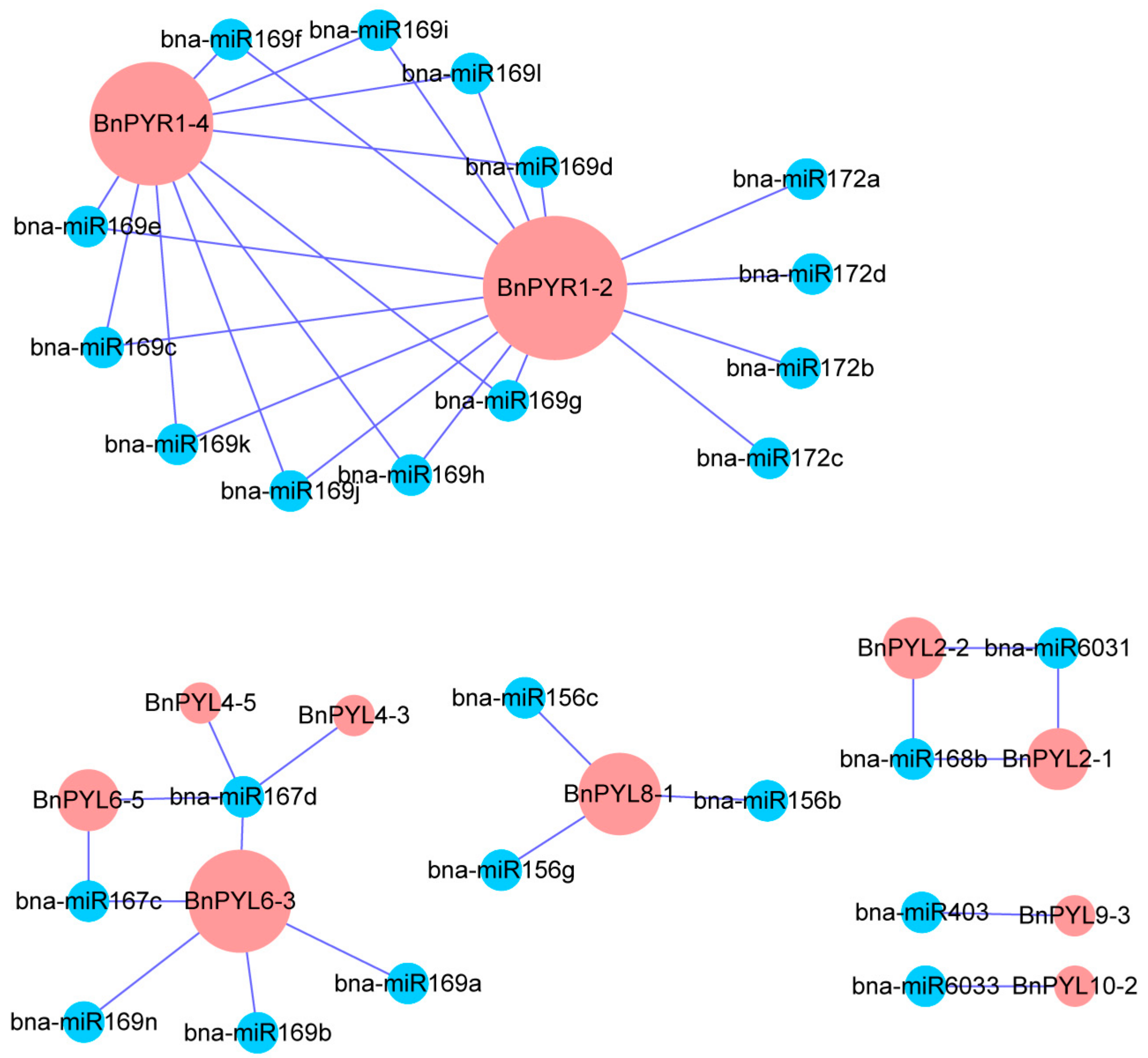
3.5. Comprehensive Analysis of microRNA Targeting BnPYL Genes
3.6. Analysis of BnPYL Expression Levels in Tissues
3.7. Gene Ontology Enrichment
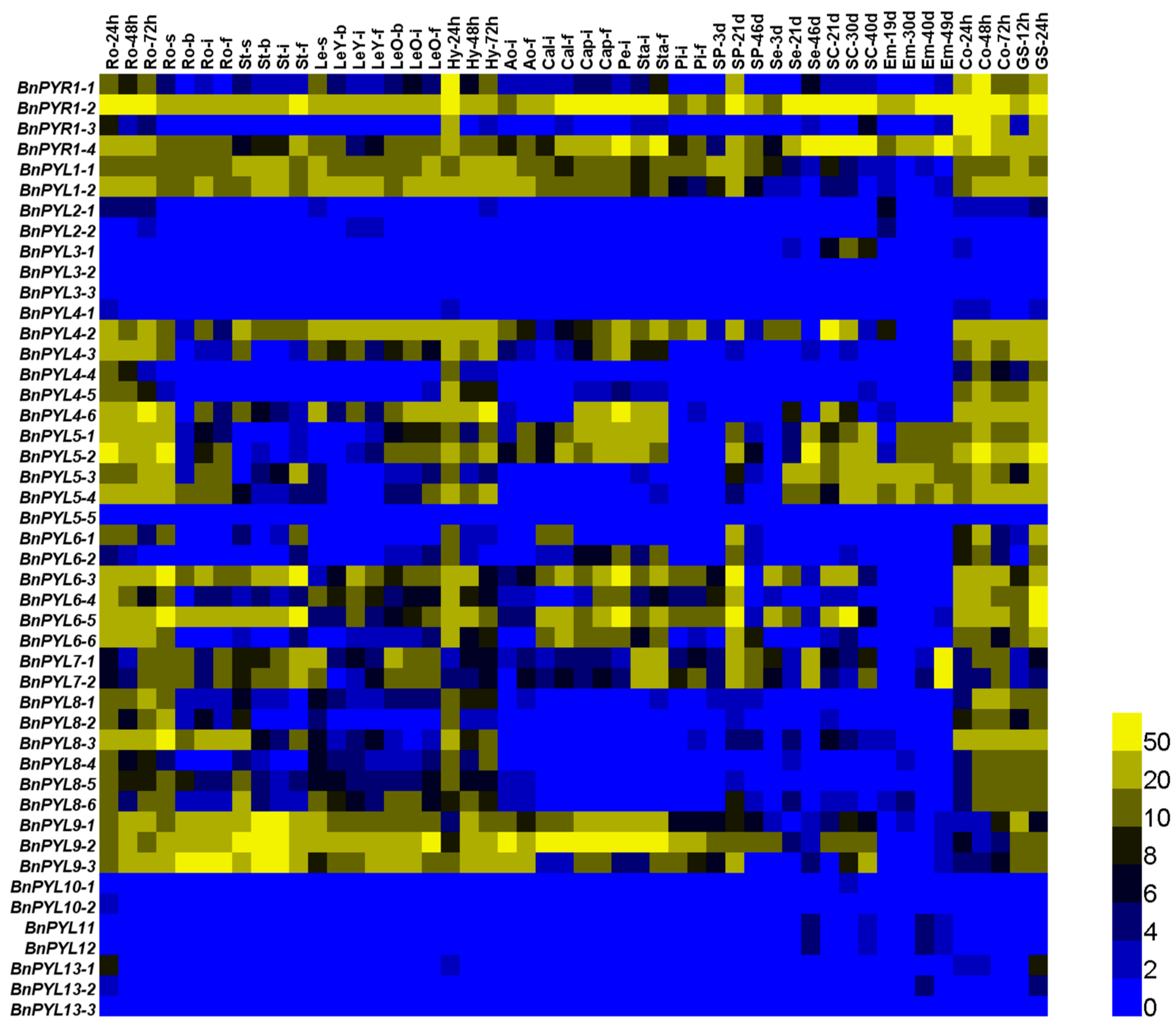
3.8. The Expression Patterns of PYLs in B. napus under Abiotic Stress
4. Discussion
4.1. Characterization of PYL Gene Family in Brassica napus
4.2. Expression Levels of BnPYLs in Various Tissues
4.3. The Expression Patterns of BnPYLs under Abiotic Stresses
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Apweiler, R.; Bairoch, A.M.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2004, 32, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Lumba, S.; Cutler, S.; McCourt, P. Plant nuclear hormone receptors: A role for small molecules in protein-protein interactions. Annu. Rev. Cell Dev. Biol. 2010, 26, 445–469. [Google Scholar] [CrossRef] [PubMed]
- Nambara, E.; Marionpoll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Zhu, J. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Miyazono, K.; Miyakawa, T.; Sawano, Y.; Kubota, K.; Kang, H.; Asano, A.; Miyauchi, Y.; Takahashi, M.; Zhi, Y.; Fujita, Y. Structural basis of abscisic acid signalling. Nature 2009, 462, 609–614. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Soon, F.F.; Ng, L.; Zhou, X.E.; West, G.M.; Kovach, A.; Tan, M.H.E.; Suinopowell, K.; He, Y.; Xu, Y.; Chalmers, M.J. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 2012, 335, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Verslues, P.E.; Zhu, J. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in arabidopsis. Plant Cell 2007, 19, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Murata, M.; Minami, H.; Yamamoto, S.; Kagaya, Y.; Hobo, T.; Yamamoto, A.; Hattori, T. Abscisic acid-activated SnRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 2005, 44, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Verslues, P.E.; Agarwal, M.; Katiyaragarwal, S.; Zhu, J.; Zhu, J. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006, 45, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.; Dupeux, F.; Betz, K.; Antoni, R.; Gonzalezguzman, M.; Rodriguez, L.; Marquez, J.A.; Rodriguez, P.L. Structural insights into PYR/PYL/RCAR ABA receptors and PP2Cs. Plant Sci. 2012, 182, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.; Lafuente, M.T.; Rodrigo, M.J. The citrus ABA signalosome: Identification and transcriptional regulation during sweet orange fruit ripening and leaf dehydration. J. Exp. Bot. 2012, 63, 4931–4945. [Google Scholar] [CrossRef] [PubMed]
- Boneh, U.; Biton, I.; Zheng, C.; Schwartz, A.; Benari, G. Characterization of potential ABA receptors in Vitis vinifera. Plant Cell Rep. 2012, 31, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Bai, G.; Yang, D.; Zhao, Y.; Ha, S.; Yang, F.; Ma, J.; Gao, X.; Wang, Z.; Zhu, J. Interactions between soybean ABA receptors and type 2C protein phosphatases. Plant Mol. Biol. 2013, 83, 651–664. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Hao, Q.; Li, W.; Yan, C.; Yan, N.; Yin, P. Identification and characterization of ABA receptors in Oryza sativa. PLoS ONE 2014, 9, e95246. [Google Scholar] [CrossRef] [PubMed]
- Gonzalezguzman, M.; Rodriguez, L.; Lorenzoorts, L.; Pons, C.; Sarrionperdigones, A.; Fernandez, M.A.; Peiratsllobet, M.; Forment, J.; Morenoalvero, M.; Cutler, S.R. Tomato PYR/PYL/RCAR abscisic acid receptors show high expression in root, differential sensitivity to the abscisic acid agonist quinabactin, and the capability to enhance plant drought resistance. J. Exp. Bot. 2014, 65, 4451–4464. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Zhou, Y.; Li, H.L.; Zhu, J.H.; Wang, Y.; Chen, X.T.; Peng, S.Q. Identification and characterization of the abscisic acid (ABA) receptor gene family and its expression in response to hormones in the rubber tree. Sci. Rep. 2017, 7, 45157. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, D.; Sun, C.; Hu, X.; Mu, X.; Hu, J.; Yang, Y.; Zhang, Y.; Xie, C.G.; Zhou, X. Molecular characterization of an atPYL1-like protein, brPYL1, as a putative ABA receptor in Brassica rapa. Biochem. Biophys. Res. Commun. 2017, 487, 684–689. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Feng, L.; Wei, N.; Liu, Z.H.; Hu, S.; Li, X.B. Overexpression of cotton PYL genes in Arabidopsis enhances the transgenic plant tolerance to drought stress. Plant Physiol. Biochem. 2017, 115, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Pizzio, G.A.; Rodriguez, L.; Antoni, R.; Gonzalezguzman, M.; Yunta, C.; Merilo, E.; Kollist, H.; Albert, A.; Rodriguez, P.L. The PYL4 A194T mutant uncovers a key role of PYR1-LIKE4/PROTEIN PHOSPHATASE 2CA interaction for abscisic acid signaling and plant drought resistance. Plant Physiol. 2013, 163, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, K.; Hwang, H.; Bhatnagar, N.; Kim, D.Y.; Yoon, I.S.; Byun, M.; Kim, S.T.; Jung, K.; Kim, B. Overexpression of PYL5 in rice enhances drought tolerance, inhibits growth, and modulates gene expression. J. Exp. Bot. 2014, 65, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Zhao, Y.; Gao, J.; Xiang, C.; Zhu, J. The ABA receptor PYL9 together with PYL8 plays an important role in regulating lateral root growth. Sci. Rep. 2016, 6, 27177. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef] [PubMed]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B. Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.C.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhao, P.X. Psrnatarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.A.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with tophat and cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Wang, Y.; Liu, Z.; Cheng, H.; Xue, Y. HEMI: A toolkit for illustrating heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, K.D.; Tatusova, T.; Brown, G.; Maglott, D. NCBI reference sequences (RefSeq): Current status, new features and genome annotation policy. Nucleic Acids Res. 2012, 40, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.L.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Krylov, D.M.; Makarova, K.S.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; Rao, B.S. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004, 5, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Oono, Y.; Zhu, J.; He, X.; Wu, J.; Iida, K.; Lu, X.; Cui, X.; Jin, H.; Zhu, J. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 2008, 20, 2238–2251. [Google Scholar] [CrossRef] [PubMed]
- Cavell, A.C.; Lydiate, D.J.; Parkin, I.A.P.; Dean, C.; Trick, M. Collinearity between a 30-centimorgan segment of Arabidopsis thaliana chromosome 4 and duplicated regions within the Brassica napus genome. Genome 1998, 41, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Luan, S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 2012, 35, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Shinozaki, K.; Yamaguchishinozaki, K. Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 2011, 52, 1569–1582. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Merlot, S.; Gosti, F.; Guerrier, D.; Vavasseur, A.; Giraudat, J. The ABI1 and ABIi2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 2001, 25, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.; Rodrigues, A.; Saez, A.; Rubio, S.; Antoni, R.; Dupeux, F.; Park, S.; Marquez, J.A.; Cutler, S.R.; Rodriguez, P.L. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade a PP2Cs. Plant J. 2009, 60, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S. ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 2014, 202, 35–49. [Google Scholar] [CrossRef] [PubMed]

| Gene ID | Gene Name | Position (bp) | Gene Length (bp) | CDS Length (bp) | Exon | Peptide Residues | MW (kDa) | pI |
|---|---|---|---|---|---|---|---|---|
| BnaCnng65400D | BnPYR1-1 | 65121492-65122292 | 801 | 576 | 1 | 191 | 21.69 | 6.39 |
| BnaC07g34880D | BnPYR1-2 | 37423726-37424722 | 997 | 576 | 1 | 191 | 21.40 | 5.98 |
| BnaAnng35310D | BnPYR1-3 | 40055219-40055779 | 561 | 561 | 1 | 186 | 21.11 | 6.39 |
| BnaA03g43410D | BnPYR1-4 | 21808356-21809284 | 929 | 576 | 1 | 191 | 21.37 | 5.98 |
| BnaC07g19450D | BnPYL1-1 | 26187392-26188931 | 1540 | 648 | 1 | 215 | 24.38 | 5.29 |
| BnaA06g40360D | BnPYL1-2 | 200261-2003467 | 854 | 633 | 1 | 210 | 23.91 | 5.20 |
| BnaA09g40690D | BnPYL2-1 | 28565054-28565746 | 693 | 567 | 1 | 188 | 20.83 | 5.49 |
| BnaC08g33170D | BnPYL2-2 | 31754064-31754755 | 692 | 576 | 1 | 188 | 21.03 | 5.86 |
| BnaC02g21700D | BnPYL3-1 | 18634088-18634705 | 618 | 618 | 1 | 205 | 22.91 | 8.87 |
| BnaC03g23260D | BnPYL3-2 | 12925626-12926243 | 618 | 618 | 1 | 205 | 22.91 | 8.88 |
| BnaA02g16230D | BnPYL3-3 | 9682751-9683368 | 618 | 618 | 1 | 205 | 22.88 | 8.91 |
| BnaAnng13200D | BnPYL4-1 | 14172422-14173048 | 627 | 627 | 1 | 208 | 22.57 | 7.08 |
| BnaA04g21960D | BnPYL4-2 | 16651226-16652092 | 867 | 615 | 1 | 204 | 21.99 | 6.22 |
| BnaA03g17720D | BnPYL4-3 | 8353447-8354313 | 867 | 624 | 1 | 207 | 22.24 | 6.43 |
| BnaC04g07010D | BnPYL4-4 | 5159788-5160411 | 624 | 624 | 1 | 207 | 22.42 | 7.08 |
| BnaC03g21240D | BnPYL4-5 | 11473885-11474748 | 864 | 624 | 1 | 207 | 22.29 | 6.43 |
| BnaC04g56560D | BnPYL4-6 | 4191879-4192790 | 912 | 615 | 1 | 204 | 21.94 | 6.04 |
| BnaA10g24990D | BnPYL5-1 | 16198838-16199452 | 615 | 615 | 1 | 204 | 22.78 | 6.02 |
| BnaC09g49910D | BnPYL5-2 | 48035079-48036202 | 1124 | 615 | 1 | 204 | 22.77 | 5.82 |
| BnaAnng40650D | BnPYL5-3 | 46553930-46554584 | 655 | 612 | 1 | 203 | 22.71 | 5.80 |
| BnaC03g02130D | BnPYL5-4 | 994403-995014 | 612 | 612 | 1 | 203 | 22.68 | 5.91 |
| BnaAnng01330D | BnPYL5-5 | 811157-811750 | 594 | 594 | 1 | 197 | 22.02 | 5.91 |
| BnaA05g05420D | BnPYL6-1 | 2798876-2799514 | 639 | 639 | 1 | 212 | 23.48 | 6.52 |
| BnaA03g19030D | BnPYL6-2 | 9003640-9004337 | 698 | 639 | 1 | 212 | 23.52 | 6.56 |
| BnaA04g29300D | BnPYL6-3 | 1303160-1304112 | 953 | 618 | 1 | 205 | 22.77 | 6.09 |
| BnaC03g22610D | BnPYL6-4 | 12499410-12500051 | 642 | 642 | 1 | 213 | 23.71 | 6.38 |
| BnaC04g47050D | BnPYL6-5 | 46155948-46156559 | 612 | 612 | 1 | 203 | 22.50 | 6.10 |
| BnaC04g04830D | BnPYL6-6 | 3526371-3527009 | 639 | 639 | 1 | 212 | 23.50 | 6.66 |
| BnaC03g31730D | BnPYL7-1 | 19523342-19524503 | 1162 | 582 | 1 | 193 | 21.69 | 6.05 |
| BnaA03g26790D | BnPYL7-2 | 13185542-13186722 | 1181 | 582 | 1 | 193 | 21.80 | 6.24 |
| BnaC02g14540D | BnPYL8-1 | 10060041-10061474 | 1434 | 567 | 3 | 188 | 21.32 | 6.71 |
| BnaA03g12450D | BnPYL8-2 | 5667935-5670048 | 2114 | 552 | 4 | 183 | 20.67 | 6.24 |
| BnaC03g15210D | BnPYL8-3 | 7539236-7541464 | 2229 | 555 | 3 | 184 | 20.80 | 6.24 |
| BnaA02g10420D | BnPYL8-4 | 5349136-5350394 | 1259 | 567 | 3 | 188 | 21.26 | 6.24 |
| BnaA10g06520D | BnPYL8-5 | 4967767-4969159 | 1393 | 555 | 3 | 184 | 21.03 | 6.07 |
| BnaCnng37890D | BnPYL8-6 | 36425463-36426990 | 1528 | 555 | 3 | 184 | 20.93 | 6.07 |
| BnaC05g00620D | BnPYL9-1 | 331808-333031 | 1224 | 564 | 3 | 187 | 21.03 | 5.98 |
| BnaC05g17260D | BnPYL9-2 | 10921616-10922842 | 1227 | 561 | 3 | 186 | 21.02 | 5.98 |
| BnaA10g00540D | BnPYL9-3 | 270030-271246 | 1217 | 567 | 3 | 188 | 21.20 | 6.06 |
| BnaA01g16740D | BnPYL10-1 | 8730925-8731696 | 772 | 558 | 3 | 185 | 20.78 | 5.61 |
| BnaCnng68710D | BnPYL10-2 | 68363157-68363922 | 766 | 558 | 3 | 185 | 20.73 | 5.71 |
| BnaA06g40220D | BnPYL11 | 1928871-1929359 | 489 | 489 | 1 | 162 | 17.82 | 5.40 |
| BnaCnng60010D | BnPYL12 | 59845750-59846238 | 489 | 489 | 1 | 162 | 17.83 | 5.40 |
| BnaC01g11020D | BnPYL13-1 | 6873982-6874482 | 501 | 501 | 1 | 166 | 18.41 | 5.12 |
| BnaA01g09460D | BnPYL13-2 | 4636999-4637499 | 501 | 501 | 1 | 166 | 18.40 | 5.26 |
| BnaC07g48850D | BnPYL13-3 | 1477983-1478483 | 501 | 501 | 1 | 166 | 18.41 | 5.26 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di, F.; Jian, H.; Wang, T.; Chen, X.; Ding, Y.; Du, H.; Lu, K.; Li, J.; Liu, L. Genome-Wide Analysis of the PYL Gene Family and Identification of PYL Genes That Respond to Abiotic Stress in Brassica napus. Genes 2018, 9, 156. https://doi.org/10.3390/genes9030156
Di F, Jian H, Wang T, Chen X, Ding Y, Du H, Lu K, Li J, Liu L. Genome-Wide Analysis of the PYL Gene Family and Identification of PYL Genes That Respond to Abiotic Stress in Brassica napus. Genes. 2018; 9(3):156. https://doi.org/10.3390/genes9030156
Chicago/Turabian StyleDi, Feifei, Hongju Jian, Tengyue Wang, Xueping Chen, Yiran Ding, Hai Du, Kun Lu, Jiana Li, and Liezhao Liu. 2018. "Genome-Wide Analysis of the PYL Gene Family and Identification of PYL Genes That Respond to Abiotic Stress in Brassica napus" Genes 9, no. 3: 156. https://doi.org/10.3390/genes9030156
APA StyleDi, F., Jian, H., Wang, T., Chen, X., Ding, Y., Du, H., Lu, K., Li, J., & Liu, L. (2018). Genome-Wide Analysis of the PYL Gene Family and Identification of PYL Genes That Respond to Abiotic Stress in Brassica napus. Genes, 9(3), 156. https://doi.org/10.3390/genes9030156








