Abstract
Small RNAs have been studied in detail in domains Bacteria and Eukarya but, in the case of the domain Archaea, the knowledge is scarce and the physiological function of these small RNAs (sRNAs) is still uncertain. To extend the knowledge of sRNAs in the domain Archaea and their possible role in the regulation of the nitrogen assimilation metabolism in haloarchaea, Haloferax mediterranei has been used as a model microorganism. The bioinformatic approach has allowed for the prediction of 295 putative sRNAs genes in the genome of H. mediterranei, 88 of which have been verified by means of RNA-Sequencing (RNA-Seq). The secondary structure of these sRNAs and their possible targets have been identified. Curiously, some of them present as possible target genes relating to nitrogen assimilation, such as glutamate dehydrogenase and the nitrogen regulatory PII protein. Analysis of RNA-Seq data has also revealed differences in the expression pattern of 16 sRNAs according to the nitrogen source. Consequently, RNomic and bioinformatic approaches used in this work have allowed for the identification of new sRNAs in H. mediterranei, some of which show different expression patterns depending on the nitrogen source. This suggests that these sRNAs could be involved in the regulation of nitrogen assimilation and can constitute an important gene regulatory network.
1. Introduction
Small RNAs (sRNAs) play an essential role in the post-transcriptional regulation of many cellular processes in all domains of life, i.e., Eukarya, Bacteria, and Archaea. In eukaryotes, there are different classes of these, the best studied being microRNA (miRNAs), small interference RNA (siRNAs), and piwi-interacting RNAs (piRNAs). These are approximately 20–30 nucleotides (nt) in length and are involved in development, cellular activities, and different physiology processes [1,2,3]. In Archaea and Bacteria, sRNAs are much longer than eukaryotic small non-coding RNAs, which range in length from 50 to 500 nt [4,5,6,7]. Different mechanisms of action of sRNAs have been described, most of which affect the translation of the target messenger RNA and/or its stability [4]. Hence, they seem to be involved in the post-transcriptional regulation of metabolism, stress response, virulence processes, and so on. Depending on the location of their targets, sRNAs are classified into two groups: trans-encoded and cis-encoded sRNAs [8]. Trans-encoded sRNAs are those that are encoded within intergenic regions of the genome: they show a stable secondary structure and act on target sequences located at different positions in the genome. The complementarity between the sRNA and its target sequence is not complete and, for this reason, they require the presence of RNA chaperones to facilitate nucleotide binding [9]. In contrast, cis-encoded sRNAs originate in the nonsense strand of an open reading frame (ORF). Generally, that ORF corresponds to the sRNA target which presents complete complementarity to the cis-encoded sRNA. In addition, there have also been other types of RNA identified in Archaea: small nucleolar RNAs (snoRNAs) involved in the modifications of ribosomal RNA, whose presence was originally believed to be restricted to eukaryotic organisms [10]; sRNAs involved in the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas prokaryotes immune system called crRNAs [11]; and sRNAs that derive from transfer RNAs called tRFs [12].
In Eukarya and Bacteria, many sRNAs have been characterised in detail. However, in domain Archaea, though there has been considerable progress in recent years, the number of sRNAs characterised is considerably lower than those in the other two domains [5,13,14]. The application of bioinformatic approaches and high-throughput sequencing systems for the analysis of complementary DNA (cDNA) libraries, RNA-Sequencing (RNA-Seq), have made it easier to understand the transcriptome and discover new sRNAs. However, to date, the identification of sRNAs using RNA-Seq analysis has only been carried out on seven species of Archaea under specific conditions: Archaeoglobus fulgidus [15], Sulfolobus solfataricus [16], Pyrococcus abyssi [10], Methanosarcina mazei [17], Haloferax volcanii [13], Pyrobaculum sp. [18], and Thermococcus kodakaraensis [19]. In these studies, it has been shown that the majority of sRNAs identified in Archaea are not conserved, even within species of the same genus [5]. This fact has also been observed in Bacteria, so it appears that the evolution of sRNA genes in prokaryotic organisms is greater than that of genes which encode proteins. Despite the large number of sRNAs identified in Archaea in recent years, very little is known about their biological functions and mechanisms of interaction, with their possible targets still an unknown and unexplored area of research.
Haloferax mediterranei is an extremely halophilic archaeon which belongs to the lineage of Euryarchaeota. This microorganism grows optimally at 2.5 M NaCl [20] in a defined medium with glucose as a carbon source and nitrate, nitrite, amino acids, or ammonium as sole nitrogen sources under aerobic conditions [21,22]. Most of the studies focused on nitrogen metabolism in halophilic archaea have been conducted using H. mediterranei as a haloarchaeal model. Specifically, biochemical, physiological, and transcriptomic studies of the assimilatory pathway in the presence of different nitrogen sources have been previously performed [22,23,24,25], as well as the development of molecular biology tools in H. mediterranei [26]. These results have revealed that the assimilatory pathway is highly regulated at a transcriptional level. However, little is known about the global regulatory networks that allow for the survival of this microorganism under stress or nitrogen starvation conditions. Besides transcriptional regulators analyses, the number of studies which show that sRNA is involved in a great variety of adaptive cellular responses to different stresses have surprisingly increased in the recent years.
In this work, we present an analysis of putative sRNAs in H. mediterranei expressed under two different nitrogen sources, nitrate and ammonium, using a combination of RNA-Seq and bioinformatic approaches. The main aim of this work is to extend the understanding of the global regulation network of the assimilatory pathway in H. mediterranei, specifically, and in domain Archaea, generally.
2. Materials and Methods
2.1. Strains and Growth Conditions
H. mediterranei strain R4 (ATCC 33500T) was grown at 42 °C with aeration at 225 rpm, contained in a 25% (w/v) mixture of inorganic salts (25% salt water) [27]. The pH value was adjusted to 7.3. H. mediterranei was grown in two different nitrogen sources, in a defined medium which contained 40 mM KNO3 or 40mM NH4Cl and supplemented with 5 g/L glucose, 0.0005 g/L FeCl3, and 0.5 g/L KH2PO4. Three independent biological replicates of each condition were employed.
2.2. Bioinformatic Prediction of small RNAs
The library of candidate sRNAs was constructed from putative sRNAs obtained in four different species of Archaea: H. volcanii [28], M. mazei [17], S. solfataricus [16], and A. fulgidus [15]. All putative sRNA sequences predicted from these four species were compared with the genome of H. mediterranei strain R4 (Genbank numbers: CP001868.2, CP001869.1, CP001870.1, and CP001871.1) using BLASTn [25]. The sequences obtained using the candidate sRNAs of other species (E-value < 0.05, p-value < 0.05) were prioritised using a function of identity, lower number of gaps (maximum 3), and lower number of mismatches.
2.3. RNA Isolation
For RNA isolation, H. mediterranei was grown in the presence of two different nitrogen sources, nitrate and ammonium, to mid-exponential phase. The mid-exponential growth phase was reached at different times and values of OD600 nm depending on the nitrogen source [23]. RNA was isolated with the mirVanaTM miRNA isolation kit (Ambion, Thermo Fisher Scientific, Waltham, MA, USA) following product specifications. Afterwards, the RNA samples were treated with Turbo DNase (Ambion, Thermo Fisher Scientific, Waltham, MA, USA). RNA concentration was analysed by means of a Nanodrop ND-100 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and the quality was analysed using the Small RNA Analysis Kit on Agilent 2200 Tapestation (Agilent Technologies, Santa Clara, CA, USA), respectively.
2.4. Library of Complementary DNA Preparation and Sequencing
Library preparation and sequencing were performed by the Bioarray, S.L. company (Alicante, Spain). sRNA libraries were constructed using TruSeq Small RNA Library Prep (Illumina, San Diego, CA, USA) and sequenced on an llumina HiSeq 2500 system using a 50-base pair (bp) read length. The corresponding FASTQ files were obtained as a result.
2.5. RNA-Sequencing Bioinformatic Analysis
Raw reads from each FASTQ file were aligned into BAM files—compressed binary file used to represent aligned sequences up to 128Mb [29] for each sample of each condition—through bowtie2 2.3.0 [30] using the H. mediterranei ATCC 33500 genome (Genbank numbers: GCF_000306765.2_ASM30676v2; Assembly: GCA_000306765.2) as a reference. Seven ammonium BAM files and eight nitrogen BAM files were viewed in the Integrative Genomics Viewer (IGV) program [31,32], alongside the annotated H. mediterranei genome.
The raw readings, which aligned at the positions of the library of candidate sRNAs obtained from the different species against the H. mediterranei genome, were analysed manually using the Integrative Genomics Viewer (IGV). Only sequences with a score of at least 20 (frequency of reads in high-throughput sequencing) and that were present in the six samples of each condition were selected as sRNAs in H. mediterranei. The candidate sRNAs were classified as cis-encoded sRNA if they were encoded in the reverse direction of ORF, as trans-encoded sRNA if they were in intergenic regions, and as crRNA if they were in a CRISPR array. Since the size of the readings was generally greater than the homology shown in the different species analysed, the BAM files of each condition were pooled, thus, increasing the coverage of the readings, allowing more precise limiting of the sRNAs location in the H. mediterranei genome. Once the positions were obtained, the approximate sizes of the sRNAs were calculated. In addition, the genetic environment of each sRNA was analysed manually.
The counts of the sRNAs of H. mediterranei were obtained using the intersection nonempty feature from the HT-Seq program [33]. In this way, for each position i in the read, a set S(i) is defined as the set of all features overlapping the i position. Then, if S contains precisely one feature, the read is counted for this feature. If S is empty, the read is counted as no feature. Finally, if S contains more than one feature, the read is considered as ambiguous and is not counted for any features.
Differential expression analysis in the function of the nitrogen source (nitrate/ammonium) was performed using the DESeq2 library from the Bioconductor 3.5 package [34] obtaining the fold-change of each sRNA (p-value < 0.01 and p-adj < 0.05). Through this method, the variance-mean dependence in count data was estimated and tested for differential expression; this was based on a model using the negative binomial distribution between nitrate and ammonium group samples.
Both raw (FASTAQ files) and processed data (normalised counts) are available on the Gene Expression Omnibus (GEO) database (Series entry number: GSE108616) [35].
Mfold was used to predict the secondary structure of sRNAs obtained [36]. Potential gene targets for each sRNA were identified using TargetRNA2 [37]. Moreover, the cis-encoded sRNAs antisense of characterised ORF were analysed using IntaRNA [38]. BLASTn [25] was used to search homology regions in another organism.
2.6. Validation of sRNAs Using Reverse Transcription Polymerase Chain Reaction
The validation of 20 sRNAs was performed by reverse transcription polymerase chain reaction (RT-PCR), prioritising those with differential expression based on the nitrogen source and/or target genes of known functions. The RNA was isolated as described before. Between 0.5 and 0.8 µg of DNA-free RNA was used for the synthesis of cDNA using M-MuLV Reverse Transcriptase (Thermo Fisher Scientific, Waltham, MA, USA) and random hexamer primer (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Negative controls for reverse transcription polymerase chain reaction (RT-PCR) were prepared by omitting reverse transcriptase and cDNA. The oligonucleotides used to carry out the RT-PCR were designed based on the sequence of sRNA candidates (Table S1). Amplified products were analysed using 3% agarose gel electrophoresis with GeneRulerTM Low Range DNA Ladder (Thermo Fisher Scientific, Waltham, MA, USA) run in parallel. PCR products were purified with the IllustraTM GFXTM PCR DNA and Gel Band Purification Kit (GE Healthcare, Little Chalfont, UK) and confirmed by Sanger sequencing (Stabvida, Caparica, Portugal).
3. Results
3.1. Identification of Putative small RNAs in Haloferax mediterranei
The library of candidate sRNAs was generated from putative sRNAs obtained in four different species of domain Archaea: H. volcanii [28], M. mazei [17], S. solfataricus [16], and A. fulgidus [15]. Since the candidate sRNAs of H. volcanii aligned at many positions in the genome of H. mediterranei (E-value < 0.05, p-value < 0.05), the results obtained were prioritised by a function of identity, lower number of gaps (maximum 3), and lower number of mismatches. Given the evolutionary distance, the candidate sRNAs of the remaining species aligned in fewer positions (E-value < 1) than H. volcanii in the genome of H. mediterranei, so it was not necessary to prioritise in the same way. From the bioinformatic analysis, a library of 295 candidate sRNAs in H. mediterranei were obtained (Table 1). The number of mismatches allowed depends on the length of the sequence, permitting a greater number of mismatches in larger sequences with a high identity percentage. The results obtained after alignment with the candidate sRNAs of other species are shown in detail in the supplementary material (Tables S2–S5).

Table 1.
Preliminary library of 295 sRNAs in H. mediterranei obtained by a bioinformatic approach based on candidate small RNAs of other species.
3.2. Verification of Predicted 295 sRNAs Using RNA-Sequencing
The 295 sRNAs that were bioinformatically predicted were verified with the RNA-Seq results of the H. mediterranei ATCC33500. The raw aligned readings obtained from the analysis of RNA-Seq results and the 295 candidate sRNA sequences obtained by means of the bioinformatic approach were manually verified using the high-performance visualisation tool, IGV [31,32]. The transcripts expressed from intergenic regions, antisense to the characterised ORFs, and in the CRISPR array were classified in trans-encoded sRNA, cis-encoded sRNA, and crRNA, respectively. Due to the size of the readings being generally greater than the homology shown in different species being analysed, the BAM files of each condition were pooled, thus increasing the coverage of the readings and allowing for more accurate localization of the sRNAs in the H. mediterranei genome. Once the positions were obtained, the approximate size of the sRNAs was calculated. They were in the range of 20–500 nt. The results of the genetic environment analysis of the 88 verified sRNAs in H. mediterranei and their sequences are shown in Table 2 and Table S6, respectively.

Table 2.
Compilation of positive predicted sRNAs in H. mediterranei using RNA-Sequencing.
3.3. Bioinformatic Analysis of Putative Small RNAs in H. mediterranei
3.3.1. Identification and Classification of small RNAs Verified by RNA-Sequencing
The results of the localisation and classification of the sRNAs verified by RNA-Seq reveals that 58 sRNAs of the 116 sRNAs predicted by homology with H. volcanii (Table 1 and Table S2) could be assigned as sRNAs in H. mediterranei. There were 56 sRNAs on the chromosome and two on the plasmid pHM500. According to the classification established above, 51 sRNAs are intergenic sRNAs, six are antisense sRNAs, and one is crRNA. The homology analysis performed with M. mazei results in 55 predicted sRNAs (Table 1 and Table S3), of which 16 could be sRNAs in H. mediterranei. Twelve sRNAs are located on the chromosome, three on the plasmid pHM500, and one on the plasmid pHM100. Related to the classification according to the location of the target, 11 sRNAs are intergenic sRNAs and five antisense. Of the 47 sRNAs predicted by homology with S. solfataricus (Table 1 and Table S4), only eight sRNAs were identified in the results of RNA-Seq. These are located on the chromosome, and are classified as four intergenic sRNAs and four antisense sRNAs. From the analysis with A. fulgidus (Table 1 and Table S5), only six candidate sRNAs from a total of 74 sRNAs predicted by homology could be assigned as sRNAs. As in the analysis of S. solfataricus, all of them are located on the chromosome, with four being intergenic sRNAs and two being antisense sRNAs. From a total of 88 sRNAs verified in H. mediterranei by RNA-Seq, 93.18% were located on the chromosome, with most of them (79.54%) being intergenic (trans-encoded sRNA).
3.3.2. Structure and Targets of 88 small RNAs
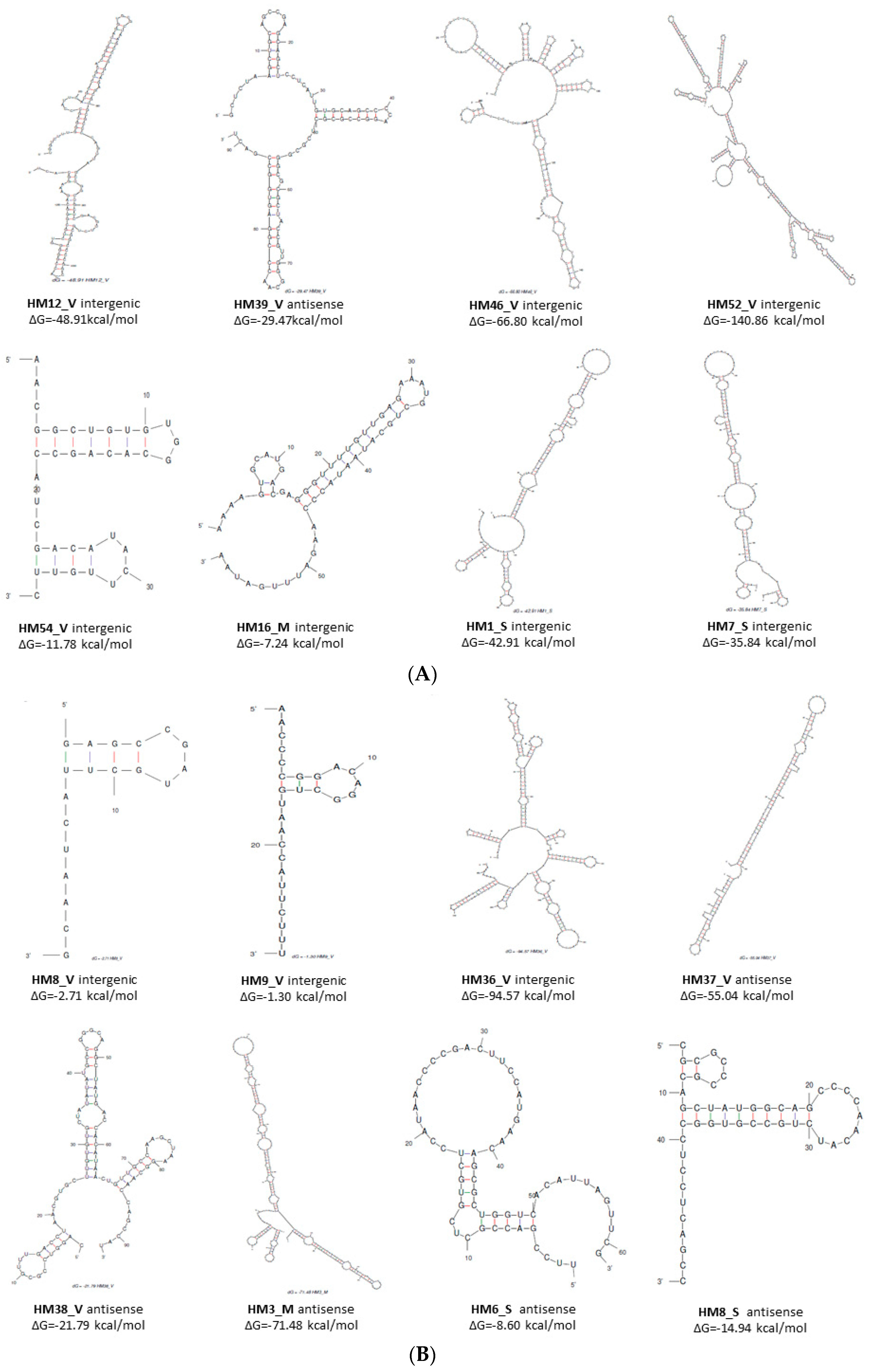
Usually, sRNAs are characterised by stable secondary structures. In order to analyse the secondary structure of the 88 putative sRNAs, Mfold software [36] was used with default parameters set and with the temperature modified to 42 °C. The core algorithm predicts a minimum free energy, ΔG°, as well as minimum free energies for folding that must contain any particular base pair. All of the sRNAs were shown to have significant predicted secondary structures since they presented with ΔG < 0. However, only the secondary structures of 16 sRNAs with differential expression according to the nitrogen source (nitrate/ammonium) are shown in Figure 1. Furthermore, most secondary structures of sRNAs revealed highly structured molecules, including more than one hairpin loop and high stability (ΔG < 0). It is noteworthy that the structures of HM39_V, HM54_V, HM38_V, and HM8_S sRNAs present predominant structures with two or three hairpin loops and are similar to the structures of other characterised sRNAs [39,40].
Figure 1.
Prediction of the secondary structure of 16 sRNAs with differential expression (nitrate/ammonium) in Haloferax mediterranei using Mfold [36]. (A) sRNAs overexpressed in nitrate. (B) sRNAs overexpressed in ammonium.
The identification of potential sRNA targets was carried out via a bioinformatic approach using TargetRNA2 [37]. Gene targets were found for 71.59% of the sRNA verified, of which the first five are most likely due to their energy values (Table S8). Taking the five gene targets for each sRNA analysed into account, the most commonly predicted targets correspond to hypothetical proteins with unknown functions. Many examples of transcriptional regulators such as the ArsR family, nitrogen regulatory PII protein, transporters (including metal and ATP-binding cassette (ABC) transporters), proteins related to RNA (i.e., H/ACA RNA-protein complex component Gar1, ribonuclease P subunit p30, 50S ribosomal protein L24e, 50S ribosomal protein L37Ae, DNA-directed RNA polymerase subunit B″, DNA-directed RNA polymerase subunit F, 30S ribosomal protein S8P, etc.), and proteins related to DNA metabolism (i.e., DNA lyase, DNA ligase, DNA double-strand break repair protein mre11, DNA primase large subunit) were also identified. To go in depth with this analysis, the cis-encoded sRNAs antisense of characterised ORF were analysed using IntaRNA [38] (Table 3). The results show that nine of the 17 cis-encoded sRNAs analysed present high interaction energy and full complementarity with the ORF where they were identified. The combination of these software programs has allowed the identification of possible gene targets for more than 70% of the 88 possible sRNAs identified by RNA-Seq. However, there are still 26 sRNAs whose possible targets could not be found with the software employed.

Table 3.
Analysis of possible targets of antisense sRNAs using IntaRNA [38].
3.3.3. Conservation of small RNAs Verified in H. mediterranei
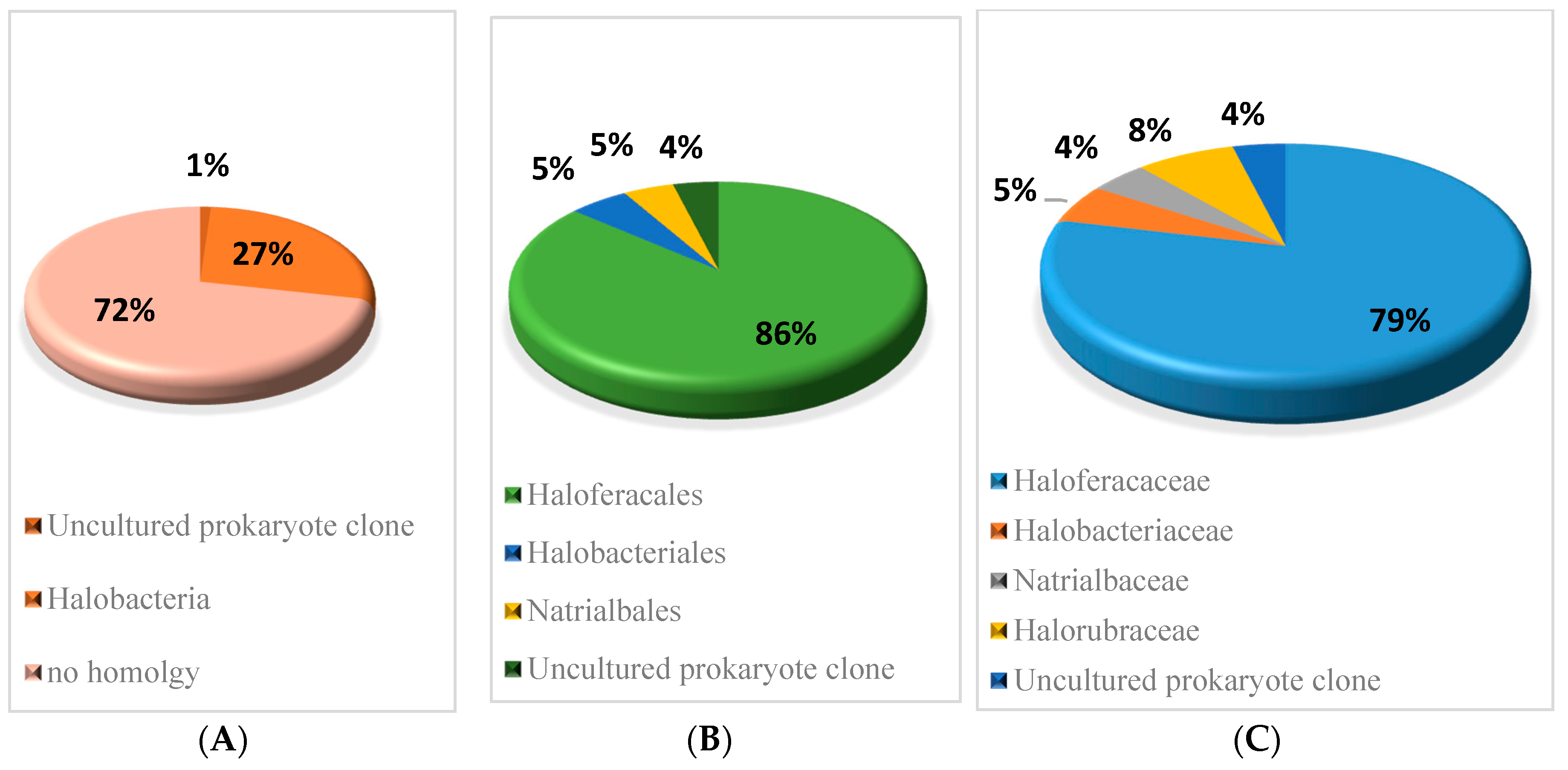
The sequence conservation of the sRNAs predicted in H. mediterranei was analysed using BLASTn [25], comparing each sequence to all sequenced archaeal genomes (E value 10 × 10−6). Only hits with a nucleotide identity higher than 60% combined with a coverage between the query and subject sequence higher than 80% were considered as conserved. The results of homology are shown in Figure 2 and Table S7. It was found that 27% of verified sRNAs present conserved sequences in other Halobacteria, mainly in the Haloferacales order (86%) and Haloferacaceae family (79%). The remaining 63 sRNAs showed no sequence homology with any other Archaea.
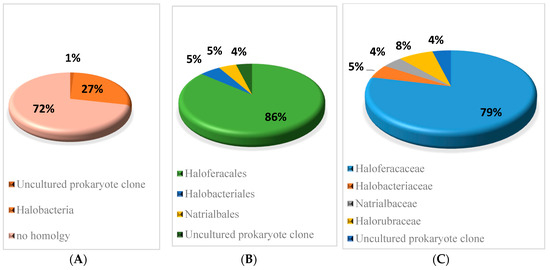
Figure 2.
Conservation percentage of the sequences of sRNAs across different (A) classes, (B) orders, and (C) families. BLASTn, homology (E value 10 × 10−6); query covered at least 80% and sequence identity covered at least 60%.
3.4. Expression Analysis According to the Nitrogen Source
The expression analysis of the sRNAs in H. mediterranei according to the nitrogen source was performed using the HT-Seq program, which counts the readings of the sRNAs verified by RNA-Seq and DESeq2, which carries out the differential expression analysis (nitrate/ammonium). From the 88 candidate sRNAs analysed, 16 sRNAs met the statistical criteria with a p-value and p-adj lower than 0.02 and 0.05, respectively (Table 4). These sRNAs show differences in their expression pattern. Eight of them are overexpressed in the presence of nitrate as a sole nitrogen source (log2-fold-change between 0.519 and 8.699), whereas the other eight present a decrease in their transcriptional level in presence of ammonium (log2-fold-change between −0.478 and 1.325).

Table 4.
Summary of the characteristics of sRNAs with differential expression (nitrate/ammonium).
3.5. Validation of Expression of Small RNAs Using Reverse Transcription Polymerase Chain Reaction
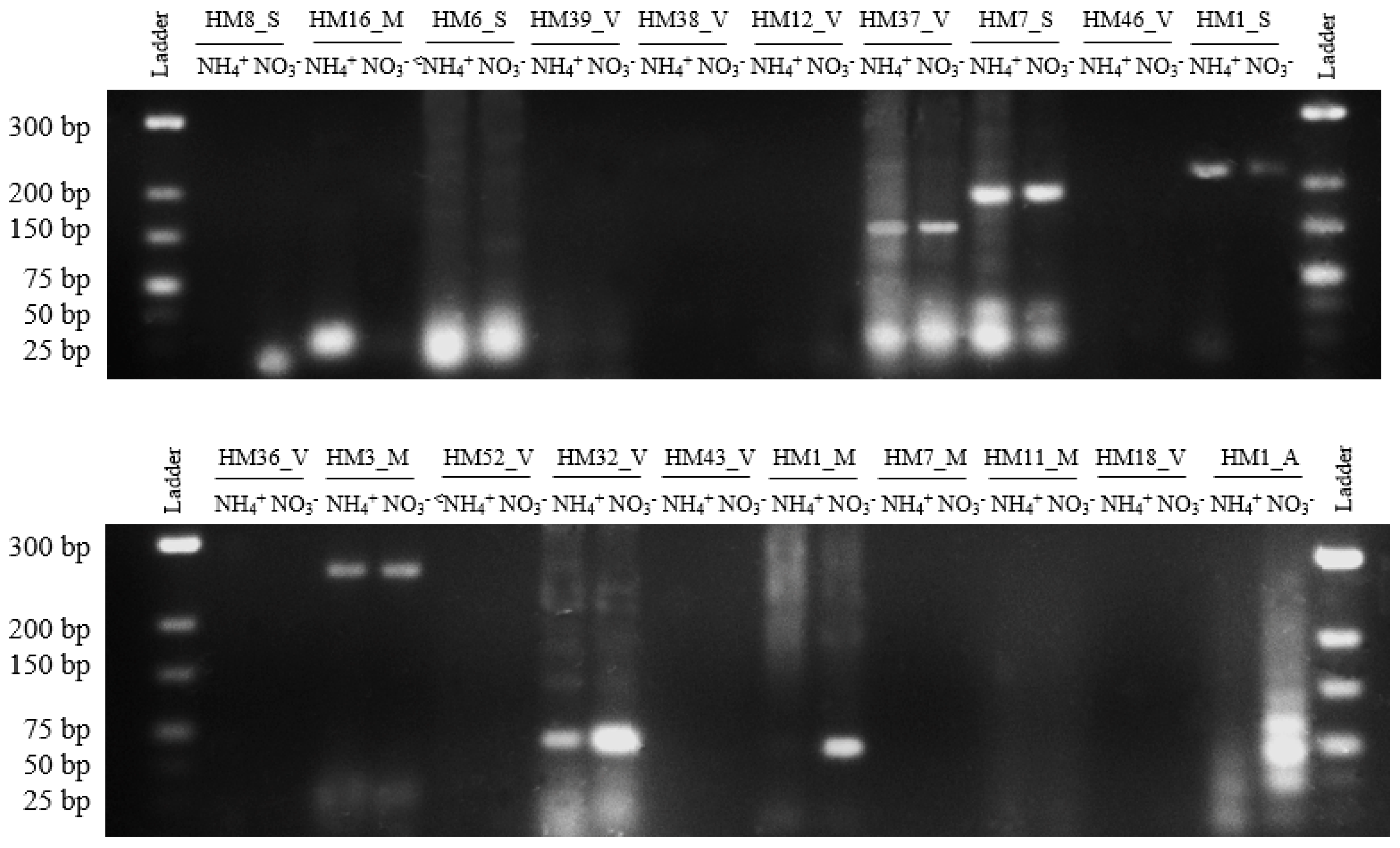
To confirm the presence of 20 sRNAs predicted by RNA-Seq we used RT-PCR [41,42,43,44]. Of 20 predicted sRNAs tested, nine (40%) gave positive results of the expected size as shown in Figure 3. These PCR products were subsequently verified using Sanger sequencing. This proportion of successfully validated sRNAs (45%) agrees with the results obtained in other works, where validation is often successful around 40–50% [39,42].
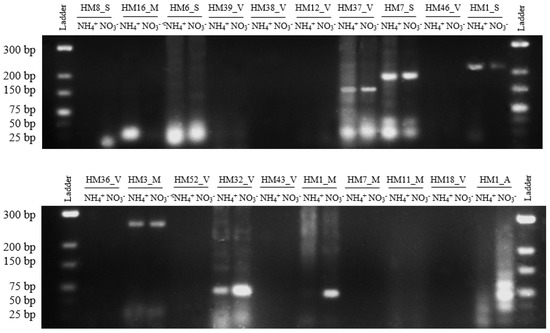
Figure 3.
Reverse transcription polymerase chain reaction (RT-PCR) validation of 20 sRNAs. Electrophoresis of PCR products of 20 sRNAs on 3% agarose gel. Lanes 1 and 12: GeneRuler Low Range DNA Ladder (Thermo Scientific, Waltham, MA, USA). Lanes 2–11: PCR products of sRNAs.
The majority of sRNAs (HM6_S, HM37_V, HM7_S, HM1_S, HM3_M, and HM32_V) are expressed in both conditions, in the presence of nitrate and ammonium. However, three sRNAs show a different expression pattern according to the nitrogen source; HM8_S and HM1_M sRNAs are expressed exclusively in the presence of nitrate and HM16_M sRNA is expressed in the presence of ammonium.
4. Discussion
Two different approaches, bioinformatic and RNomic, have allowed the identification of 88 sRNAs in H. mediterranei. Most of them are located on the chromosome, as in the case of H. volcanii. However, there is a difference between these two halophilic microorganisms. H. mediterranei sRNAs are predominantly intergenic (trans-encoded sRNA), whereas H. volcanii sRNAs are mostly antisense instead [28,45].
The 88 sRNAs verified by RNA-Seq were analysed using different strategies to obtain further information about their characteristics and their possible involvement in different cellular processes. Some of the predicted secondary structures of these sRNAs remain similar to other sRNA structures which have been analysed in detail (Figure 1) [39,40]. According to Gaimster et al. [39], these results are significant because they imply that many of these sRNAs could have the potential to form complex conformations like those commonly associated with many other directly-acting RNA transcripts, including known bacterial sRNAs. Moreover, additional molecular mechanisms of sRNA functions in bacteria have been described, including the destabilisation or stabilisation of the target mRNAs. These actions are conducted via specific binding to a protein or full complementarity to their target mRNA [5]. Thus, putative gene targets of all the sRNAs candidates in H. mediterranei were predicted using TargetRNA2 [37]. As it has been observed in other works [39], the results obtained reveal that the most commonly predicted targets matched to hypothetical proteins. This information could potentially be useful in eventually assigning a function to these unknown proteins. It has also been identified to target some transcriptional regulators such as the ArsR family, as well as the nitrogen regulatory PII protein. Interestingly, the expression of the ArsR and PII proteins in H. mediterranei is closely related to the nitrogen source [24], so the existence of a regulatory network where the sRNAs act by activating or repressing the expression of these proteins is highly likely. In other bacterial studies, it has been observed that global regulators are subject to regulation by multiple Hfq-dependent sRNAs [46]. In the case of domain Archaea, similar interactions have been described between sRNAs and Like Sm (Lsm) homologous proteins, instead of the bacterial Hfq [47]. H. mediterranei contains one gene encoding the Lsm1 protein (HFX_2733) in its genome that shares 99% sequence identity with the H. volcanii RNA-binding Lsm protein (E value 6 × 10−51). These results suggest that the interaction between Lsm proteins and sRNAs and their participation in the RNA metabolism are also possible in H. mediterranei. Transporters are also included in the different targets predicted, specifically metal and ABC transporters, which have been observed in other analyses of bacterial sRNAs such as Paracoccus denitrificans [39] and Ruegeria pomeroyi [48], as well as in Archaeal sRNA such as S. solfataricus [49] and H. volcanii [50]. Based on these results, it could be possible that the regulation of transporters is not a characteristic only of the Bacteria domain. This suggests that antisense sRNA might be a common regulatory mechanism for such genes. It has also been identified as a target of different proteins related to RNA and DNA metabolism, which support the concept that sRNAs play an essential role in transcriptional and post-transcriptional regulation. Furthermore, analysis of co-purification with ribosomal protein L7Ae in S. solfataricus allowed for the identification of one sRNA in that microorganism which suggests a direct interaction of these proteins with sRNAs [51]. In parallel, the cis-encoded sRNAs antisense of ORF characterised in H. mediterranei were also analysed using IntaRNA [38]. This analysis showed positive results in nine of the 17 cis-encoded sRNAs. Four of them were present in only one target and the other five could interact with other possible targets. Most sRNAs that did not exhibit interaction with RNA using IntaRNA did exhibit interaction with other mRNAs using TargetRNA2. Therefore, it is likely that some sRNAs are specific to a gene, while others have a broader range of action, being involved in several metabolic pathways (Table S8). Only 26 of the 88 sRNAs analysed did not show positive results using either IntaRNA or TargetRNA2. Hence, more than 70% of the 88 sRNAs verified by RNA-Seq present possible target genes using such software. Nevertheless, it is clear that the only way to confirm the interaction between RNA and its target is through experimental validation. Therefore, in future works we will try to characterise the targets of the sRNAs that present a differential expression in the function of the nitrogen source through the interaction with the protein Lsm1, the generation of deletion sRNA mutants, and other analyses (such as overexpression mutants) to demonstrate the influence of these sRNAs on the respective target.
BLASTn [25] analysis reveals that 27% of sRNAs identified in H. mediterranei present conserved sequences in other Halobacteria. Consequently, it is likely that these conserved sRNAs may play a conserved role in closely related species. However, despite the fact that the sequences of the sRNAs are slightly conserved in Archaea, this does not imply that they are all true sRNA candidates. The remaining 72% of sRNAs identified showed no sequence homology to any other archaea. According to Babski et al. [5], many sRNA genes of Archaea are not even shared by species of the same genus. Therefore, this 72% of the sRNA could be specific to the halophilic archaea H. mediterranei.
The transcript pattern analysis of H. mediterranei sRNA according to the nitrogen source demonstrates that 16 sRNA showed higher or lower transcript levels with statistically significant parameters (p-value < 0.02 and p-adj < 0.05). Of these, eight show overexpression when H. mediterranei grows with nitrate as the sole nitrogen source, whereas the other eight show a decrease in their transcriptional level in the presence of ammonium. The majority of sRNAs validated by RT-PCR are expressed in the presence of nitrogen sources nitrate and ammonium. However, HM8_S and HM16_M sRNAs are expressed exclusively in the presence of nitrate and ammonium, respectively. Curiously, although the HM1_M sRNA transcript level does not show differences in the RNA-Seq analysis according to the nitrogen source, RT-PCR results clearly reveal that this sRNA is only expressed in the presence of nitrate (Figure 3).
Interestingly, some of the sRNAs which show differences in their expression pattern according to the nitrogen source possibly have target genes whose expression also depends on the nitrogen source. HM8_S, which is overexpressed in nitrate as the nitrogen source, has the protein glutamate dehydrogenase as a possible target (Table 3). Glutamate dehydrogenase is underexpressed in nitrate’s presence. Therefore, this sRNA could be involved in the repression of this key enzyme in the metabolism of nitrogen. Moreover, two sRNAs (i.e., HM7_S and HM54_V) are slightly more expressed in the presence of nitrate. Curiously, their possible targets are transcriptional regulators belonging to the ArsR family. Previous results revealed that the expression of different ArsR proteins depends on the nitrogen source [24]. Therefore, sRNAs could also be involved in the regulation of the expression of these transcriptional regulators, and it could be an example of a gene regulatory network related to nitrogen assimilation. Finally, HM1_A sRNA is expressed in the presence of ammonium as a nitrogen source, and it has been found to be a possible target of the amt1 gene (Table 3), which encodes an ammonium transporter. Different studies performed with H. mediterranei have confirmed that ammonium transporters are expressed in the presence of nitrate or under nitrogen starvation [24]. Hence, this sRNA could also be related to the regulation of Amt transporters expression and, consequently, be adjusting the uptake of ammonium from the medium. More work is needed to confirm these hypotheses and to find out the role of these sRNAs in nitrogen metabolism.
5. Conclusions
This work, focused on identifying sRNAs involved in nitrogen assimilation, has also increased the knowledge about sRNAs in the domain Archaea. Specifically, 88 sRNAs have been identified in H. mediterranei using bioinformatic and RNomic approaches, some of which show different expression patterns depending on the nitrogen source and/or present genes involved in the nitrogen assimilation as a potential gene target. This data suggests that some of these sRNAs could be related to the regulation of nitrogen assimilation, being able to constitute an important gene regulatory network which involves enzymes, transporters, and transcriptional regulators in this metabolism. Undoubtedly, this work constitutes an excellent starting point to elucidate the role of these sRNAs in the nitrogen metabolism of haloarchaea.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/9/2/83/s1. Table S1: Nucleotide sequences of PCR primers for amplifying sRNA genes. Table S2: Library of sRNAs obtained after alignment of H. volcanii sRNAs [28] against the H. mediterranei genome (E-value < 0.05). Table S3: Library of sRNAs obtained after alignment of M. mazei sRNAs [17] against the H. mediterranei genome (E-value < 1). Table S4: Library of sRNAs obtained after alignment of S. solfataricus sRNAs [16] against the H. mediterranei genome (E-value < 1). Table S5: Library of sRNAs obtained after alignment of A. fulgidus sRNAs [15] against the H. mediterranei genome (E-value < 1). Table S6: Sequences of 88 candidates sRNAs identified in H. mediterranei by RNA-Seq. Table S7: Conservation of the sequences of sRNAs across different orders, classes, and species. BLASTn, homology (E value 10 × 10−6, query cover at least 80% and sequence identity at least 60%). Table S8: Prediction of putative gene targets of 88 sRNA in H. mediterranei using TargetRNA2 [36].
Acknowledgments
We would like to thank Jörg Soppa (Goethe-University, Frankfurt, Germany) for useful comments and assistance in this work. This work was supported by grant BIO2013-42921-P from the Spanish Ministry of Economy and Competitiveness. Funds from “Programa Propio para el fomento de la I+D+i del Vicerrectorado de Investigación y Transferencia de Conocimiento” de la Universidad de Alicante are also acknowledged.
Author Contributions
J.E. and M.-J.B. conceived and designed the experiments; G.P., J.E., V.B., and M.C. performed most of the experiments; N.C.-F. and L.A.A. carried out specific experimental parts; G.P., J.E., N.C.-F., and M.-J.B. analysed the data; M.-J.B. supervised the experimental work; G.P., J.E., and M.-J.B. wrote the paper. V.B. and M.C. contributed to the writing of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chu, C.Y.; Rana, T.M. Small RNAs: Regulators and guardians of the genome. J. Cell. Physiol. 2007, 213, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Choudhuri, S. Lesser known relatives of miRNA. Biochem. Biophys. Res. Commun. 2009, 388, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Paroo, Z. Biochemical principles of small RNA pathways. Annu. Rev. Biochem. 2010, 79, 295–319. [Google Scholar] [CrossRef] [PubMed]
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by small RNAs in bacteria: Expanding frontiers. Mol. Cell. 2011, 43, 880–891. [Google Scholar] [CrossRef] [PubMed]
- Babski, J.; Maier, L.K.; Heyer, R.; Jaschinski, K.; Prasse, D.; Jäger, D.; Randau, L.; Schmitz, R.A.; Marchfelder, A.; Soppa, J. Small regulatory RNAs in Archaea. RNA Biol. 2014, 11, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Michaux, C.; Hartke, A.; Martini, C.; Reiss, S.; Albrecht, D.; Budin-Verneuil, A.; Sanguinetti, M.; Engelmann, S.; Hain, T.; Verneuil, N.; et al. Involvement of Enterococcus faecalis small RNAs in stress response and virulence. Infect. Immun. 2014, 82, 3599–3611. [Google Scholar] [CrossRef] [PubMed]
- Durand, S.; Tomasini, A.; Braun, F.; Condon, C.; Romby, P. sRNA and mRNA turnover in Gram-positive bacteria. FEMS Microbiol. Rev. 2015, 39, 316–330. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, S.; Storz, G. Bacterial small RNA regulators: Versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef] [PubMed]
- Moll, I.; Leitsch, D.; Steinhauser, T.; Bläsi, U. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 2003, 4, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Muller, S.; Leclerc, F.; Behm-Ansmant, I.; Fourmann, J.B.; Charpentier, B.; Branlant, C. Combined in silico and experimental identification of the Pyrococcus abyssi H/ACA sRNAs and their target sites in ribosomal RNAs. Nucleic Acids Res. 2008, 36, 2459–2475. [Google Scholar] [CrossRef] [PubMed]
- Phok, K.; Moisan, A.; Rinaldi, D.; Brucato, N.; Carpousis, A.J.; Gaspin, C.; Clouet-d’Orval, B. Identification of CRISPR and riboswitch related RNAs among novel noncoding RNAs of the euryarchaeon Pyrococcus abyssi. BMC Genom. 2011, 12, 312. [Google Scholar] [CrossRef] [PubMed]
- Heyer, R.; Dörr, M.; Jellen-Ritter, A.; Späth, B.; Babski, J.; Jaschinski, K.; Soppa, J.; Marchfelder, A. High throughput sequencing reveals a plethora of small RNAs including tRNA derived fragments in Haloferax volcanii. RNA Biol. 2012, 9, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Soppa, J.; Straub, J.; Brenneis, M.; Jellen-Ritter, A.; Heyer, R.; Fischer, S.; Granzow, M.; Voss, B.; Hess, W.R.; Tjaden, B.; et al. Small RNAs of the halophilic archaeon Haloferax volcanii. Biochem. Soc. Trans. 2009, 37, 133–136. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.G.; Romby, P. Small RNAs in bacteria and archaea: Who they are, what they do, and how they do it. Adv. Genet. 2015, 90, 133–208. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.H.; Bachellerie, J.P.; Rozhdestvensky, T.; Bortolin, M.L.; Huber, H.; Drungowski, M.; Elge, T.; Brosius, J.; Hüttenhofer, A. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. USA 2002, 99, 7536–7541. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.-H.; Polacek, N.; Zywicki, M.; Huber, H.; Brugger, K.; Garrett, R.; Bachellerie, J.P.; Hüttenhofer, A. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol. Microbiol. 2005, 55, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Jäger, D.; Sharma, C.M.; Thomsen, J.; Ehlers, C.; Vogel, J.; Schmitz, R.A. Deep sequencing analysis of the Methanosarcina mazei Gö1 transcriptome in response to nitrogen availability. Proc. Natl. Acad. Sci. USA 2009, 106, 21878–21882. [Google Scholar] [CrossRef] [PubMed]
- Bernick, D.L.; Dennis, P.P.; Höchsmann, M.; Lowe, T.M. Discovery of Pyrobaculum small RNA families with atypical pseudouridine guide RNA features. RNA 2012, 18, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Jäger, D.; Förstner, K.U.; Sharma, C.M.; Santangelo, T.J.; John, N.; Reeve, J.N. Primary transcriptome map of the hyperthermophilic archaeon Thermococcus kodakarensis. BMC Genom. 2014, 15, 684. [Google Scholar] [CrossRef] [PubMed]
- Torreblanca, M.; Rodriguez-Valera, F.; Juez, G.; Ventosa, A.; Masahiro, K.; Morris, K. Classification of non-alkaliphilic halobacteria based on numerical taxonomy and polar lipid composition, and description of Haloarcula gen. nov. and Haloferax gen. nov. Syst. Appl. Microbiol. 1986, 8, 89–99. [Google Scholar] [CrossRef]
- Rodríguez-Valera, F.; Juez, G.; Kushner, D.J. Halobacterium mediterranei spec, nov., a new carbohydrate-utilizing extreme halophile. Syst. Appl. Microbiol. 1983, 4, 369–381. [Google Scholar] [CrossRef]
- Bonete, M.J.; Martínez-Espinosa, R.M.; Pire, C.; Zafrilla, B.; Richardson, D.J. Nitrogen metabolism in haloarchaea. Saline Syst. 2008, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- Esclapez, J.; Bravo-Barrales, G.; Bautista, V.; Pire, C.; Camacho, M.; Bonete, M.J. Effects of nitrogen sources on the nitrate assimilation in Haloferax mediterranei: Growth kinetics and transcriptomic analysis. FEMS Microbiol. Lett. 2014, 350, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Esclapez, J.; Pire, C.; Camacho, M.; Bautista, V.; Martínez-Espinosa, R.M.; Zafrilla, B.; Vegara, A.; Alcaraz, L.A.; Bonete, M.J. Transcriptional profiles of Haloferax mediterranei based on nitrogen availability. J. Biotechnol. 2015, 193, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Esclapez, J.; Camacho, M.; Pire, C.; Bautista, V.; Vegara, A.; Pedro-Roig, L.; Pérez-Pomares, F.; Martínez-Espinosa, R.M.; Bonete, M.J. Recent Advances in The Nitrogen Metabolism in Haloarchaea and Its Biotechnological Applications. In Biotechnology of Extremophiles: Advances and Challenges; Rampelotto, P., Ed.; Springer: Cham, Switzerland, 2016; Volume 1, pp. 273–301. ISBN 978-3-319-13520-5. [Google Scholar]
- Pedro-Roig, L.; Lange, C.; Bonete, M.J.; Soppa, J.; Maupin-Furlow, J. Nitrogen regulation of protein-protein interactions and transcript levels of GlnK PII regulator and AmtB ammonium transporter homologs in Archaea. Microbiologyopen 2013, 2, 826–840. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Valera, F.; Ruiz-Berraquero, F.; Ramos-Cormenzana, A. Behaviour of mixed populations of halophilic bacteria in continuous cultures. Can. J. Microbiol. 1980, 26, 1259–1263. [Google Scholar] [CrossRef] [PubMed]
- Babski, J.; Tjaden, B.; Voss, B.; Jellen-Ritter, A.; Marchfelder, A.; Hess, W.R.; Soppa, J. Bioinformatic prediction and experimental verification of sRNAs in the haloarchaeon Haloferax volcanii. RNA Biol. 2011, 8, 806–816. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative genomics viewer (IGV): High-performance genomics. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002 30, 207–210. [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Kery, M.B.F.M.; Livny, J.; Tjaden, B. TargetRNA2: Identifying targets of small regulatory RNAs in bacteria. Nucleic Acids Res. 2014, 42, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Mann, M.; Wright, P.R.; Backofen, R. IntaRNA 2.0: Enhanced and customizable prediction of RNA–RNA interactions. Nucleic Acids Res. 2017, 45, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Gaimster, H.; Chalklen, L.; Alston, M.; Munnoch, J.T.; Richardson, D.J.; Gates, A.J.; Rowley, G. Genome-wide discovery of putative sRNAs in Paracoccus denitrificans expressed under nitrous oxide emitting conditions. Front. Microbiol. 2016, 7, 1806. [Google Scholar] [CrossRef] [PubMed]
- Prasse, D.; Förstner, K.U.; Jäger, D.; Backofen, R.; Schmitz, R.A. sRNA154 a newly identified regulator of nitrogen fixation in Methanosarcina mazei strain Gö1. RNA Biol. 2017, 14, 1544–1558. [Google Scholar] [CrossRef] [PubMed]
- Pánek, J.; Bobek, J.; Mikulík, K.; Basler, M.; Vohradský, J. Biocomputational prediction of small non-coding RNAs in Streptomyces. BMC Genom. 2008, 9, 217. [Google Scholar] [CrossRef] [PubMed]
- Khoo, J.S.; Chai, S.F.; Mohamed, R.; Nathan, S.; Firdaus-Raih, M. Computational discovery and RT-PCR validation of novel Burkholderia conserved and Burkholderia pseudomallei unique sRNAs. BMC Genom. 2012, 13. [Google Scholar] [CrossRef]
- Panda, G.; Tanwer, P.; Ansari, S.; Khare, D.; Bhatnagar, R. Regulation and RNA-binding properties of Hfq-like RNA chaperones in Bacillus anthracis. Biochim. Biophys. Acta 2015, 1850, 1661–1668. [Google Scholar] [CrossRef] [PubMed]
- Kwenda, S.; Gorshkov, V.; Ramesh, A.M.; Naidoo, S.; Rubagotti, E.; Birch, P.R.; Moleleki, L.N. Discovery and profiling of small RNAs responsive to stress conditions in the plant pathogen Pectobacterium atrosepticum. BMC Genom. 2016, 12, 47. [Google Scholar] [CrossRef] [PubMed]
- Babski, J.; Haas, K.A.; Näther-Schindler, D.; Pfeiffer, F.; Förstner, K.U.; Hammelmann, M.; Hilker, R.; Becker, A.; Sharma, C.M.; Marchfelder, A.; Soppa, J. Genome-wide identification of transcriptional start sites in the haloarchaeon Haloferax volcanii based on differential RNA-Seq (dRNA-Seq). BMC Genom. 2016, 17, 629. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Gottesman, S. sRNA roles in regulating transcriptional regulators: Lrp and SoxS regulation by sRNAs. Nucleic Acids Res. 2016, 44, 6907–6923. [Google Scholar] [CrossRef] [PubMed]
- Fischer, S.; Benz, J.; Späth, B.; Maier, L.K.; Straub, J.; Granzow, M.; Raabe, M.; Urlaub, H.; Hoffmann, J.; Brutschy, B.; et al. The archaeal Lsm protein binds to small RNAs. J. Biol. Chem. 2010, 285, 45, 34429–34438. [Google Scholar] [CrossRef] [PubMed]
- Rivers, A.R.; Burns, A.S.; Chan, L.K.; Moran, M.A. Experimental identification of small non-coding RNAs in the model marine bacterium Ruegeria pomeroyi DSS-3. Front. Microbiol. 2016, 29, 380. [Google Scholar] [CrossRef] [PubMed]
- Wurtzel, O.; Sapra, R.; Chen, F.; Zhu, Y.; Simmons, B.A.; Sorek, R. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010, 20, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Straub, J.; Brenneis, M.; Jellen-Ritter, A.; Heyer, R.; Soppa, J.; Marchfelder, A. Small RNAs in haloarchaea: Identification, differential expression and biological function. RNA Biol. 2009, 6, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Zago, M.A.; Dennis, P.P.; Omer, A.D. The expanding world of small RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 2005, 55, 1812–1828. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).