Enhancer Trapping and Annotation in Zebrafish Mediated with Sleeping Beauty, piggyBac and Tol2 Transposons
Abstract
1. Introduction
2. Materials and Methods
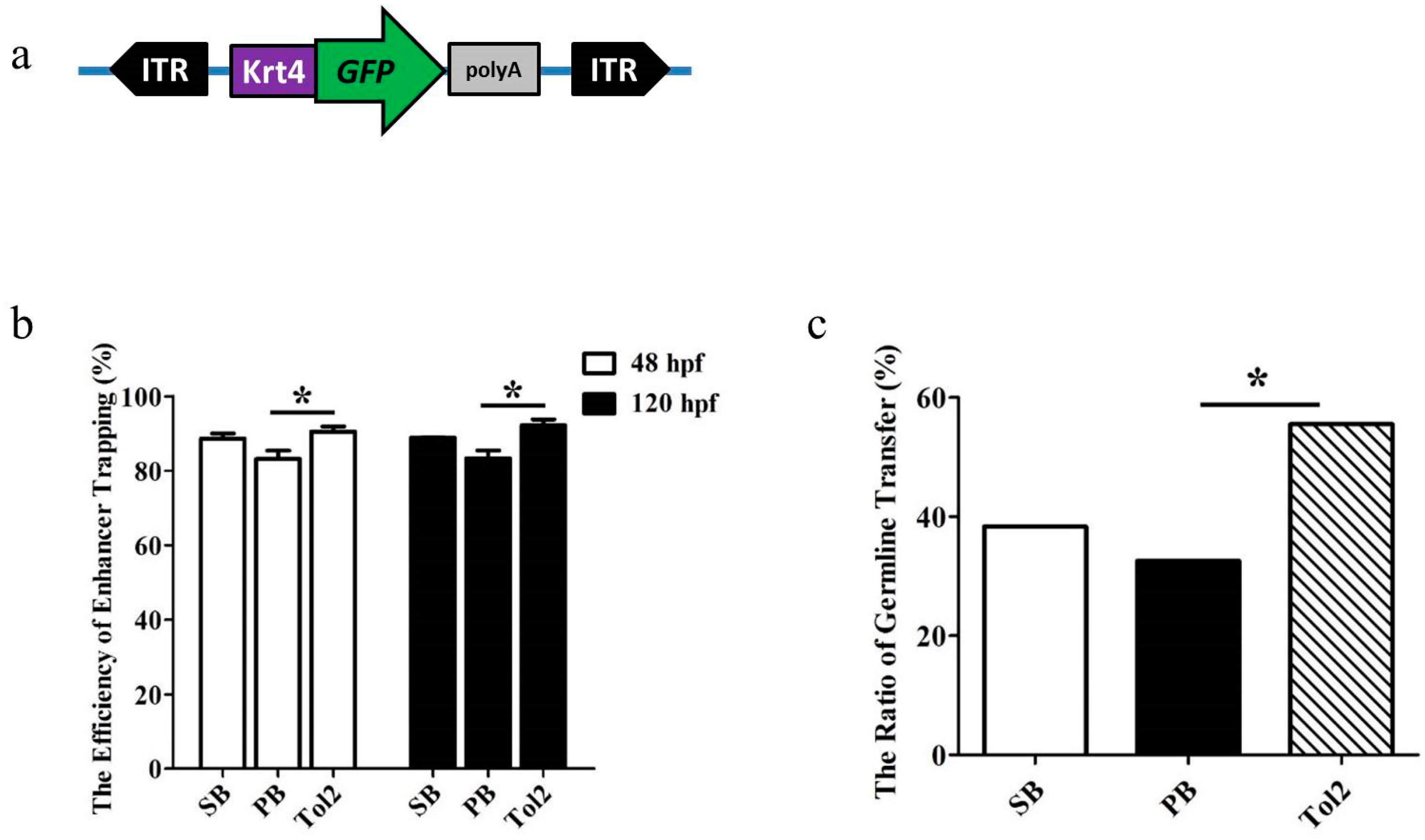
2.1. Vector Construction, Injection and Transgenic Screening
2.2. Injection of Fertilized Eggs
2.3. Amplification of Transposon Insertion Sites
2.4. Computational Analysis of Genomic Sequences
2.5. Whole-Mount RNA In Situ Hybridization and Transcription Analysis of Identified Ens
2.6. Enhancer Activity Analysis In Vivo
3. Results
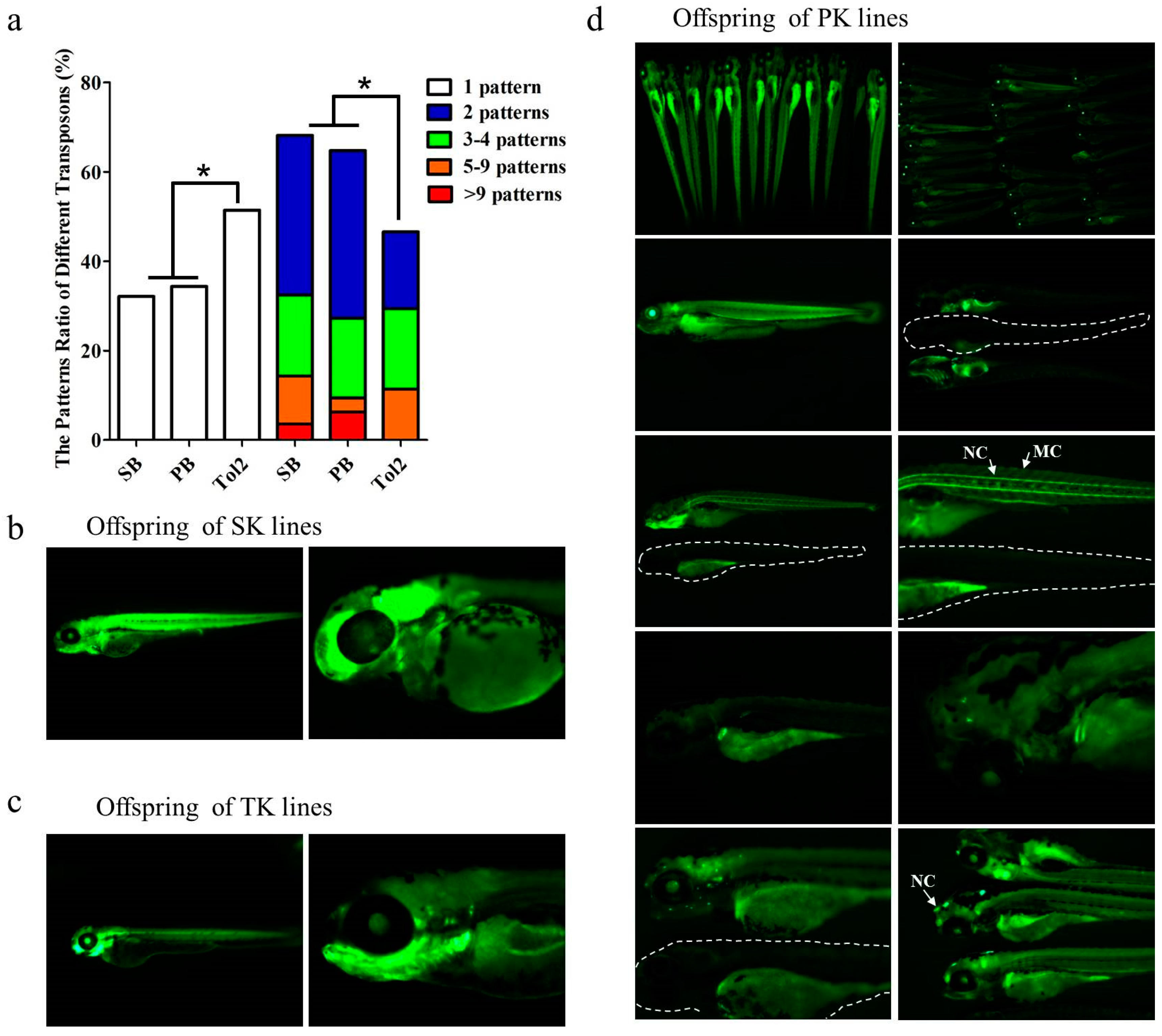
3.1. Comparative Analysis of the Enhancer Trapping Efficiency of PB, SB and Tol2 Transposons
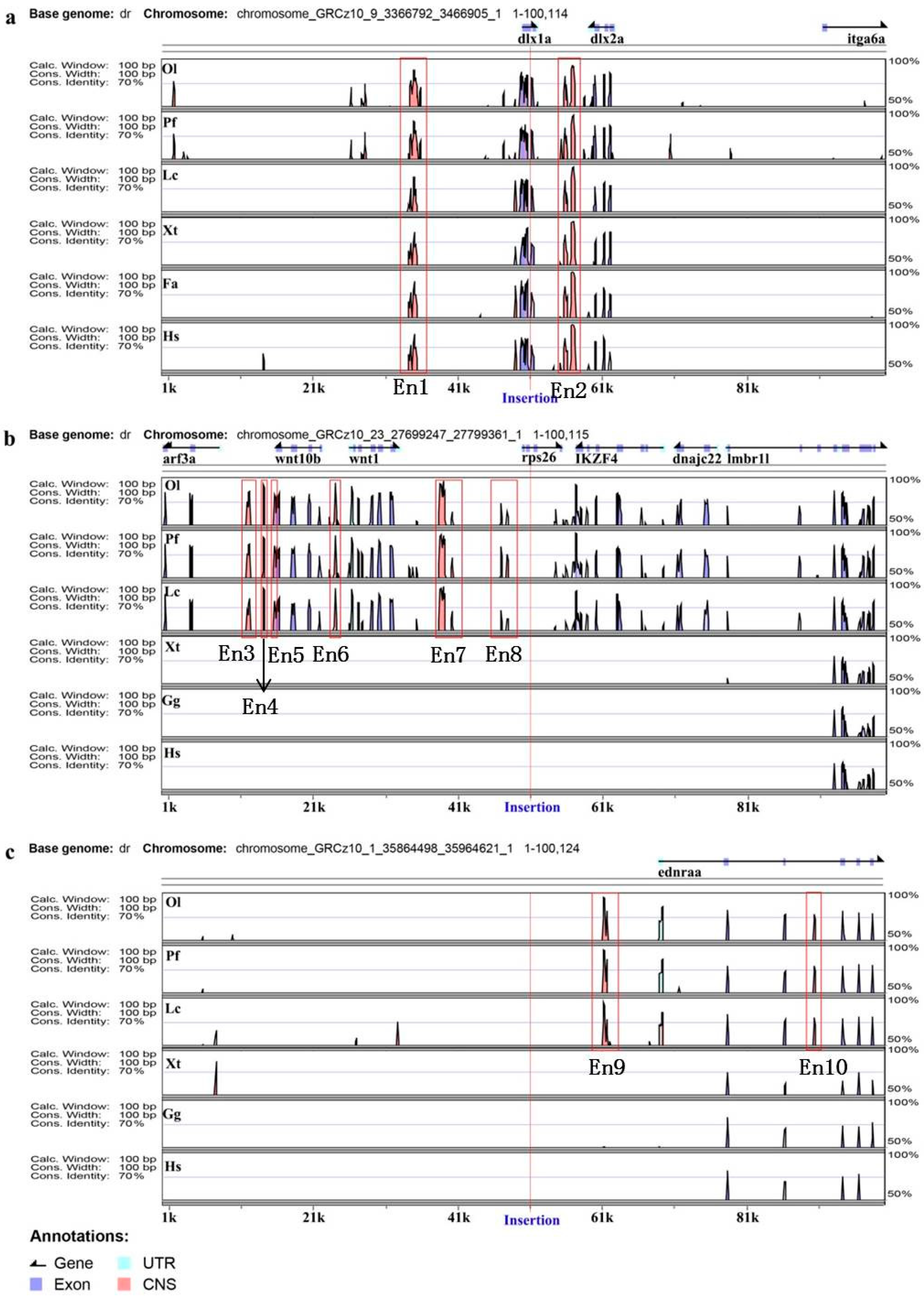
3.2. Annotation of Trapped Enhancers
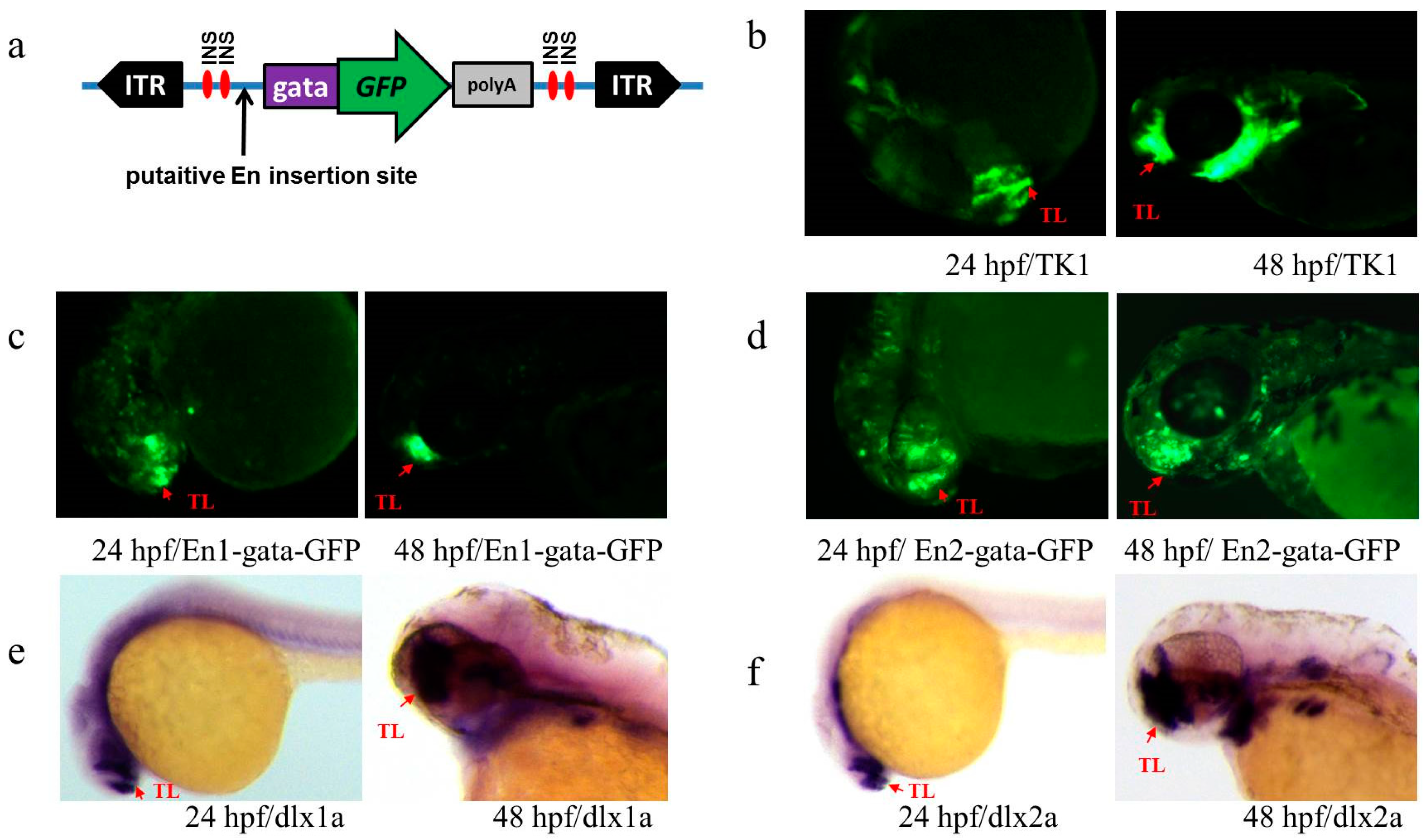
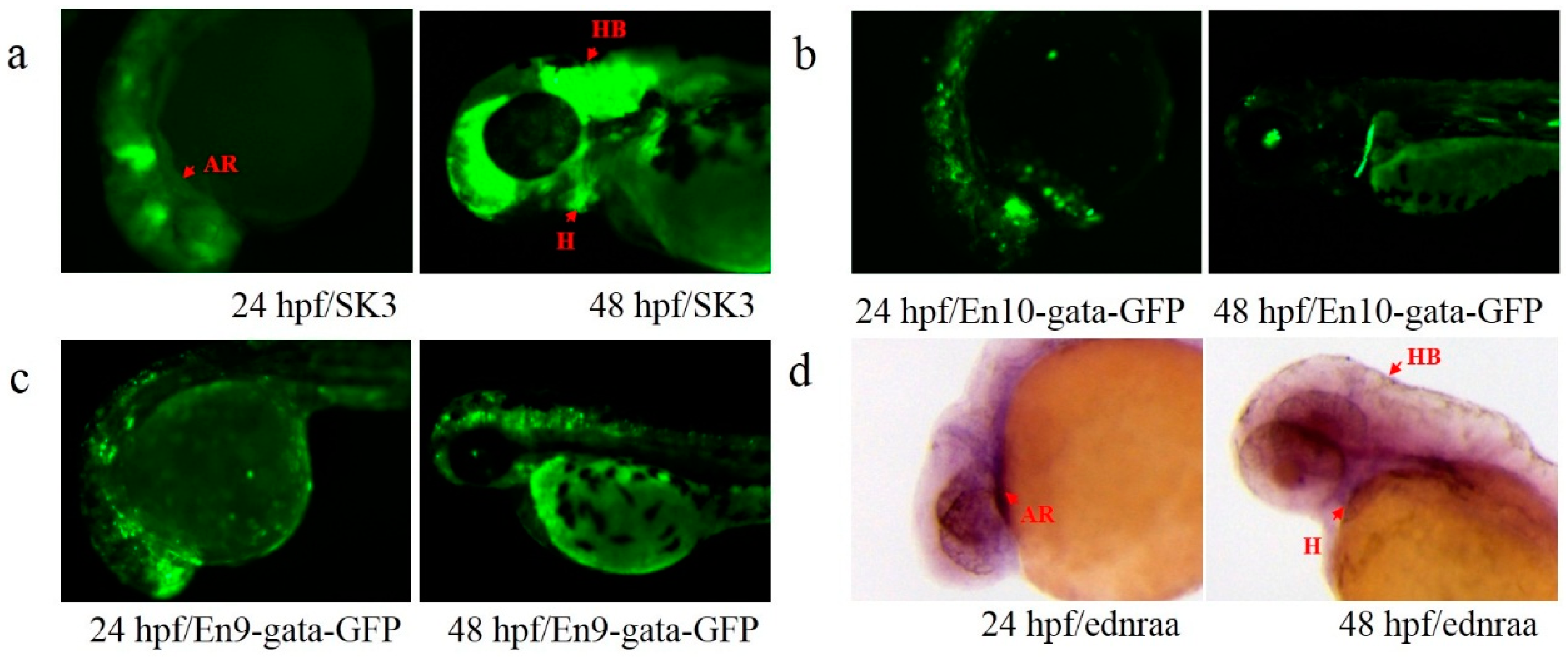
3.3. Activity Test of Identified Enhancers
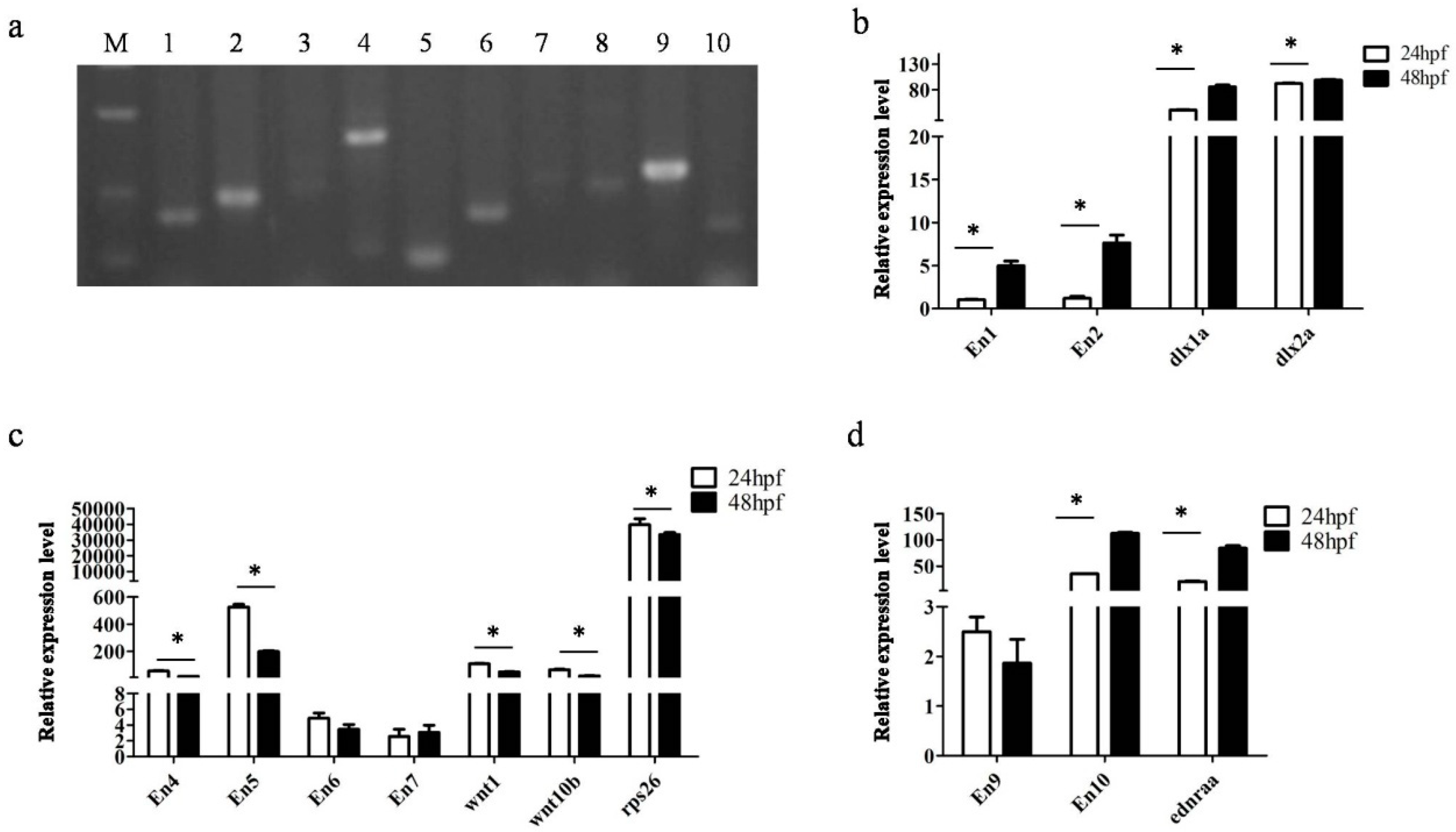
3.4. Transcription Detection of Putative Enhancers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Banerji, J.; Rusconi, S.; Schaffner, W. Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell 1981, 27, 299–308. [Google Scholar] [CrossRef]
- Furlong, E.E.M.; Levine, M. Developmental enhancers and chromosome topology. Science 2018, 361, 1341–1345. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Notani, D.; Rosenfeld, M.G. Enhancers as non-coding RNA transcription units: Recent insights and future perspectives. Nat. Rev. Genet. 2016, 17, 207–223. [Google Scholar] [CrossRef] [PubMed]
- Soldner, F.; Stelzer, Y.; Shivalila, C.S.; Abraham, B.J.; Latourelle, J.C.; Barrasa, M.I.; Goldmann, J.; Myers, R.H.; Young, R.A.; Jaenisch, R. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature 2016, 533, 95. [Google Scholar] [CrossRef] [PubMed]
- Weedon, M.N.; Cebola, I.; Patch, A.M.; Flanagan, S.E.; De, F.E.; Caswell, R.; Rodríguezseguí, S.A.; Shawsmith, C.; Cho, C.H.; Lango, A.H. Recessive mutations in a distal PTF1A enhancer cause isolated pancreatic agenesis. Nat. Genet. 2014, 46, 61. [Google Scholar] [CrossRef] [PubMed]
- Bellen, H.J. Ten Years of Enhancer Detection: Lessons from the Fly. Plant Cell 1999, 11, 2271. [Google Scholar] [CrossRef]
- Bier, E.; Vaessin, H.; Shepherd, S.; Lee, K.; Mccall, K.; Barbel, S.; Ackerman, L.; Carretto, R.; Uemura, T.; Grell, E. Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes Dev. 1989, 3, 1273–1287. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.; Pearson, R.K.; Bellen, H.J.; O’Kane, C.J.; Grossniklaus, U.; Gehring, W.J. P-element-mediated enhancer detection: An efficient method for isolating and characterizing developmentally regulated genes in Drosophila. Genes Dev. 1989, 3, 1301–1313. [Google Scholar] [CrossRef]
- O’Kane, C.J.; Gehring, W.J. Detection in situ of genomic regulatory elements in Drosophila. Proc. Natl. Acad. Sci. USA 1987, 84, 9123–9127. [Google Scholar] [CrossRef]
- Dunne, C.R.; Cillo, A.R.; Glick, D.R.; John, K.; Johnson, C.; Kanwal, J.; Malik, B.T.; Mammano, K.; Petrovic, S.; Pfister, W. Structured inquiry-based learning: Drosophila GAL4 enhancer trap characterization in an undergraduate laboratory course. PLoS Biol. 2014, 12, e1002030. [Google Scholar] [CrossRef]
- Liu, C.; Song, G.; Mao, L.; Long, Y.; Li, Q.; Cui, Z. Generation of an enhancer-trapping vector for insertional mutagenesis in Zebrafish. PLoS ONE 2015, 10, e0139612. [Google Scholar] [CrossRef] [PubMed]
- Grabher, C.; Henrich, T.; Sasado, T.; Arenz, A.; Wittbrodt, J.; Furutani-Seiki, M. Transposon-mediated enhancer trapping in medaka. Gene 2003, 322, 57–66. [Google Scholar] [CrossRef]
- Balciunas, D.; Davidson, A.E.; Sivasubbu, S.; Hermanson, S.B.; Welle, Z.; Ekker, S.C. Enhancer trapping in zebrafish using the Sleeping Beauty transposon. BMC Genom. 2004, 5, 62. [Google Scholar] [CrossRef] [PubMed]
- Scott, E.K.; Mason, L.; Arrenberg, A.B.; Ziv, L.; Gosse, N.J.; Xiao, T.; Chi, N.C.; Asakawa, K.; Kawakami, K.; Baier, H. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat. Methods 2007, 4, 323–326. [Google Scholar] [CrossRef]
- Parinov, S.; Kondrichin, I.; Korzh, V.; Emelyanov, A. Tol2 transposon-mediated enhancer trap to identify developmentally regulated zebrafish genes in vivo. Dev. Dyn. 2004, 231, 449–459. [Google Scholar] [CrossRef]
- Kawakami, K.; Abe, G.; Asada, T.; Asakawa, K.; Fukuda, R.; Ito, A.; Lal, P.; Mouri, N.; Muto, A.; Suster, M.L. zTrap: Zebrafish gene trap and enhancer trap database. BMC Dev. Biol. 2010, 10, 105. [Google Scholar] [CrossRef] [PubMed]
- Koelzer, S.; Kölsch, Y.; Panfilio, K.A. Visualizing Late Insect Embryogenesis: Extraembryonic and mesodermal enhancer trap expression in the beetle Tribolium castaneum. PLoS ONE 2014, 9, e103967. [Google Scholar] [CrossRef] [PubMed]
- Quach, H.N.; Tao, S.; Vrljicak, P.; Joshi, A.; Ruan, H.; Sukumaran, R.; Varshney, G.K.; Lafave, M.C. The Ds Screen Team; Burgess, S.M. A Multifunctional mutagenesis system for analysis of gene function in zebrafish. G3 Genes Genomes Genet. 2015, 5, 1283–1299. [Google Scholar]
- O’Brochta, D.A.; Pilitt, K.L.; Harrell, R.A.; Channa, A.; Alford, R.T. Gal4-based enhancer-trapping in the malaria mosquito Anopheles stephensi. G3 Genes Genomes Genet. 2012, 2, 1305–1315. [Google Scholar] [CrossRef]
- Shima, Y.; Sugino, K.; Hempel, C.M.; Shima, M.; Taneja, P.; Bullis, J.B.; Mehta, S.; Lois, C.; Nelson, S.B. A mammalian enhancer trap resource for discovering and manipulating neuronal cell types. Elife 2016, 5, e13503. [Google Scholar] [CrossRef]
- Ruf, S.; Symmons, O.; Uslu, V.V.; Dolle, D.; Hot, C.; Ettwiller, L.; Spitz, F. Large-scale analysis of the regulatory architecture of the mouse genome with a transposon-associated sensor. Nat. Genet. 2011, 43, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Asakawa, K.; Suster, ML.; Mizusawa, K.; Naqayoshi, S.; Kotani, T.; Urasaki, A.; Kishimoto, Y.; Hibi, M.; Kawakami, K. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc. Natl. Acad. Sci. USA 2008, 105, 1255–1260. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, M.; Mechaly, A.S.; Becker, T.S.; Rinkwitz, S. Using zebrafish transgenesis to test human genomic sequences for specific enhancer activity. Methods 2013, 62, 216–225. [Google Scholar] [CrossRef] [PubMed]
- García-Lecea, M.; Gasanov, E.; Jedrychowska, J.; Kondrychyn, I.; Teh, C.; You, M.S.; Korzh, V. Development of circumventricular organs in the mirror of zebrafish enhancer-trap transgenics. Front. Neuroanat. 2017, 11, 114. [Google Scholar] [CrossRef] [PubMed]
- Lal, P.; Tanabe, H.; Suster, M.L.; Ailani, D.; Kotani, Y.; Muto, A.; Itoh, M.; Iwasaki, M.; Wada, H.; Yaksi, E. Identification of a neuronal population in the telencephalon essential for fear conditioning in zebrafish. Bmc Biol. 2018, 16, 45. [Google Scholar] [CrossRef] [PubMed]
- Urasaki, A.; Asakawa, K.; Kawakami, K. Efficient transposition of the Tol2 transposable element from a single-copy donor in zebrafish. Proc. Natl. Acad. Sci. USA 2008, 105, 19827–19832. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Sahaihernandez, P.; Bohm, R.A.; Welch, W.P.; Zhang, B.; Nystul, T. Enhancer-trap flippase lines for clonal analysis in the Drosophila ovary. G3 Genes Genomes Genet. 2014, 4, 1693–1699. [Google Scholar] [CrossRef]
- Abidi, S.; Smithbolton, R.K. Cell fate changes induced by a Distal-less enhancer-trap transgene in the Drosophila antennal imaginal disc. Sci. Rep. 2018, 8, 4950. [Google Scholar] [CrossRef]
- Otsuna, H.; Hutcheson, D.A.; Duncan, R.N.; Mcpherson, A.D.; Scoresby, A.N.; Gaynes, B.F.; Tong, Z.; Fujimoto, E.; Kwan, K.M.; Chien, C.B. High-resolution analysis of CNS expression patterns in zebrafish Gal4enhancer-trap lines. Dev. Dyn. 2015, 244, 785–796. [Google Scholar] [CrossRef]
- Grabundzija, I.; Irgang, M.L.; Belay, E.; Matrai, J.; Gogol, D.A.; Kawakami, K.; Chen, W.; Ruiz, P.; Chuah, M.K.; Vandendriessche, T. Comparative analysis of transposable element vector systems in human cells. Mol. Ther. J. Am. Soc. Gene Ther. 2010, 18, 1200. [Google Scholar] [CrossRef]
- Mátés, L.; Chuah, M.K.; Belay, E.; Jerchow, B.; Manoj, N.; Acosta-Sanchez, A.; Grzela, D.P.; Schmitt, A.; Becker, K.; Matrai, J. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 2009, 41, 753–761. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Kong, J.; Stalker, J.; Bradley, A. Chromosomal mobilization and reintegration of Sleeping Beauty and PiggyBac transposons. Genesis 2009, 47, 404. [Google Scholar] [CrossRef] [PubMed]
- Li, M.A.; Pettitt, S.J.; Eckert, S.; Ning, Z.; Rice, S.; Cadiñanos, J.; Yusa, K.; Conte, N.; Bradley, A. The piggyBac transposon displays local and distant reintegration preferences and can cause mutations at noncanonical integration sites. Mol. Cell. Biol. 2013, 33, 1317–1330. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Lin, C.; Lu, D.; Ning, Z.; Cox, T.; Melvin, D.; Wang, X.; Bradley, A.; Liu, P. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 2008, 105, 9290–9295. [Google Scholar] [CrossRef] [PubMed]
- Dupuy, A.J.; Akagi, K.; Largaespada, D.A.; Copeland, N.G.; Jenkins, N.A. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature 2005, 436, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Thisse, C.; Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 2008, 3, 59–69. [Google Scholar] [CrossRef]
- Goody, M.F.; Kelly, M.W.; Reynolds, C.J.; Khalil, A.; Crawford, B.D.; Henry, C.A. NAD+ biosynthesis ameliorates a Zebrafish model of muscular dystrophy. PLoS Biol. 2012, 10, e1001409. [Google Scholar] [CrossRef]
- Kim, T.K.; Hemberg, M.; Gray, J.M. Enhancer RNAs: A class of long noncoding RNAs synthesized at enhancers. Cold Spring Harb. Perspect. Biol. 2015, 7, a018622. [Google Scholar] [CrossRef]
- Ivics, Z.; MA, L.; Mátés, L.; Boeke, J.D.; Nagy, A.; Bradley, A.; Izsvák, Z. Transposon-mediated genome manipulation in vertebrates. Nat. Methods 2009, 6, 415–422. [Google Scholar] [CrossRef]
- Ivics, Z.; Mates, L.; Yau, T.Y.; Landa, V.; Zidek, V.; Bashir, S.; Hoffmann, O.I.; Hiripi, L.; Garrels, W.; Kues, W.A.; et al. Germline transgenesis in rodents by pronuclear microinjection of Sleeping Beauty transposons. Nat. Protoc. 2014, 9, 773–793. [Google Scholar] [CrossRef]
- Ivics, Z.; Garrels, W.; Mates, L.; Yau, T.Y.; Bashir, S.; Zidek, V.; Landa, V.; Geurts, A.; Pravenec, M.; Rulicke, T.; et al. Germline transgenesis in pigs by cytoplasmic microinjection of Sleeping Beauty transposons. Nat. Protoc. 2014, 9, 810–827. [Google Scholar] [CrossRef] [PubMed]
- Katter, K.; Geurts, A.M.; Hoffmann, O.; Mátés, L.; Landa, V.; Hiripi, L.; Moreno, C.; Lazar, J.; Bashir, S.; Zidek, V. Transposon-mediated transgenesis, transgenic rescue, and tissue-specific gene expression in rodents and rabbits. Faseb J. 2013, 27, 930–941. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Wilkinson, M.N.; Chen, X.; Sankararaman, S.; Mayhew, D.; Mitra, R.D. An optimized, broadly applicable piggyBac transposon induction system. Nucleic Acids Res. 2017, 45, 1–13. [Google Scholar] [CrossRef]
- Chung, J.H.; Whiteley, M.; Felsenfeld, G. A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 1993, 74, 505–514. [Google Scholar] [CrossRef]
- Bessa, J.; Tena, J.J.; Elisa, D.L.C.; Naranjo, S.; Fernández, A.; Montoliu, L.; Akalin, A.; Lenhard, B.; Casares, F. Zebrafish enhancer detection (ZED) vector: A new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Dev. Dyn. 2009, 238, 2409. [Google Scholar] [CrossRef] [PubMed]
- Ellingsen, S.; Laplante, M.A.; König, M.; Kikuta, H.; Furmanek, T.; Hoivik, E.A.; Becker, T.S. Large-scale enhancer detection in the zebrafish genome. Development 2005, 132, 3799. [Google Scholar] [CrossRef] [PubMed]
- Schaffner, W. Enhancers, enhancers—From their discovery to today’s universe of transcription enhancers. Biol. Chem. 2015, 396, 311–327. [Google Scholar] [CrossRef]
- Ghanem, N.; Yu, M.; Long, J.; Hatch, G.; Rubenstein, J.L.R.; Ekker, M. Distinct cis-regulatory elements from the Dlx1/Dlx2 locus mark different progenitor cell populations in the ganglionic eminences and different subtypes of adult cortical interneurons. J. Neurosci. Off. J. Soc. Neurosci. 2007, 27, 5012. [Google Scholar] [CrossRef]
- Zerucha, T.; Stühmer, T.; Hatch, G.; Park, B.K.; Long, Q.; Yu, G.; Gambarotta, A.; Schultz, J.R.; Rubenstein, J.L.; Ekker, M. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J. Neurosci. Off. J. Soc. Neurosci. 2000, 20, 709–721. [Google Scholar] [CrossRef]
- Ghanem, N.; Jarinova, O.; Amores, A.; Long, Q.; Hatch, G.; Park, B.K.; Rubenstein, J.L.R.; Ekker, M. Regulatory roles of conserved intergenic domains in vertebrate Dlx bigene clusters. Genome Res. 2003, 13, 533–543. [Google Scholar] [CrossRef]
- Harmston, N.; Ing-Simmons, E.; Tan, G.; Perry, M.; Merkenschlager, M.; Lenhard, B. Topologically associating domains are ancient features that coincide with Metazoan clusters of extreme noncoding conservation. Nat. Commun. 2017, 8, 441. [Google Scholar] [CrossRef] [PubMed]
- Lettice, L.A.; Williamson, I.; Devenney, P.S.; Kilanowski, F.; Dorin, J.; Hill, R.E. Development of five digits is controlled by a bipartite long-range cis-regulator. Development 2014, 141, 1715–1725. [Google Scholar] [CrossRef]
- Kvon, E.Z.; Kazmar, T.; Stampfel, G.; Yáñezcuna, J.O.; Pagani, M.; Schernhuber, K.; Dickson, B.J.; Stark, A. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature 2005, 512, 91–95. [Google Scholar] [CrossRef]
- Cannavò, E.; Koelling, N.; Harnett, D.; Garfield, D.; Casale, F.P.; Ciglar, L.; Gustafson, H.E.; Viales, R.R.; Marcoferreres, R.; Degner, J.F. Genetic variants regulating expression levels and isoform diversity during embryogenesis. Nature 2016, 541, 402. [Google Scholar] [CrossRef] [PubMed]
- Ghavi-Helm, Y.; Klein, F.A.; Pakozdi, T.; Ciglar, L.; Noordermeer, D.; Huber, W.; Furlong, E.E.M. Enhancer loops appear stable during development and are associated with paused polymerase. Nature 2014, 512, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.K.; Al, E. Widespread transcription at neuronal activity-regulated enhancers. Nature 2010, 465, 182–187. [Google Scholar] [CrossRef]
- Lam, M.T.Y.; Han, C.; Lesch, H.P.; Gosselin, D.; Heinz, S.; Tanakaoishi, Y.; Benner, C.; Kaikkonen, M.U.; Kim, A.S.; Kosaka, M. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature 2013, 498, 511. [Google Scholar] [CrossRef]
- Li, W.; Notani, D.; Ma, Q.; Tanasa, B.; Nunez, E.; Chen, A.Y.; Merkurjev, D.; Zhang, J.; Ohgi, K.; Song, X. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 2013, 498, 516–520. [Google Scholar] [CrossRef]
- Melo, C.A.; Drost, J.; Wijchers, P.J.; van de Werken, H.; de Wit, E.; Oude Vrielink, J.A.; Elkon, R.; Melo, S.A.; Léveillé, N.; Kalluri, R. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol. Cell 2013, 49, 524–535. [Google Scholar] [CrossRef]

| ET Lines | Nearest Gene | Other Genes within 50 kb Upstream and Downstream Regions | Chromosome | Insertion Position Relative to Gene | Orientation in Genome |
|---|---|---|---|---|---|
| TK1 | ENSDARG00000013125 (dlx1a) | dlx2a, itga6a | 9 | Exon 1 | Forward |
| TK4 | ENSDARG00000037071(rps26) | arf3a, wnt10b, wnt1, IKZF4, dnajc22, lmbr1l | 23 | Intron 1 | Forward |
| SK1 | ENSDARG00000020699 (slc9a8) | FP102158.1 | 23 | Intron 8 | Reverse |
| SK3 | ENSDARG00000011876 (ednraa) | – | 1 | 20 kb (upstream) | Forward |
| SK6 | ENSDARG00000077863 (mettl22) | abat, CU464120.3 | 3 | Exon 12 | Reverse |
| SK12 | ENSDARG00000018272 (wdr33) | si:ch1073-184j22.1, sft2d3, dusp28, proca | 2 | Intron 10 | Reverse |
| PK0 | ENSDARG00000006314 (itgav) | si:dkey-69o16.5, zc3h15 | 9 | Intron 4 | Forward |
| Name | ET Lines | Location | En Length (bp) | En Position Relative to the Insertion |
|---|---|---|---|---|
| En1 | TK1 | chromosome_GRCz10_9_3366792_3466905_1:34392-35394 | 1003 | Upstream |
| En2 | TK1 | chromosome_GRCz10_9_3366792_3466905_1:55781-57232 | 1452 | Downstream |
| En3 | TK4 | chromosome_GRCz10_23_27699247_27799361_1:11781-12216 | 436 | Upstream |
| En4 | TK4 | chromosome_GRCz10_23_27699247_27799361_1:14057-14217 | 161 | Upstream |
| En5 | TK4 | chromosome_GRCz10_23_27699247_27799361_1:15551-15703 | 153 | Upstream |
| En6 | TK4 | chromosome_GRCz10_23_27699247_27799361_1:23944-24162 | 219 | Upstream |
| En7 | TK4 | chromosome_GRCz10_23_27699247_27799361_1:38416-39198 | 783 | Upstream |
| En8 | TK4 | chromosome_GRCz10_23_27699247_27799361_1:46874-47866 | 993 | Upstream |
| En9 | SK3 | chromosome_GRCz10_1_35864498_35964621_1:61133-61728 | 596 | Downstream |
| En10 | SK3 | chromosome_GRCz10_1_35864498_35964621_1:90391-90590 | 200 | Downstream |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, D.; Xue, S.; Chan, S.; Sang, Y.; Wang, S.; Wang, Y.; Chen, C.; Gao, B.; Mueller, F.; Song, C. Enhancer Trapping and Annotation in Zebrafish Mediated with Sleeping Beauty, piggyBac and Tol2 Transposons. Genes 2018, 9, 630. https://doi.org/10.3390/genes9120630
Shen D, Xue S, Chan S, Sang Y, Wang S, Wang Y, Chen C, Gao B, Mueller F, Song C. Enhancer Trapping and Annotation in Zebrafish Mediated with Sleeping Beauty, piggyBac and Tol2 Transposons. Genes. 2018; 9(12):630. https://doi.org/10.3390/genes9120630
Chicago/Turabian StyleShen, Dan, Songlei Xue, Shuheng Chan, Yatong Sang, Saisai Wang, Yali Wang, Cai Chen, Bo Gao, Ferenc Mueller, and Chengyi Song. 2018. "Enhancer Trapping and Annotation in Zebrafish Mediated with Sleeping Beauty, piggyBac and Tol2 Transposons" Genes 9, no. 12: 630. https://doi.org/10.3390/genes9120630
APA StyleShen, D., Xue, S., Chan, S., Sang, Y., Wang, S., Wang, Y., Chen, C., Gao, B., Mueller, F., & Song, C. (2018). Enhancer Trapping and Annotation in Zebrafish Mediated with Sleeping Beauty, piggyBac and Tol2 Transposons. Genes, 9(12), 630. https://doi.org/10.3390/genes9120630






