Abstract
The molecular mechanisms of translation are highly conserved in all organisms indicative of a single evolutionary origin. This includes the molecular interactions of tRNAs with their cognate aminoacyl-tRNA synthetase, which must be precise to ensure the specificity of the process. For many tRNAs, the anticodon is a major component of the specificity. This is not the case for the aminoacylation of alanine and serine to their cognate tRNAs. Rather, aminoacylation relies on other features of the tRNA. For tRNASer, a key specificity feature is the variable arm, which is positioned between the anticodon arm and the T-arm. The variable arm is conserved from yeast to human. This work was initiated to determine if the structure/function of tRNASer has been conserved from Saccharomyces cerevisiae to human. We did this by detecting mistranslation in yeast cells with tRNASer derivatives having the UGA anticodon converted to UGG for proline. Despite being nearly identical in everything except the acceptor stem, human tRNASer is less active than yeast tRNASer. A chimeric tRNA with the human acceptor stem and other sequences from the yeast molecule acts similarly to the human tRNASer. The 3:70 base pair in the acceptor stem (C:G in yeast and A:U in humans) is a prime determinant of the specificity. Consistent with the functional difference of yeast and human tRNASer resulting from subtle changes in the specificity of their respective SerRS enzymes, the functionality of the human and chimeric tRNASerUGG molecules was enhanced when human SerRS was introduced into yeast. Residues in motif 2 of the aminoacylation domain of SerRS likely participated in the species-specific differences. Trp290 in yeast SerRS (Arg313 in humans) found in motif 2 is proximal to base 70 in models of the tRNA-synthetase interaction. Altering this motif 2 sequence of hSerRS to the yeast sequence decreases the activity of the human enzyme with human tRNASer, supporting the coadaptation of motif 2 loop–acceptor stem interactions.
1. Introduction
A fundamental property of all cells is the conversion of DNA sequence into protein via an RNA intermediate. The genetic code, the mechanism of translation, and the machinery involved are highly conserved in all domains of life. Fidelity during translation is maintained at two steps. First, specific aminoacyl-tRNA synthetases (aaRS) are responsible for ligating the correct amino acid onto their cognate tRNAs. Secondly, decoding at the ribosome ensures the correct pairing between a tRNA anticodon and the mRNA codon. Errors at either step can result in the incorporation of the wrong amino acid into the growing polypeptide chain, known as mistranslation [1].
Each aaRS selectively recognizes its cognate tRNAs through individual bases and base pairs, known as identity elements. In two dimensions, the canonical tRNA structure consists of an acceptor stem, where the amino acid is ligated, and three stem-loop structures (reviewed in Reference [2]). From 5′ to 3′ these are the D-arm, the anticodon stem-loop that interacts with the mRNA at the ribosome, and the T-arm, which contributes to the interaction with elongation factor thermo unstable (EF-Tu) to recruit the tRNA to the ribosome. Serine, leucine, and bacterial tyrosine tRNAs have an addition arm, i.e., the variable arm, between the anticodon and T-arm. In three dimensions, all tRNAs fold into a L-shape (reviewed in Reference [3]). In this structure, the acceptor stem and T-arm interact to form one part of the L and the anticodon stem and D-arm interact to form the other. Many aaRS enzymes span the entire length of the tRNA with both the anticodon and acceptor stem being major determinants for recognition [4,5,6]. In contrast, the anticodon is not a determinant for seryl- and alanyl-tRNA synthetases [6]. For the alanyl-tRNA synthetase (AlaRS), the major identity element is a G3:U70 base pair in the acceptor stem [7,8,9,10]. A major determinant for seryl-tRNA synthetase (SerRS) is the variable arm [11,12,13].
All aaRSs catalyze the attachment of an amino acid onto the 3′ end of a tRNA in a two-step reaction. In the first step, an amino acid is recognized and activated with ATP to form an aminoacyl-adenylate. In the second step, the activated amino acid is transferred to the terminal 3′ adenosine of the tRNA (reviewed in Reference [14]). The aaRSs are divided into two classes based on their catalytic domains [14,15,16]. Class I aaRS enzymes contain a Rossmann fold with two consensus motifs (HIGH and KMSKS) that interact with ATP [15,17,18]. In class II enzymes, a unique seven sheet antiparallel beta fold [19] containing the highly degenerate motif 2 (FRXE) and motif 3 (GXGXGF[D/E]R) are responsible for binding ATP [15,20,21].
Class II enzymes are composed of a catalytic domain, often an editing domain and, in all members except AlaRS and SerRS, an anticodon binding domain [22]. Like other class II enzymes, SerRS approaches the tRNA acceptor stem from the major groove side [12,23]. In the catalytic site, beta sheets form the base of a pocket with two sides composed of large hairpins [22]. A conserved phenylalanine near the end of motif 2 stacks with the adenine ring of ATP, while a conserved arginine interacts with the alpha-phosphate [12]. The hairpin loop between two beta-strands of motif 2 contacts the upper bases of the acceptor stem [12,22,23,24,25]. The main recognition element of SerRS is the unique long variable arm, which is bound by an N-terminal tRNA binding domain comprising two alpha helices in all organisms except methanogens [12,26]. The enzyme functions as a homo-dimer, where the tRNA binding domain of one subunit binds the variable arm and positions the 3′ end of the tRNA in the active site of the other subunit [12,27,28]. Additional in vitro and in vivo experiments have identified other weak identity elements in the acceptor stem of bacterial tRNASer [29,30].
Translation is a fundamental process where both the mechanism and the molecules involved in steps such as aminoacylation of tRNAs are conserved. This is evident from the ability of heterologous aaRS to complement deletions of native synthetase genes. For example, the human LysRS rescues a deletion of Escherichia coli LysRS [31], and the human AlaRS can replace the native yeast enzyme [32]. More recently, complementation of MetRS in yeast by the human enzyme was used to investigate disease-related variants [33]. However, coevolution of tRNA and synthetase pairs often results in decreased or abolished tRNA function when expressed in non-native organisms. For example, Burke et al. [34] found that human tRNAPro is not aminoacylated by E. coli ProRS due to differences in the acceptor stem and D-arm of the tRNA. Edwards et al. [35] used a tyrosine tRNA suppressor from E. coli to suppress stop codons in yeast and found it was not aminoacylated by yeast TyrRS but rather was aminoacylated with leucine. In addition, approaches to encode noncanonical amino acids at stop codons rely on orthogonal tRNA:aaRS pairs, often transplanted from other organisms, that do not interact with endogenous tRNAs or aaRSs (reviewed in Reference [36]).
This work was initiated to determine if the structure/function of tRNASer has been conserved from Saccharomyces cerevisiae to human. We analyzed human tRNASer function in S. cerevisiae by detecting mistranslation and suppression with tRNASer derivatives having the UGA anticodon converted to UGG for proline. We found that despite the conservation of tRNASer, differences in the acceptor stem result in reduced functionality of the human tRNA in yeast. The 3:70 base pair in the acceptor stem (C:G in yeast and A:U in humans) is a prime determinant of the specificity. Toxicity of the humanized tRNAs increases when human SerRS is introduced into yeast, suggesting a role for aminoacylation in the differential function.
2. Materials and Methods
2.1. Yeast Strains
Yeast strains are derivatives of the wild-type haploid strain BY4742 (Table S1; Reference [37]). The tti2 disruption strain complemented by tti2-L187P on a LEU2 centromeric plasmid (CY7020) has been previously described [10]. The met22Δ strain (CY7640) was derived from a spore colony of the yeast magic marker strain in the BY4743 diploid background [38].
Yeast strains were grown in yeast peptone media containing 2% glucose or synthetic media supplemented with nitrogenous bases and amino acids at 30 °C. Selections for CY7020-derived strains were on yeast peptone dextrose (YPD) plus 5% (v/v) ethanol. Cells were grown in dropout medium to stationary phase, 10-fold serially diluted, and spotted onto the indicated medium.
Growth curves were generated by diluting saturated cultures to OD600 ~ 0.1 in minimal media and incubating at 30 °C. OD600 was measured every 15 min using a BioTek Epoch 2 (BioTek, Winooski, VT, USA) microplate spectrophotometer for 24 h.
2.2. Yeast Transformation
Approximately 106 cells were pelleted and washed twice in an equivalent volume of 100 mM lithium acetate, 1.0 mM ethylenediaminetetraacetic acid, and 10 mM TrisHCl (pH 7.5; LiAc) and suspended in 200 μl of LiAc. Five milligrams of denatured calf thymus DNA and ~1.0 μg of plasmid DNA was incubated with 100 μl of cells for 15 min at 30 °C. Then, 1.0 mL of 40% polyethylene glycol 4000 was added, and the cells were incubated 15 min at 30 °C. Dimethyl sulfoxide was added to 7%, and the cells were incubated at 42 °C for 15 min. Cells were pelleted, plated on selective medium, and grown at 30 °C.
2.3. Plasmid Constructs
SUP17 [SctRNASerUGA] including ~300 bp upstream and downstream and either the wild-type anticodon (UGA; pCB3076) or the proline anticodon (UGG; pCB3082) in the URA3 centromeric plasmid YCplac33 have been previously described [39]. The G26A derivative of pCB3082 (pCB4023) and A4G derivative of pCB3082 (pCB4097) were identified by selection [39]. DNA containing the sequence encoding human tRNASer (chr6.trna172-SerUGA) with proline anticodon UGG inserted directly in place of SUP17 (HstRNASerUGG; see Figure S1) was purchased from Invitrogen (Pleasanton, CA, USA). This was digested with PstI and EcoRI and inserted into YCplac33 (pCB4062). The SUP17-human acceptor stem chimera (chtRNASerUGG) with proline anticodon was constructed by two-step PCR using the outside primers UG5953 and UG5954, internal primers VD5993 and VD5994 to alter the 3’ end of the acceptor stem, VD5578 and VD5579 to alter the 5′ end of the acceptor stem, and pCB4062 as template. The molecule was cloned as a HindIII-EcoRI fragment into YCplac33 (pCB4122). (See Table S2 for oligonucleotides.) The A4G:U69C derivative of pCB3082 (pCB4145) was similarly constructed using pCB4097 as template and the inside primers VF8687 and VF8688. The A3C:T70G derivative of the chimeric tRNASerUGG (pCB4336) was similarly constructed using outside primers UG5953 and UG5954, inside primers WE0866 and WE0867 to alter the 3’ end, WE0868 and WE0869 to alter the 5’ end, and pCB4122 as the template.
The cloned coding region of human seryl-tRNA synthetase (SARS) was purchased from NovoPro Biosciences Inc. (Cat# 715151-2, Shanghai, China; RefSeq NM_006513). This was amplified by PCR using oligonucleotides VD5232 and VD5803 and cloned as a NotI-EcoRI fragment into YCplac111 containing a DED1 promoter and myc9-tag (pCB4329; Hoffman et al. [40]) and YEplac181 containing a DED1 promoter and myc-tag (pCB4355). Note that codon 435, which codes for C in the purchased clone, was converted by two-step PCR to an R codon to resemble those more commonly found in mammalian SerRS. A derivative of SARS with mutations T312A and R313W was constructed by ligating an NcoI-EcoRI fragment synthesized by PCR with oligonucleotides WF1846 and VD5803 into pCB4355 to give pCB4357. S. cerevisiae SES1 was similarly inserted into YCplac111 (pCB4342) and YCplac181 (pCB4356) after PCR with oligonucleotides WF1727 and UK0551.
The centromeric plasmid containing HSE-eGFP has previously been described [41] and was kindly provided by Martin Duennwald.
2.4. Western Blot Assay
Yeast extract prepared by grinding with glass beads [42] was separated by SDS-PAGE and transferred to PVDF membrane (Roche Applied Science, Penzberg, Germany). Anti-myc (9E10, Sigma-Aldrich, St. Louis, MO, USA) was used at a ratio of 1:5000. Secondary antibody (anti-Mouse IgG HRP, Promega, Madison, WI, USA) was used at a ratio of 1:10,000 and was detected using SuperSignal West Pico Chemiluminiscent Substrate (Thermo Scientific, Waltham, MA, USA).
2.5. Fluorescence Heat Shock Reporter
Yeast strains containing the heat shock response element (HSE)-eGFP reporter were grown to stationary phase in a medium lacking leucine and uracil, diluted 1:50 in the same medium, and grown for 6 h at 30 °C. Cell densities were normalized to OD600 before measuring fluorescence. Fluorescence was measured with a BioTek Synergy H1 microplate reader at an emission wavelength of 528 nm using Gen5 2.08 software. The mean relative fluorescence units were calculated across three technical and three biological replicates for each strain.
2.6. Mutual Information Analysis
Coevolving residues within SerRS were identified following the protocol outlined by Dickson and Gloor [43]. Briefly, crystal structures of SerRS from Candida albicans (3QNE; [44]), Aquifex aeolicus (2DQ3), Pyrococcus horikoshii (2ZR2; [45]), Homo sapiens (4L87; [46]), Naegleria fowleri (6BLJ), and Thermus thermophilus (1SET; [47]) from the Protein Data Bank were used to construct a structural alignment. ASN.1 files for each structure were obtained from the Molecular Modeling Database, and the structures were aligned in Cn3D [48] to obtain block structures for sequence alignment. Seryl-tRNA synthetase sequences were obtained from the UniProt Reference Clusters 50 (UniRef50) dataset as FASTA files (Supplemental File S1), imported into Cn3D and block aligned with the SerRS structures. Sequences that were not annotated as cytoplasmic SerRS, as well as hypothetical proteins, were removed. Coevolution scores were calculated with MIpToolset [49].
3. Results and Discussion
The processes involved in translation are conserved across all species. Not surprisingly, there is significant similarity in the key molecules involved [50,51]. In this work, our goal was to investigate the cross-species similarities and differences in tRNA function by analyzing the activity of human tRNASer in S. cerevisiae. To begin the analysis, we aligned the tRNASerUGA isodecoders from S. cerevisiae and human (Figure 1A). Of the 83 nucleotides, 60 are conserved in both sets of tRNAs. Fifteen positions are always different between the three identical yeast tRNAs and the four human tRNAs. The variable arms are conserved in length and G:C content.

Figure 1.
Human (Homo sapiens) tRNASerUGA functions inefficiently in Saccharomyces cerevisiae. (A) Sequence alignment of genes encoding tRNASerUGA isodecoders from S. cerevisiae and humans. The red box highlights the anticodon. Sequences were obtained from the GtRNAdb ([52]; http://gtrnadb.ucsc.edu/index.html) and aligned with MUSCLE (https://www.ebi.ac.uk/Tools/msa/muscle/). (B) URA3 centromeric plasmid (YCplac33) expressing S. cerevisiae (SUP17) or human (tRNA-Ser-TGA-3-1) tRNASer genes with their UGA anticodon converted to UGG for proline [SctRNASerUGG and HstRNASerUGG] were transformed into the wild-type yeast strain BY4742 using a lithium acetate protocol and grown for 3 days at 30 °C. (C) The growth of BY4742 containing a centromeric plasmid expressing HstRNASerUGG was compared to strains containing wild-type SctRNASerUGA and the mistranslating SctRNASerUGG-G26A. Strains were grown in medium lacking uracil for 40 h, then 10-fold serially diluted, spotted on minimal plates lacking uracil, and grown for 2 days at 30 °C. (D) Plasmids expressing SctRNASerUGA, HstRNASerUGG, or SctRNASerUGG-G26A were transformed into the tti2-L187P strain CY7020, streaked for individual colonies, grown in minimal medium lacking uracil to stationary phase, then 10-fold serially diluted, spotted on minimal plates lacking uracil (-URA) or yeast peptone dextrose (YPD) containing 5% ethanol, and grown at 30 °C for 2 days (for -URA) or 3 days (for YPD + 5% ethanol).
3.1. Mistranslation Assay for tRNASer Function
We previously identified a stress-sensitive allele (L187P) of the co-chaperone protein TTI2 that is suppressed by mistranslation of L187P with serine [39]. A gene expressing yeast SUP17 (SctRNASer) with the UGA anticodon of the tRNA changed to UGG (to decode proline codons) cannot be introduced into yeast [39] due to high levels of mistranslation of serine at proline codons. Dampening the function of the tRNA with secondary mutations allows the introduction of tRNASerUGG variants. If the tRNASerUGG retains function, it is aminoacylated with serine and inserts (mistranslates) serine for proline at a level sufficient to suppress tti2-L187P. If the tRNASerUGG variant is not functional, it can be introduced into yeast but does not mistranslate and thus does not suppress tti2-L187P. In our assay tRNASerUGG competes with endogenous tRNAPro for the decoding of tti2-L187P mutation. We used this system to analyze the activity of human tRNASerUGA in S. cerevisiae after converting the anticodon to UGG. A gene encoding human tRNASer (chr2.trna21) with the UGA anticodon converted to UGG was inserted into the SUP17 locus with ~300 bp of 5′ and 3′ flanking yeast sequence and cloned into the yeast URA3 centromeric plasmid YCplac33 (see Figure S1 for the gene sequence). As shown in Figure 1B, the human gene expressing tRNASerUGG (HstRNASerUGG) could be transformed into the wild-type strain BY4742, in contrast to SctRNASerUGG [39]. The growth of this strain on a minimal plate was slightly slower than a strain containing YCplac33 alone and slightly faster than a strain containing SctRNASerUGG with a G26A mutation (Figure 1C; see Reference [39]). The reduced toxicity of the human tRNASerUGG indicates that its activity in yeast is diminished relative to S. cerevisiae tRNASerUGG. To determine if the human tRNASer functions in yeast, HstRNASerUGG was transformed into CY7020, which contains the crippled tti2-L187P allele. As shown by growth on medium containing 5% ethanol (Figure 1D), HstRNASerUGG suppressed the stress sensitive growth caused by tti2-L187P. The human tRNA thus has partial function in yeast.
Because 7 of the 15 differences always found between yeast and human tRNASer are in the acceptor stem (Figure 1A), we engineered a chimeric tRNA with the human acceptor stem substituted on the S. cerevisiae tRNA and with a UGG anticodon (chtRNASerUGG; Figure 2A). The chimeric tRNASerUGG mimicked HstRNASerUGG in its ability to be transformed into BY4742 (Figure 2B). As shown in Figure 2C, chtRNASerUGG was more toxic than HstRNASerUGG, suggesting that at least one other difference between S. cerevisiae and human tRNASer exists outside the acceptor stem (discussed in Section 3.4). The chimeric tRNA was also functional as determined by its ability to suppress tti2-L187P when transformed into CY7020 (Figure 2D).

Figure 2.
The human tRNASer acceptor stem decreases the functionality of yeast tRNASer. (A) Secondary structures of tRNASerUGG derivatives used in these studies. The chimeric molecule contains the human acceptor stem on the yeast tRNA. Differences with the S. cerevisiae tRNASerUGG (SUP17UGG) are in red. (B) URA3 centromeric plasmids expressing the chimeric tRNA (chtRNASerUGG) with a UGG anticodon or SctRNASer (SUP17) with a UGG anticodon were transformed into BY4742 and grown at 30 °C for 2 days on a minimal plate. (C) BY4742 expressing SctRNASerUGA, HstRNASerUGG, or chtRNASerUGG were grown overnight in minimal media, serially diluted, plated on media lacking uracil, and grown at 30 °C for 2 days. (D) Plasmids expressing SctRNASerUGA, HstRNASerUGG, or chtRNASerUGG were transformed into the tti2-L187P strain CY7020, streaked for individual colonies, grown in minimal medium for 40 h, 10-fold serially diluted, spotted on minimal plates lacking uracil (-URA) or yeast peptone dextrose (YPD) containing 5% ethanol, and grown at 30 °C for 2 days (for -URA) or 3 days (for YPD + 5% ethanol). (E) A reporter plasmid containing GFP expressed from a Hsf1-activated promoter was transformed into BY4742 also containing centromeric plasmids expressing wild-type SctRNASerUGA, HstRNASerUGG, or chtRNASerUGG. Starter cultures were grown in minimal medium, diluted into fresh selective medium, and grown for eight hours. Induction of the Hsf1-GFP was measured by fluorescence at 528 nm. Numbers shown are the average of five biological replicates performed in duplicate with the standard deviation shown.
The mistranslation experiments we performed were a balance between improved growth due to suppression of tti2-L187P and toxicity due to proteome-wide mistranslation. To provide another indication of the toxicity arising from mistranslation due to each of the tRNAs, we measured heat shock induction in the strains using a reporter plasmid containing GFP expressed from an Hsf1-activated promoter (Figure 2E). HstRNASerUGG and chtRNASerUGG resulted in a heat shock response 2.1- and 2.8-fold, respectively, greater than the control. SctRNASerUGG-G26A resulted in a 4.8-fold increase in the heat shock response. These results suggest that the toxicity of human and chimeric tRNAs is the result of a loss of proteostasis due to mistranslation.
3.2. Evaluating Mechanisms for the Reduced Functionality of Human tRNASer
Differences between yeast and human tRNASer could be due to any step leading to the functionality of the tRNA. Included in this are factors that contribute to the cellular concentration of the tRNA. Although it is difficult to compare the levels of the mistranslating tRNAs because the genomically encoded copies of yeast tRNASerUGA differ from SctRNASerUGG by only one nucleotide, we addressed a possible role for the rapid tRNA decay (RTD) pathway in the function of HstRNASerUGG. HstRNASerUGG was transformed into a met22Δ strain where the RTD pathway is inhibited [53,54]. If the mistranslating human tRNA was being turned over by the RTD pathway, we would expect it to be more toxic in a met22Δ strain, where it would accumulate at higher levels and result in increased levels of mistranslation. The toxicity of HstRNASerUGG was not increased in the met22Δ strain (Figure 3A), suggesting the human tRNA is not being turned over by the RTD pathway.

Figure 3.
Evaluating mechanisms of decreased function of human tRNASerUGG in yeast. (A) BY4742 (MET22) and CY7641 (met22Δ) containing either wild-type (WT) SctRNASerUGA or HstRNASerUGG were grown to stationary phase in media lacking uracil before cells were spotted in 10-fold serial dilutions onto a plate lacking uracil and grown at 30°. (B) Expression of human SerRS, SARS, increases the functionality and thus the toxicity of HstRNASerUGG and chtRNASerUGG in yeast. Yeast strain BY4742 containing a URA3 centromeric plasmid expressing SctRNASerUGA, SctRNASerUGG-G26A. HstRNASerUGG or chtRNASerUGG and the LEU2 multicopy plasmid YCplac181 containing myc-tagged yeast SES1 (ySerRS) or human SARS (hSerRS) were grown to stationary phase in media lacking uracil and leucine. Cell densities were normalized, and cultures were spotted in 10-fold serial dilutions on the same media.
If the functionality and resulting mistranslation of chtRNASerUGG was reduced relative to SctRNASer because of decreased aminoacylation, we would expect that toxicity would increase if human SerRS, SARS, was also introduced into yeast cells. We amplified the coding region of SARS by PCR and cloned it 3’ of the yeast DED1 promoter and a myc tag. The myc-tagged SARS is expressed in S. cerevisiae strain BY4742 at a level comparable to S. cerevisiae Ses1 (Figure S2). Centromeric plasmids expressing SctRNASerUGA, SctRNASerUGG-G26A, HstRNASerUGG, and chtRNASerUGG were then transformed into strains expressing SARS and Ses1 on 2μ plasmids. Their growth was compared on minimal media lacking uracil and leucine. As shown in Figure 3B, the presence of human SARS reduces the growth of the strain containing either HstRNASerUGG or chtRNASerUGG but not the strain containing SctRNASerUGG-G26A. The increased toxicity of HstRNASerUGG and chtRNASerUGG in the presence of SARS further indicates that the acceptor stem contributes to differences in the aminoacylation of human and yeast tRNASer.
3.3. The tRNASer 3:70 Base Pair Contributes to Species Specificity
To begin to map the key bases within the acceptor stem that could account for the functional difference of human tRNASer in yeast, we first compared sequences of the S. cerevisiae tRNASer isoacceptors. We hypothesized that key bases for yeast SerRS should be conserved. Five bases in the acceptor stem are conserved in all yeast serine tRNAs. These are the G1:C72 and C3:G70 bases pairs and U69 (Figure 4A; see Figure S3 for the sequence of the tRNAs). Figure 4B shows the bases that differ between yeast and human tRNASerUGA isodecoders. Only G1:C72 is conserved for all these tRNAs (see sequences in Figure 1A). U69 and the 3:70 base pair are thus strong candidates for a difference determinant between S. cerevisiae and human tRNASer. We evaluated the importance of U69 with a variant of SctRNASerUGG containing the human G4:C69 base pair. A gene expressing this variant could not be transformed into yeast (Figure S4), indicating that it is fully functional and suggesting that U69 does not signal the specificity difference between yeast and human tRNASer. To test the 3:70 base pair, we constructed a variant of chtRNASerUGG with the yeast C3:G70 base pair (chtRNASerUGG-C3:G70). Unlike the unmodified chimeric gene, chtRNASerUGG = C3:G70 gave rise to extremely slow growing colonies when transformed into BY4742 (Figure 4C). The C3:G70 mutation thus increases the functionality and toxicity of the chimeric tRNA in S. cerevisiae.

Figure 4.
The acceptor stem 3:70 base pair is a determinant of yeast versus human tRNASer specificity. (A) Variation in the acceptor stem of the S. cerevisiae tRNASer isoacceptors. The acceptor stem of the tRNA encoded by SUP17 is shown with the positions that vary in the different cytoplasmic tRNASer isoacceptors colored in red. (B) Variation in the acceptor stems of tRNASerUGA between S. cerevisiae and humans. The acceptor stem of the tRNA encoded by SUP17 is shown with the bases always different between yeast and human colored in red and those sometimes-different colored in blue. (C) BY4742 expressing chtRNASerUGG or chtRNASerUGG-C3:G70 on a URA3 centromeric plasmid were transformed into BY4742 and grown at 30 °C for 2 days.
The aminoacylation domain of the class II aaRS enzymes contains antiparallel β sheets. The loop between strands 2 and 3 comprise the conserved motif 2, which interacts with the tRNA acceptor arm [34]. The crystal structures of SerRS from several organisms have been solved. Included in these are the human enzyme and a structure of the T. thermophilus enzyme in complex with tRNASer. Unfortunately, in the latter, the acceptor stem is not fully resolved. Models for the interaction of SerRS with tRNASer position motif 2 close to base pairs 3:70 and 4:69 of the acceptor stem (for example see Figure 5A and Reference [25]). To begin to identify what may account for the different acceptor stem preferences of the SerRS enzymes, we performed a sequence alignment of motif 2 from the human, T. thermophilus, C. albicans, and S. cerevisiae enzymes. The human and T. thermophilus enzymes prefer a A3:U70 base pair in the acceptor stem, whereas the two yeast enzymes prefer C3:G70. Two residues in motif 2 are conserved in the yeast enzymes that differ in the human and T. thermophilus enzymes (Figure 5B). Trp290 and Ala297 of the S. cerevisiae enzyme align with Arg and Gln, respectively, in T. thermophilus and human enzymes. Ala297 is not well conserved across yeast and fungal species containing C3:G70 tRNASer (Figure S5); in contrast, the 290 position is conserved in yeast and is proximal to the base at position 70 (Figure 5A). We constructed a version of hSerRS, where residues T312 and R313 in motif 2 were converted to the corresponding yeast residues A and W. The mutated hSerRS was introduced into BY4742 containing wild-type SctRNASerUGA, SctRNASerUGG-G26A, HstRNASerUGG, or chtRNASerUGG (Figure 5C). The wild-type human SerRS decreased the growth rate in strains containing the human or chimeric tRNASer but did not affect the strains containing the yeast tRNAs. In contrast, the hSerRS TR to AW mutant did not alter the growth rate in these strains when compared to the yeast SerRS, suggesting that T312 and R313 are important for the function of the human SerRS on the human tRNA.
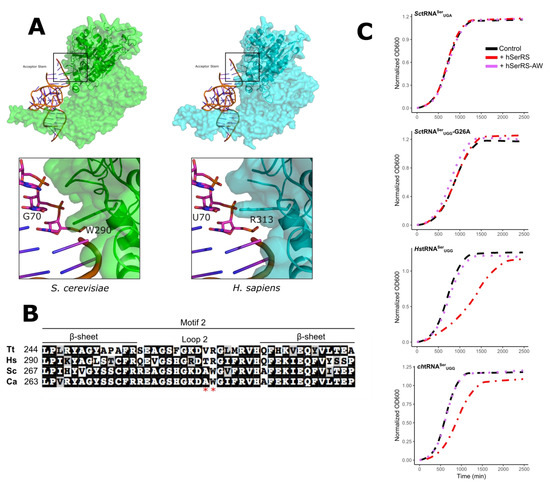
Figure 5.
Sequence differences in Motif 2 of SerRS. (A) Model of motif 2 SerRS:tRNASer interactions. The yeast (green) and human (cyan) SerRS were modeled using SWISS-MODEL [55] onto the Thermus thermophilus synthetase:tRNA co-crystal structure (PDB: 1SER; [12]). The region around motif 2 is boxed and expanded in the lower images. (B) Sequence alignment of SerRS motif 2 sequences from T. thermophilus (WP_024119136), H. sapiens (NP_006504), S. cerevisiae (NP_010306), and Candida albicans (XP_719967) using the default settings of MUSCLE (https://www.ebi.ac.uk/Tools/msa/muscle/). The red stars mark the residues mutated to the corresponding yeast residues. (C) Yeast strain BY4742 containing a URA3 centromeric plasmid expressing SctRNASerUGA, HstRNASerUGG, chtRNASerUGG. or SctRNASerUGG-G26A and the LEU2 multicopy plasmid YEplac181 containing myc-tagged ySerRS (black), hSerRS (red), or mutant hSerRS-AW (purple) were grown to stationary phase in media lacking uracil and leucine, diluted to an OD600 ~ 0.1, and grown for 40 h at 30 °C. OD600 was measured every 15 min to generate growth curves.
Though it is difficult to exclude contributions from all other mechanisms, such as the exact cellular levels of each tRNA and their interactions, we suggest that aminoacylation is a component of the different functionality of human tRNASer in S. cerevisiae. For class II enzymes, motif 2 contacts the acceptor stem [23,56,57,58]. Interactions between SerRS and tRNASer suggest that these interactions influence aminoacylation by S. cerevisiae SerRS [12,25]. In addition, the toxicity, and thus the inferred functionality, of both HstRNASerUGG and chtRNASerUGG was enhanced when human SerRS was introduced into yeast. Furthermore, altering the motif 2 sequence of hSerRS to resemble the yeast sequence decreases the activity of the human enzyme with human tRNASer. We do note that in our analysis, we compared one yeast serine isodecoders with one version of the same human isodecoders. Sequence differences in isodecoders families may result in different cross-species functionality. For tRNASer, the major identity element is the variable arm, both its length and its sequence [6,13,59]. Our data support the argument that the acceptor stem of tRNASer also has a role in its aminoacylation and that these changes are species-specific. The role of the acceptor stem of tRNASer parallels that seen for many other aaRS enzymes [6]. Interestingly, Shiba et al. [31] observed that the positioning of identity elements for specific tRNA isoacceptors is generally common across species, but the exact nature of these sequences is often species-specific.
The possibility of a shared importance of the 3:70 base pair for aminoacylation by the class II enzymes AlaRS and SerRS is noteworthy. G3:U70 is unique for tRNAAla and highly conserved. The significance of the G3:U70 identity element for AlaRS has likely led to its parallel exclusion from other tRNAs. In fact, G3:C70 and A3:U70 show a somewhat limited distribution in non-alanine tRNAs in eukaryotic species (Figure S6), likely because of the mistranslation that would result from a single base mutation generating a G3:U70. These sequence limitations may have led to the 3:70 base pair’s role in recognition arising in some other tRNAs in some species. For many yeast and fungal species, the C3:G70 base pair is conserved in tRNASer-Ser. It is also conserved in T. thermophilus as A:U in all tRNA isoacceptors, which is interesting because of all the tRNAs, the four serine tRNAs are the only ones where A3:U70 is found.
3.4. Unique Aspects of the SerRS Motif 2
It is interesting that the loop sequence in motif 2 in SerRS is distinct from other aaRS (Figure S7). The placement of Trp in the motif 2 loop of yeast SerRS was previously observed by Lenhard et al. [60]. They demonstrated that mutations to the loop region affect functionality. Our model proposes that Trp290 of yeast SerRS is a discriminating factor in the sequence preferences of the enzyme for the tRNA. Arginine in motif 2 of SerRS is frequently found with tRNASer containing the A3:U70 base pair and in species containing a broader range of acceptor stem sequences, including humans, whereas tryptophan is frequently found where C3:G70 is the sole or predominant sequence as seen in yeast and fungi. Eichert et al. [25] examined the structure of minihelices with the T. thermophilus SerRS enzyme. They found that the side chain of Arg267 of the T. thermophilus enzyme (the equivalent of Trp290 in S. cerevisiae Ses1) interacts with the phosphate backbone of bases C69 and U70 via a water network. The exact positioning of Trp290 in the C. albicans structure is difficult to predict because of low electron density in this region; however, the hydrophobic Trp290 side chain almost certainly will have different interactions than a polar arginine. Tryptophan is infrequently found in base interactions [61], but hydrophobic residues occur in nucleic acid interactions through base stacking. For example, a tryptophan in the Tn5 transpose base stacks with a thymine of its substrate DNA [62]. This interaction requires distortion of the DNA duplex. Similar hydrophobic interactions occurring after base flipping are observed for other enzymes, including methyltransferases and endonucleases [63]. Further experiments will be required to determine if correct positioning of the tRNA acceptor stem of the Trp290 enzymes involves distortion of the base pairing. Alternatively, the Trp290 containing enzymes may compensate for the lack of interaction through additional contacts. To address this possibility, we analyzed the mutual information between each pair of positions within the aminoacylation domain of the cytoplasmic SerRS family of proteins to determine residues coevolving with Trp290 (Figure S8; Supplemental File S2). No residue was found to specifically coevolve with Trp290. We do note that although the role of the distinct motif 2 sequence found in SerRS may be in tRNA recognition, motif 2 more generally plays a role in ATP binding. This function could be indirectly affected by our mutations.
The chimeric S. cerevisiae tRNASer with the human acceptor stem was somewhat more toxic and induced a slightly greater heat shock response in yeast than the human tRNASer. This suggests that some of the other base differences between yeast and human tRNASer molecules contribute a partial effect to tRNA function. These may include the differences in the anticodon stem, specifically at position 29 (A in S. cerevisiae, G in humans); through random selection, we have identified a G mutation in the yeast tRNA as having reduced function [64]. It is unlikely that this mutation affects aminoacylation given the lack of involvement of the anticodon stem-loop in the reaction. There are also differences in the T-arms of yeast and human tRNAs, which may influence interactions of the tRNA with the elongation factor EF-1α.
3.5. Applications for Mistranslation
tRNAs that mistranslate have roles in synthetic biology and potentially therapeutically. The latter arises because most diseases are the result of missense mutations that could be corrected through mistranslation. The synthetic biology applications include expanding the diversity of expressed products (statistical proteins) and controlling expression while transferring DNA between species. All these cases require tRNAs that mistranslate at a level below a toxic threshold. Our results indicate that a simple approach to achieve this is to use the inherent differences in function seen in the tRNAs from different species.
Translation requires multiple independent yet connected processes. Overall, these are highly conserved across species. It is, however, the differences that allow the enzymes involved in translation to be the targets of antibiotic and antimicrobial agents. The subtle differences seen for SerRS and the species specificity of its interactions with tRNASer suggest its potential as a therapeutic target.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/9/12/612/s1. Figure S1: Human tRNASer sequence, Figure S2: Expression of human SerRS in yeast, Figure S3: Gene sequence of S. cerevisiae tRNASer isoacceptors for cytoplasmic translation, Figure S4: Human tRNASerUGG with G4:C69 is toxic in S. cerevisiae, Figure S5: Alignment of cytoplasmic SerRS motif 2 sequences from yeast and fungal species, Figure S6: Percent distribution of the base pair at 3:70 in the tRNAs in Saccharomyces cerevisiae, Homo sapiens, Candida albicans, Thermus thermophilus and Escherichia coli, Figure S7: Motif 2 sequences from the yeast class II aaRS enzymes Hts1, Figure S8: Co-evolution of cytoplasmic SerRS, Table S1: Yeast strains used in this study, Table S2: Oligonucleotides used in this study, File S1: SerRS Coevolution Sequences, File S2: SerRS Coevolution Mutual Information Output.
Author Contributions
M.D.B. and C.J.B. conceived and designed the experiments; M.D.B., J.G. and Y.Z. performed the experiments; M.D.B. and S.M. analyzed the data and created the figures; G.B.G. guided the co-evolution analysis; M.D.B. and C.J.B wrote the paper.
Funding
This work was supported from the Natural Sciences and Engineering Research Council of Canada [RGPIN-2015-04394 to C.J.B.] and generous donations from Graham Wright and James Robertson to M.D.B. M.D.B. holds an NSERC Alexander Graham Bell Canada Graduate Scholarship (CGS-D).
Acknowledgments
We thank Martin Duennwald for providing the HSE-eGFP plasmid and Patrick Lajoie for his help with the growth curve analyses.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hoffman, K.S.; O’Donoghue, P.; Brandl, C.J. Mistranslation: From adaptations to applications. Biochim. Biophys. Acta 2017. [Google Scholar] [CrossRef] [PubMed]
- Giegé, R.; Jühling, F.; Pütz, J.; Stadler, P.; Sauter, C.; Florentz, C. Structure of transfer RNAs: Similarity and variability. Wiley Interdiscip. Rev. RNA 2012, 3, 37–61. [Google Scholar] [CrossRef] [PubMed]
- Rich, A.; RajBhandary, U.L. Transfer RNA: Molecular structure, sequence, and properties. Annu. Rev. Biochem. 1976, 45, 805–860. [Google Scholar] [CrossRef] [PubMed]
- De Duve, C. Transfer RNAs: The second genetic code. Nature 1988, 333, 117–118. [Google Scholar] [CrossRef] [PubMed]
- Commans, S.; Lazard, M.; Delort, F.; Blanquet, S.; Plateau, P. tRNA anticodon recognition and specification within subclass IIb aminoacyl-tRNA synthetases. J. Mol. Biol. 1998, 278, 801–813. [Google Scholar] [CrossRef] [PubMed]
- Giegé, R.; Sissler, M.; Florentz, C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998, 26, 5017–5035. [Google Scholar] [CrossRef] [PubMed]
- Imura, N.; Weiss, G.B.; Chambers, R.W. Reconstitution of alanine acceptor activity from fragments of yeast tRNA-Ala II. Nature 1969, 222, 1147–1148. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.M.; Schimmel, P. Evidence that a major determinant for the identity of a transfer RNA is conserved in evolution. Biochemistry 1989, 28, 6800–6804. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.M.; Schimmel, P. A Simple structural feature is a major determinant of the identity of a transfer RNA. Nature 1988, 333, 140–145. [Google Scholar] [CrossRef]
- Hoffman, K.S.; Berg, M.D.; Shilton, B.H.; Brandl, C.J.; O’Donoghue, P. Genetic Selection for mistranslation rescues a defective co-chaperone in yeast. Nucleic Acids Res. 2017, 45, 3407–3421. [Google Scholar] [CrossRef]
- Asahara, H.; Himeno, H.; Tamura, K.; Nameki, N.; Hasegawa, T.; Shimizu, M. Escherichia coli seryl-tRNA synthetase recognizes tRNASer by its characteristics tertiary structure. J. Mol. Biol. 1994, 738–748. [Google Scholar] [CrossRef]
- Biou, V.; Yaremchuk, A.; Tukalo, M.; Cusack, S. The 2.9 Å crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNASer. Science 1994, 263, 1404–1410. [Google Scholar] [CrossRef] [PubMed]
- Himeno, H.; Yoshida, S.; Soma, A.; Nishikawa, K. Only one nucleotide insertion to the long variable arm confers an efficient serine acceptor activity upon Saccharomyces cerevisiae tRNALeu in vitro. J. Mol. Biol. 1997, 268, 704–711. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.L.J.; Poruri, K.; Martinis, S.A. tRNA synthetase: tRNA aminoacylation and beyond. Wiley Interdiscip. Rev. RNA 2014, 5, 461–480. [Google Scholar] [CrossRef] [PubMed]
- Eriani, G.; Delarue, M.; Poch, O.; Gangloff, J.; Moras, D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 1990, 347, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Ibba, M.; Soll, D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000, 69, 617–650. [Google Scholar] [CrossRef] [PubMed]
- Eriani, G.; Dirheimer, G.; Gangloff, J. Cysteinyl-tRNA synthetase: Determination of the last E. Coli aminoacyl-tRNA synthetase primary structure. Nucleic Acids Res. 1991, 19, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Sugiura, I.; Nureki, O.; Ugaji-Yoshikawa, Y.; Kuwabara, S.; Shimada, A.; Tateno, M.; Lorber, B.; Giegé, R.; Moras, D.; Yokoyama, S.; et al. The 2.0 Å crystal structure of Thermus thermophilus methionyl-tRNA synthetase reveals two RNA-binding modules. Structure 2000, 8, 197–208. [Google Scholar] [CrossRef]
- Artymiuk, P.J.; Rice, D.W.; Poirrette, A.R.; Willet, P. A tale of two synthetases. Nat. Struct. Biol. 1994, 1, 758–760. [Google Scholar] [CrossRef] [PubMed]
- Cusack, S.; Berthet-Colominas, C.; Härtlein, M.; Nassar, N.; Leberman, R. A Second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature 1990, 347, 249–255. [Google Scholar] [CrossRef]
- Cusack, S.; Härtlein, M.; Leberman, R. Sequence, structural and evolutionary relationships between class 2 aminoacyl-tRNA synthetases. Nucleic Acids Res. 1991, 19, 3489–3498. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.F.; Hartman, H. The evolution of class II aminoacyl-tRNA synthetases and the first code. FEBS Lett. 2015, 589, 3499–3507. [Google Scholar] [CrossRef] [PubMed]
- Ruff, M.; Krishnaswamy, S.; Boeglin, M.; Poterszman, A.; Mitschler, A.; Podjarny, A.; Rees, B.; Thierry, J.C.; Moras, D. Class II aminoacyl transfer RNA synthetases: Crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNAAsp. Science 1991, 252, 1682–1689. [Google Scholar] [CrossRef] [PubMed]
- Cavarelli, J.; Moras, D. Recognition of tRNAs by aminoacyl-tRNA synthetases. FASEB J. 1993, 7, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Eichert, A.; Oberthuer, D.; Betzel, C.; Geßner, R.; Erdmann, V.A.; Fürste, J.P.; Förster, C. The seryl-tRNA synthetase/tRNA ser acceptor stem interface is mediated via a specific network of water molecules. Biochem. Biophys. Res. Commun. 2011, 412, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Bilokapic, S.; Maier, T.; Ahel, D.; Gruic-Sovulj, I.; Söll, D.; Weygand-Durasevic, I.; Ban, N. Structure of the unusual seryl-tRNA synthetase reveals a distinct zinc-dependent mode of substrate recognition. EMBO J. 2006, 25, 2498–2509. [Google Scholar] [CrossRef] [PubMed]
- Vincent, C.; Borel, F.; Willison, J.C.; Laberman, R.; Härtlein, M. Seryl-tRNA synthetase from Escherichia coli: Functional evidence for cross-dimer tRNA binding during aminoacylation. Nucleic Acids Res. 1995, 23, 1113–1118. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Guo, Y.; Tian, Q.; Jia, Q.; Gao, Y.; Zhang, Q.; Zhou, C.; Xie, W. SerRS-tRNASec complex Structures reveal mechanism of the first step in selenocysteine biosynthesis. Nucleic Acids Res. 2015, 43, 10534–10545. [Google Scholar] [CrossRef]
- Normanly, J.; Ogden, R.C.; Horvath, S.J.; Abelson, J. Changing the identity of a transfer RNA. Nature 1986, 321, 213–219. [Google Scholar] [CrossRef]
- Normanly, J.; Ollick, T.; Abelson, J. Eight base changes are sufficient to convert a leucine-inserting tRNA into a serine-inserting tRNA. Proc. Natl. Acad. Sci. USA 1992, 89, 5680–5684. [Google Scholar] [CrossRef]
- Shiba, K.; Stello, T.; Motegi, H.; Noda, T.; Musier-Forsyth, K.; Schimmel, P. Human lysyl-tRNA synthetase accepts nucleotide 73 variants and rescues Escherichia coli double-defective mutant. J. Biol. Chem. 1997, 272, 22809–22816. [Google Scholar] [CrossRef] [PubMed]
- Ripmaster, T.L.; Shiba, K.; Schimmel, P. Wide cross-species aminoacyl-tRNA synthetase replacement in vivo: Yeast cytoplasmic alanine enzyme replaced by human polymyositis serum antigen. Proc. Natl. Acad. Sci. USA 1995, 92, 4932–4936. [Google Scholar] [CrossRef] [PubMed]
- Rips, J.; Meyer-Schuman, R.; Breuer, O.; Tsabari, R.; Shaag, A.; Revel-Vilk, S.; Reif, S.; Elpeleg, O.; Antonellis, A.; Harel, T. MARS variant associated with both recessive interstitial lung and liver disease and dominant Charcot-Marie-Tooth disease. Eur. J. Med. Genet. 2018. [Google Scholar] [CrossRef] [PubMed]
- Burke, B.; Yang, F.; Chen, F.; Stehlin, C.; Chan, B.; Musier-Forsyth, K. Evolutionary coadaptation of the motif 2−acceptor stem interaction in the class II prolyl-tRNA synthetase system. Biochemistry 2000, 39, 15540–15547. [Google Scholar] [CrossRef] [PubMed]
- Edwards, H.; Trézéguet, V.; Schimmel, P. An Escherichia coli tyrosine transfer RNA is a leucine-specific transfer RNA in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1991, 88, 1153–1156. [Google Scholar] [CrossRef] [PubMed]
- Davis, L.; Chin, J.W. Designer proteins: Applications of genetic code expansion in cell biology. Nat. Rev. Mol. Cell Biol. 2012, 13, 168–182. [Google Scholar] [CrossRef] [PubMed]
- Winzeler, E.A.; Davis, R.W. Functional analysis of the yeast genome. Curr. Opin. Genet. Dev. 1997, 7, 771–776. [Google Scholar] [CrossRef]
- Tong, A.H.; Evangelista, M.; Parsons, A.B.; Xu, H.; Bader, G.D.; Pagé, N.; Robinson, M.; Raghibizadeh, S.; Hogue, C.W.; Bussey, H.; et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 2001, 294, 2364–2368. [Google Scholar] [CrossRef]
- Berg, M.D.; Hoffman, K.S.; Genereaux, J.; Mian, S.; Trussler, R.S.; Haniford, D.B.; O’Donoghue, P.; Brandl, C.J. Evolving mistranslating tRNAs through a phenotypically ambivalent intermediate in Saccharomyces cerevisiae. Genetics 2017, 206, 1865–1879. [Google Scholar] [CrossRef]
- Hoffman, K.S.; Duennwald, M.L.; Karagiannis, J.; Genereaux, J.; McCarton, A.S.; Brandl, C.J. Saccharomyces cerevisiae Tti2 regulates PIKK proteins and stress response. G3 2016, 6, 1649–1659. [Google Scholar] [CrossRef]
- Brandman, O.; Stewart-Ornstein, J.; Wong, D.; Larson, A.; Williams, C.C.; Li, G.-W.; Zhou, S.; King, D.; Shen, P.S.; Weibezahn, J.; et al. A Ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 2012, 151, 1042–1054. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.; Lang, V.; Cook, R.; Brandl, C.J. Identification of native complexes containing the yeast coactivator/repressor proteins NGG1/ADA3 and ADA2. J. Biol. Chem. 1997, 272, 5571–5578. [Google Scholar] [CrossRef] [PubMed]
- Dickson, R.J.; Gloor, G.B. Bioinformatics Identification of Coevolving Residues. In Homing Endonucleases: Methods and Protocols; Edgell, D.R., Ed.; Humana Press: Totowa, NJ, USA, 2014; pp. 223–243. [Google Scholar]
- Rocha, R.; Pereira, P.J.B.; Santos, M.A.S.; Macedo-Ribeiro, S. Unveiling the structural basis for translational ambiguity tolerance in a human fungal pathogen. Proc. Natl. Acad. Sci. USA 2011, 108, 14091–14096. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y.; Sekine, S. ichi; Kuroishi, C.; Terada, T.; Shirouzu, M.; Kuramitsu, S.; Yokoyama, S. Crystallographic and mutational studies of seryl-tRNA synthetase from the archaeon Pyrococcus horikoshii. RNA Biol. 2008, 5, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Shi, Y.; Yang, X.-L. Crystal structure of human seryl-tRNA synthetase and ser-sa complex reveals a molecular lever specific to higher eukaryotes. Structure 2013, 21, 2078–2086. [Google Scholar] [CrossRef]
- Belrhali, H.; Yaremchuk, A.; Tukalo, M.; Larsen, K.; Berthet-Colominas, C.; Leberman, R.; Beijer, B.; Sproat, B.; Als-Nielsen, J.; Grübel, G. Crystal structures at 2.5 angstrom resolution of seryl-tRNA synthetase complexed with two analogs of seryl adenylate. Science 1994, 263, 1432–1436. [Google Scholar] [CrossRef]
- Wang, Y.; Geer, L.Y.; Chappey, C.; Kans, J.A.; Bryant, S.H. Cn3D: Sequence and Structure views for entrez. Trends Biochem. Sci. 2000, 25, 300–302. [Google Scholar] [CrossRef]
- Dickson, R.J.; Gloor, G.B. The MIp toolset: An efficient algorithm for calculating mutual information in protein alignments. arXiv, 2013; arXiv:1304.4573. [Google Scholar]
- Thompson, J.; Dahlberg, A.E. Testing the conservation of the translational machinery over evolution in diverse environments: Assaying Thermus thermophilus ribosomes and initiation factors in a coupled transcription—translation system from Escherichia coli. Nucleic Acids Res. 2004, 32, 5954–5961. [Google Scholar] [CrossRef] [PubMed]
- Ganoza, M.C.; Kiel, M.C.; Aoki, H. Evolutionary conservation of reactions in translation. Microbiol. Mol. Biol. Rev. 2002, 66, 460–485. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. GtRNAdb 2.0: An Expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016, 44, D184–D189. [Google Scholar] [CrossRef] [PubMed]
- Dewe, J.M.; Whipple, J.M.; Chernyakov, I.; Jaramillo, L.N.; Phizicky, E.M. The yeast rapid tRNA decay pathway competes with elongation factor 1A for substrate tRNAs and acts on tRNAs lacking one or more of several modifications. RNA 2012, 18, 1886–1896. [Google Scholar] [CrossRef]
- Chernyakov, I.; Whipple, J.M.; Kotelawala, L.; Grayhack, E.J.; Phizicky, E.M. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5’-3’ exonucleases Rat1 and Xrn1. Genes Dev. 2008, 22, 1369–1380. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Cavarelli, J.; Rees, B.; Ruff, M.; Thierry, J.C.; Moras, D. Yeast tRNAAsp recognition by its cognate class II aminoacyl-tRNA synthetase. Nature 1993, 362, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Goldgur, Y.; Mosyak, L.; Reshetnikova, L.; Ankilova, V.; Lavrik, O.; Khodyreva, S.; Safro, M. The Crystal structure of phenylalanyl-tRNA synthetase from Thermus thermophilus complexed with cognate tRNA (Phe). Structure 1997, 5, 59–68. [Google Scholar] [CrossRef]
- Eiler, S.; Dock-Bregeon, A.-C.; Moulinier, L.; Thierry, J.C.; Moras, D. Synthesis of aspartyl-tRNAAsp in Escherichia coli--A snapshot of the second step. EMBO J. 1999, 18, 6532–6541. [Google Scholar] [CrossRef] [PubMed]
- Achsel, T.; Gross, H.J. Identity determinants of human tRNASer: Sequence elements necessary for serylation and maturation of a tRNA with a long extra arm. EMBO J. 1993, 12, 3333–3338. [Google Scholar] [CrossRef]
- Lenhard, B.; Filipic, S.; Landeka, I.; Ivan, S.; So, D. Defining the active site of yeast seryl-tRNA synthetase. J. Biol. Chem. 1997, 272, 1136–1141. [Google Scholar] [CrossRef]
- Luscombe, N.M.; Laskowski, R.A.; Thornton, J.M. Amino acid—base interactions: A three-dimensional analysis of protein—DNA interactions at an atomic level. Nucleic Acids Res. 2001, 29, 2860–2874. [Google Scholar] [CrossRef]
- Davies, D.R.; Goryshin, I.Y.; Reznikoff, W.S.; Rayment, I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science 2000, 289, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.J.; Cheng, X. Base flipping. Annu. Rev. Biochem. 1998, 67, 181–198. [Google Scholar] [CrossRef] [PubMed]
- Berg, M.D. Western University, London, Canada. in preparation.
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).