Valproic Acid Inhibits Proliferation and Reduces Invasiveness in Glioma Stem Cells Through Wnt/β Catenin Signalling Activation
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture Conditions
2.2. Drug and Treatments
2.3. Bioinformatics Analysis by Ingenuity Pathway Analysis Software
2.4. RNA Extraction and cDNA Synthesis
2.5. Real Time-PCR Array
2.6. Real Time PCR
2.7. Luciferase Reporter Assay
2.8. MTT Assay
2.9. Trypan Blue Dye Exclusion Assay
2.10. Boyden Chamber Assay
2.11. Protein Extracts and Western Blotting
2.12. Immunofluorescence
2.13. Statistical Analysis
3. Results
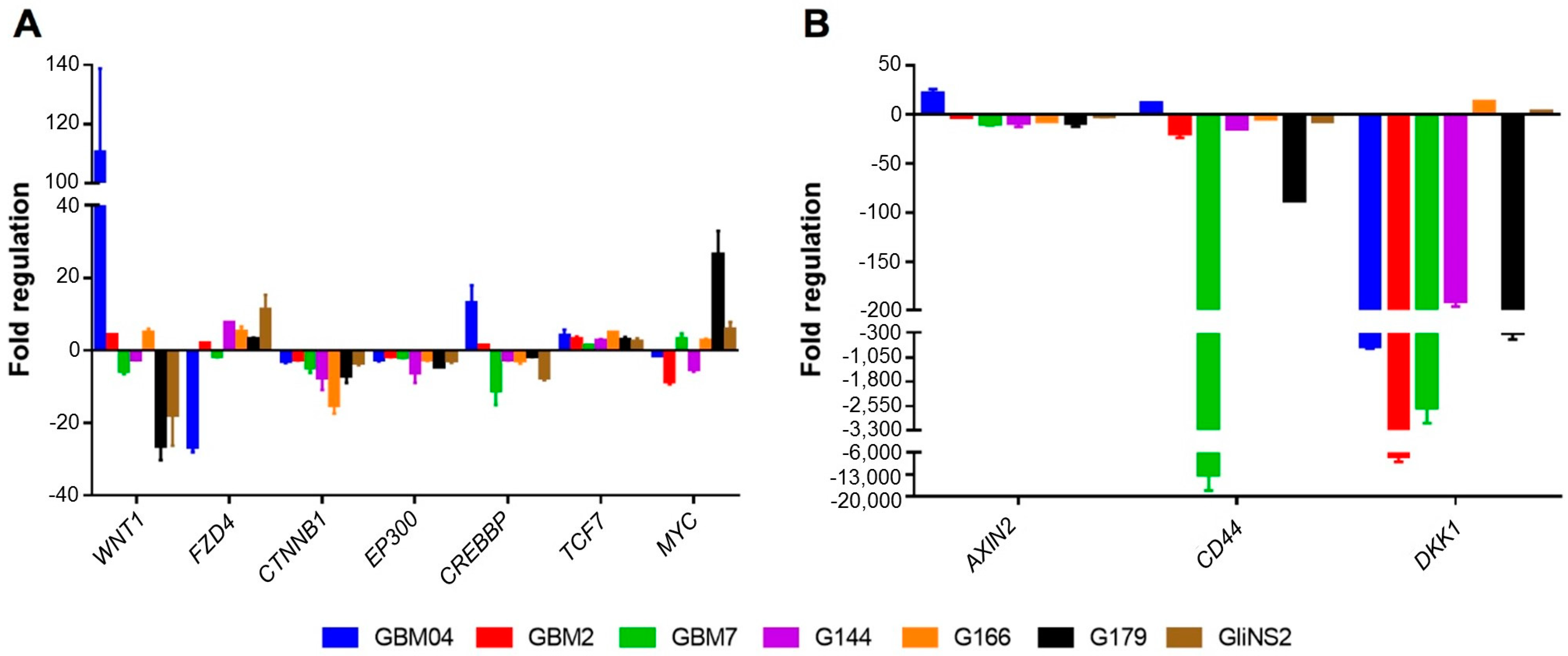
3.1. Valproic Acid Induced DNA Methylation Changes in Wnt Pathway-Related Genes
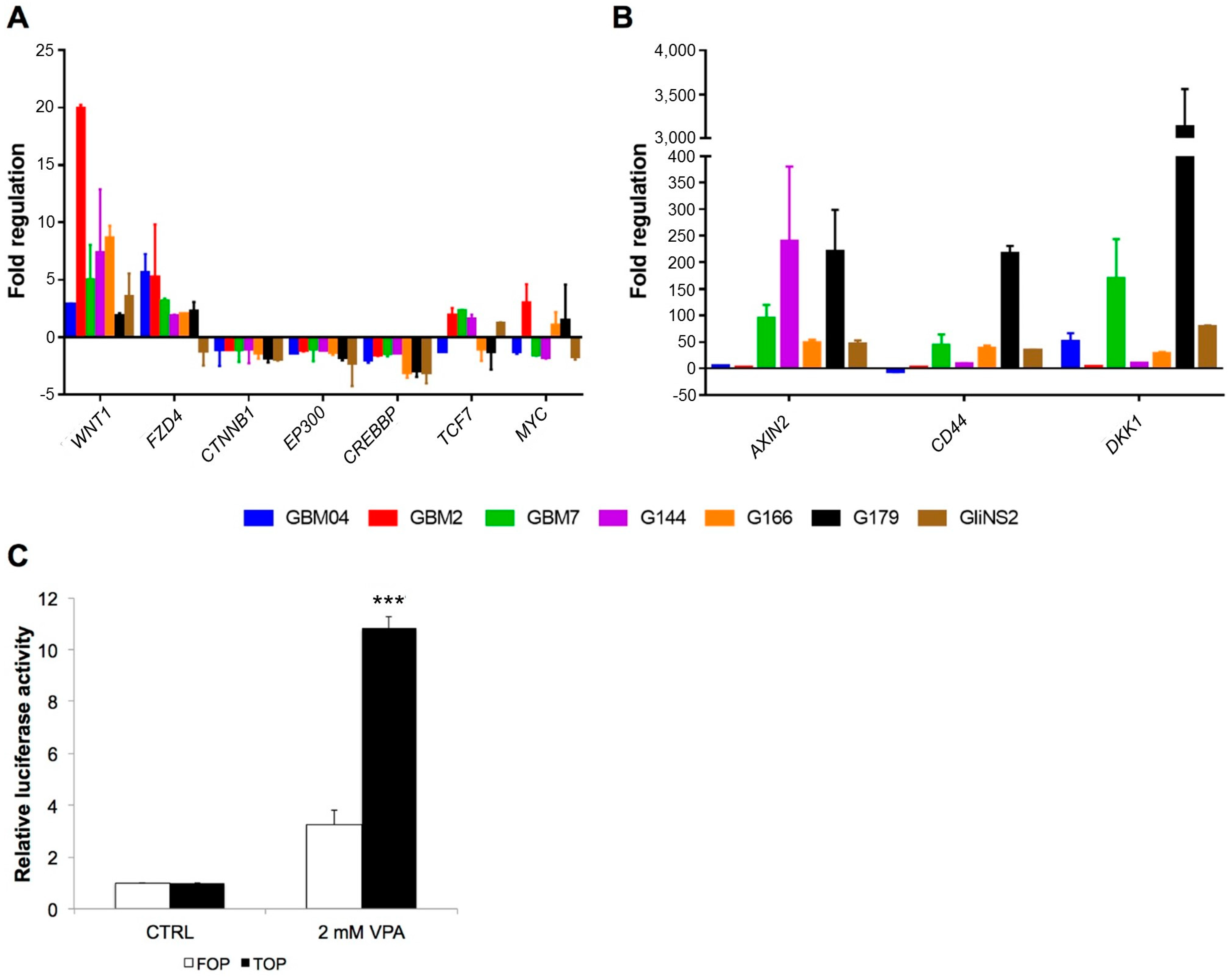
3.2. Valproic Acid Activated the Wnt Signalling Pathway in GSCs
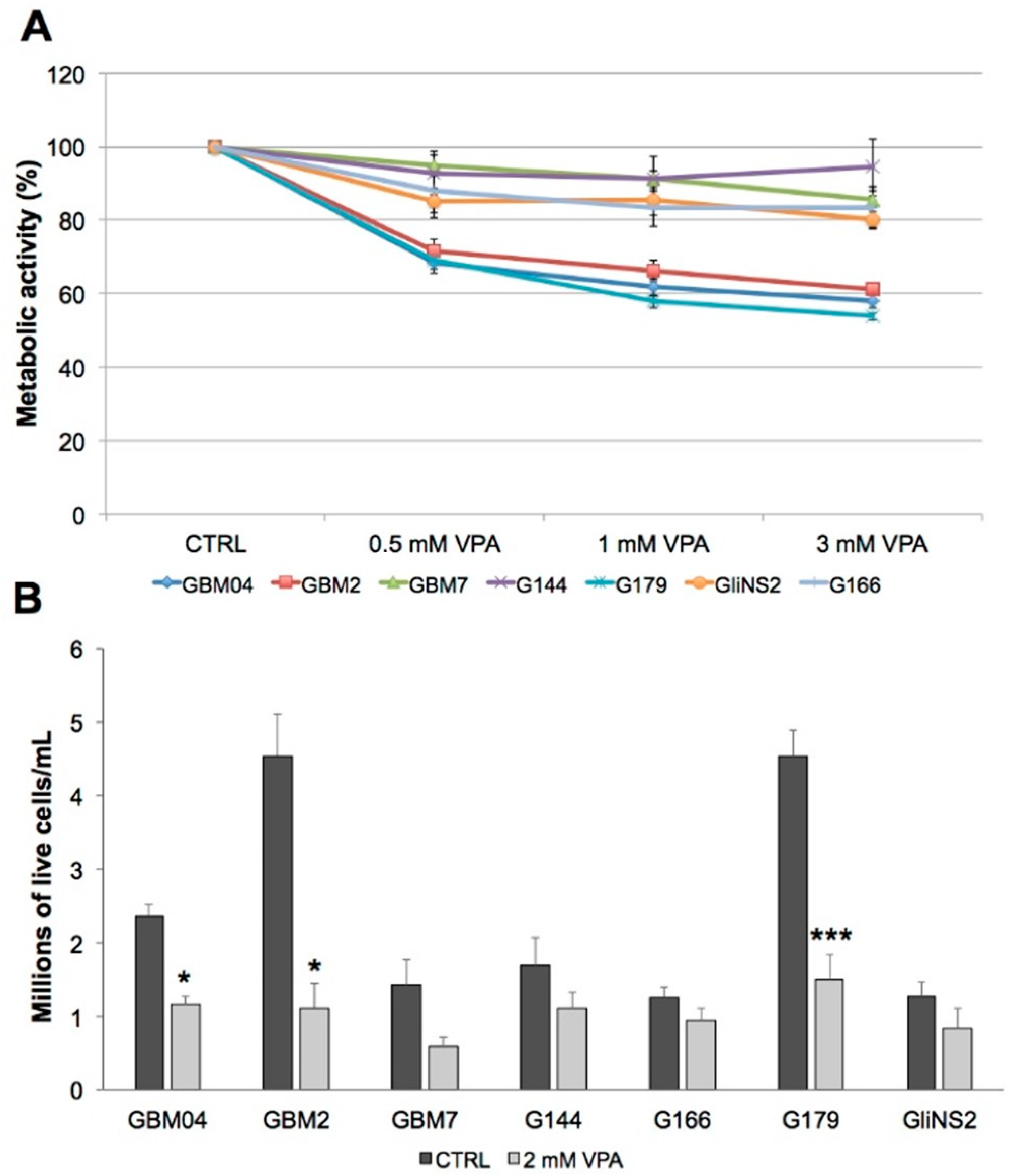
3.3. Valproic Acid Reduced Glioma Stem Cells Proliferation
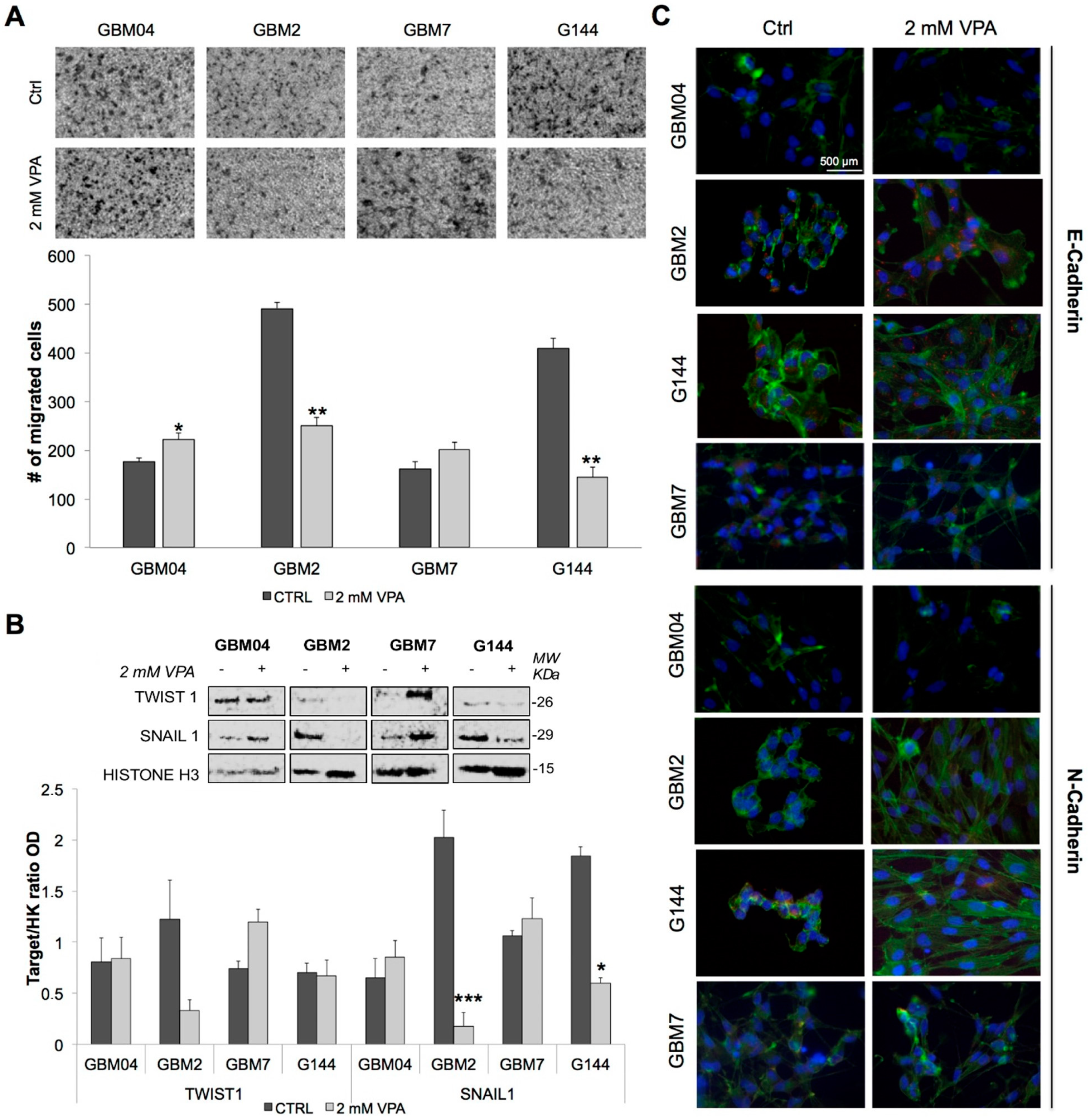
3.4. Valproic Acid Impaired Glioma Stem Cells Invasive Behaviour
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumours of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.; Hsieh, I.Y.; Huang, X.; Li, J.; Zhao, W. Glioblastoma Stem-Like Cells: Characteristics, Microenvironment, and Therapy. Front. Pharmacol. 2016, 7, 477. [Google Scholar] [CrossRef] [PubMed]
- Dirks, P.B. Brain tumour stem cells: Bringing order to the chaos of brain cancer. J. Clin. Oncol. 2008, 26, 2916–2924. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of human brain tumour initiating cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Ignatova, T.N.; Kukekov, V.G.; Laywell, E.D.; Suslov, O.N.; Vrionis, F.D.; Steindler, D.A. Human cortical glial tumours contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia 2002, 39, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef] [PubMed]
- Riva, G.; Butta, V.; Cilibrasi, C.; Baronchelli, S.; Redaelli, S.; Dalprà, L.; Lavitrano, M.; Bentivegna, A. Epigenetic targeting of glioma stem cells: Short-term and long-term treatments with valproic acid modulate DNA methylation and differentiation behavior, but not temozolomide sensitivity. Oncol. Rep. 2016. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Ryu, H.W.; Won, H.R.; Kwon, S.H. Advances in epigenetic glioblastoma therapy. Oncotarget 2017, 8, 18577–18589. [Google Scholar] [CrossRef] [PubMed]
- Barker, C.A.; Bishop, A.J.; Chang, M.; Beal, K.; Chan, T.A. Valproic acid use during radiation therapy for glioblastoma associated with improved survival. Int. J. Radiat. Oncol. Biol. Phys. 2013, 86, 504–509. [Google Scholar] [CrossRef] [PubMed]
- Rudà, R.; Pellerino, A.; Soffietti, R. Does valproic acid affect tumour growth and improve survival in glioblastomas? CNS Oncol. 2016, 5, 51–53. [Google Scholar] [CrossRef] [PubMed]
- Blaheta, R.A.; Cinatl, J. Anti-tumour mechanisms of valproate: A novel role for an old drug. Med. Res. Rev. 2002, 22, 492–511. [Google Scholar] [CrossRef] [PubMed]
- Osuka, S.; Takano, S.; Watanabe, S.; Ishikawa, E.; Yamamoto, T.; Matsumura, A. Valproic acid inhibits angiogenesis in vitro and glioma angiogenesis in vivo in the brain. Neurol. Med. Chir. (Tokyo) 2012, 52, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tsai, Y.H.; Tseng, S.H. Valproic acid affected the survival and invasiveness of human glioma cells through diverse mechanisms. J. Neurooncol. 2012, 109, 23–33. [Google Scholar] [CrossRef] [PubMed]
- van Amerongen, R.; Nusse, R. Towards an integrated view of Wnt signaling in development. Development 2009, 136, 3205–3214. [Google Scholar] [CrossRef] [PubMed]
- Nusse, R.; Fuerer, C.; Ching, W.; Harnish, K.; Logan, C.; Zeng, A.; ten Berge, D.; Kalani, Y. Wnt signaling and stem cell control. Cold Spring Harb. Symp. Quant. Biol. 2008, 73, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Duchartre, Y.; Kim, Y.M.; Kahn, M. The Wnt signaling pathway in cancer. Crit. Rev. Oncol. Hematol. 2016, 99, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2017, 36, 1461–1473. [Google Scholar] [CrossRef] [PubMed]
- Miyaki, M.; Iijima, T.; Kimura, J.; Yasuno, M.; Mori, T.; Hayashi, Y.; Koike, M.; Shitara, N.; Iwama, T.; Kuroki, T. Frequent mutation of β-catenin and APC genes in primary colorectal tumours from patients with hereditary nonpolyposis colorectal cancer. Cancer Res. 1999, 59, 4506–4509. [Google Scholar] [PubMed]
- Zeng, G.; Germinaro, M.; Micsenyi, A.; Monga, N.K.; Bell, A.; Sood, A.; Malhotra, V.; Sood, N.; Midda, V.; Monga, D.K.; et al. Aberrant Wnt/β-catenin signaling in pancreatic adenocarcinoma. Neoplasia 2006, 8, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Luis, T.C.; Ichii, M.; Brugman, M.H.; Kincade, P.; Staal, F.J. Wnt signaling strength regulates normal hematopoiesis and its deregulation is involved in leukemia development. Leukemia 2012, 26, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.Y.; Xia, W.; Wang, J.C.; Kwong, K.Y.; Spohn, B.; Wen, Y.; Pestell, R.G.; Hung, M.C. β-catenin, a novel prognostic marker for breast cancer: Its roles in cyclin D1 expression and cancer progression. Proc. Natl. Acad. Sci. USA 2000, 97, 4262–4266. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhang, J.; Han, L.; Pu, P.; Kang, C. Wnt/β-catenin signaling in glioma. J. Neuroimmune Pharmacol. 2012, 7, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Gong, A.; Huang, S. FoxM1 and Wnt/β-catenin signaling in glioma stem cells. Cancer Res. 2012, 72, 5658–5662. [Google Scholar] [CrossRef] [PubMed]
- Kahlert, U.D.; Maciaczyk, D.; Doostkam, S.; Orr, B.A.; Simons, B.; Bogiel, T.; Reithmeier, T.; Prinz, M.; Schubert, J.; Niedermann, G.; et al. Activation of canonical WNT/β-catenin signaling enhances in vitro motility of glioblastoma cells by activation of ZEB1 and other activators of epithelial-to-mesenchymal transition. Cancer Lett. 2012, 325, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Kierulf-Vieira, K.S.; Sandberg, C.J.; Grieg, Z.; Günther, C.C.; Langmoen, I.A.; Vik-Mo, E.O. Wnt inhibition is dysregulated in gliomas and its re-establishment inhibits proliferation and tumour sphere formation. Exp. Cell Res. 2016, 340, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Anastas, J.N.; Moon, R.T. WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer 2013, 13, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Kahn, M. Can we safely target the WNT pathway? Nat. Rev. Drug Discov. 2014, 13, 513–532. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, J.M.; Rank, K.B.; Zeng, Y.; Capen, A.; Yadav, V.; Manro, J.R.; Engler, T.A.; Chedid, M. Activating the Wnt/β-Catenin Pathway for the Treatment of Melanoma—Application of LY2090314, a Novel Selective Inhibitor of Glycogen Synthase Kinase-3. PLoS ONE 2015, 10, e0125028. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; DeRan, M.T.; Ignatius, M.S.; Grandinetti, K.B.; Clagg, R.; McCarthy, K.M.; Lobbardi, R.M.; Brockmann, J.; Keller, C.; Wu, X.; et al. Glycogen synthase kinase 3 inhibitors induce the canonical WNT/β-catenin pathway to suppress growth and self-renewal in embryonal rhabdomyosarcoma. Proc. Natl. Acad. Sci. USA 2014, 111, 5349–5354. [Google Scholar] [CrossRef] [PubMed]
- Duffy, D.J.; Krstic, A.; Schwarzl, T.; Halasz, M.; Iljin, K.; Fey, D.; Haley, B.; Whilde, J.; Haapa-Paananen, S.; Fey, V.; et al. Wnt signalling is a bi-directional vulnerability of cancer cells. Oncotarget 2016, 7, 60310–60331. [Google Scholar] [CrossRef] [PubMed]
- Shao, N.; Zou, J.; Li, J.; Chen, F.; Dai, J.; Qu, X.; Sun, X.; Ma, D.; Ji, C. Hyper-activation of WNT/β-catenin signaling pathway mediates anti-tumour effects of histone deacetylase inhibitors in acute T lymphoblastic leukemia. Leuk Lymphoma 2012, 53, 1769–1778. [Google Scholar] [CrossRef] [PubMed]
- Bordonaro, M.; Lazarova, D.L.; Sartorelli, A.C. The activation of β-catenin by Wnt signaling mediates the effects of histone deacetylase inhibitors. Exp. Cell Res. 2007, 313, 1652–1666. [Google Scholar] [CrossRef] [PubMed]
- Lazarova, D.L.; Bordonaro, M.; Carbone, R.; Sartorelli, A.C. Linear relationship between Wnt activity levels and apoptosis in colorectal carcinoma cells exposed to butyrate. Int. J. Cancer 2004, 110, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Pollard, S.M.; Yoshikawa, K.; Clarke, I.D.; Danovi, D.; Stricker, S.; Russell, R.; Bayani, J.; Head, R.; Lee, M.; Bernstein, M.; et al. Glioma stem cell lines expanded in adherent culture have tumour-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell 2009, 4, 568–580. [Google Scholar] [CrossRef] [PubMed]
- Griffero, F.; Daga, A.; Marubbi, D.; Capra, M.C.; Melotti, A.; Pattarozzi, A.; Gatti, M.; Bajetto, A.; Porcile, C.; Barbieri, F.; et al. Different response of human glioma tumour-initiating cells to epidermal growth factor receptor kinase inhibitors. J. Biol. Chem. 2009, 284, 7138–7148. [Google Scholar] [CrossRef] [PubMed]
- Baronchelli, S.; Bentivegna, A.; Redaelli, S.; Riva, G.; Butta, V.; Paoletta, L.; Isimbaldi, G.; Miozzo, M.; Tabano, S.; Daga, A.; et al. Delineating the cytogenomic and epigenomic landscapes of glioma stem cell lines. PLoS ONE 2013, 8, e57462. [Google Scholar] [CrossRef] [PubMed]
- Cilibrasi, C.; Riva, G.; Romano, G.; Cadamuro, M.; Bazzoni, R.; Butta, V.; Paoletta, L.; Dalprà, L.; Strazzabosco, M.; Lavitrano, M.; et al. Resveratrol Impairs Glioma Stem Cells Proliferation and Motility by Modulating the Wnt Signaling Pathway. PLoS ONE 2017, 12, e0169854. [Google Scholar] [CrossRef] [PubMed]
- Cadamuro, M.; Spagnuolo, G.; Sambado, L.; Indraccolo, S.; Nardo, G.; Rosato, A.; Brivio, S.; Caslini, C.; Stecca, T.; Massani, M.; et al. Low-Dose Paclitaxel Reduces S100A4 Nuclear Import to Inhibit Invasion and Hematogenous Metastasis of Cholangiocarcinoma. Cancer Res. 2016, 76, 4775–4784. [Google Scholar] [CrossRef] [PubMed]
- Riva, G.; Baronchelli, S.; Paoletta, L.; Butta, V.; Biunno, I.; Lavitrano, M.; Dalprà, L.; Bentivegna, A. In vitro anticancer drug test: A new method emerges from the model of glioma stem cells. Toxicol. Rep. 2014, 1, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Jho, E.H.; Zhang, T.; Domon, C.; Joo, C.K.; Freund, J.N.; Costantini, F. Wnt/β-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 2002, 22, 1172–1183. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.C.; Lin, H.; Huang, M.J.; Chow, J.M.; Lin, S.; Liu, H.E. Downregulation of c-Myc is critical for valproic acid-induced growth arrest and myeloid differentiation of acute myeloid leukemia. Leuk Res. 2007, 31, 1403–1411. [Google Scholar] [CrossRef] [PubMed]
- Iwadate, Y. Epithelial-mesenchymal transition in glioblastoma progression. Oncol. Lett. 2016, 11, 1615–1620. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [PubMed]
- Nowicki, M.O.; Dmitrieva, N.; Stein, A.M.; Cutter, J.L.; Godlewski, J.; Saeki, Y.; Nita, M.; Berens, M.E.; Sander, L.M.; Newton, H.B.; et al. Lithium inhibits invasion of glioma cells; possible involvement of glycogen synthase kinase-3. Neuro Oncol. 2008, 10, 690–699. [Google Scholar] [CrossRef] [PubMed]
- Kotliarova, S.; Pastorino, S.; Kovell, L.C.; Kotliarov, Y.; Song, H.; Zhang, W.; Bailey, R.; Maric, D.; Zenklusen, J.C.; Lee, J.; et al. Glycogen synthase kinase-3 inhibition induces glioma cell death through c-MYC, nuclear factor-kappaB, and glucose regulation. Cancer Res. 2008, 68, 6643–6651. [Google Scholar] [CrossRef] [PubMed]
- Korur, S.; Huber, R.M.; Sivasankaran, B.; Petrich, M.; Morin, P.; Hemmings, B.A.; Merlo, A.; Lino, M.M. GSK3β regulates differentiation and growth arrest in glioblastoma. PLoS ONE 2009, 4, e7443. [Google Scholar] [CrossRef] [PubMed]
- Rampazzo, E.; Persano, L.; Pistollato, F.; Moro, E.; Frasson, C.; Porazzi, P.; Della Puppa, A.; Bresolin, S.; Battilana, G.; Indraccolo, S.; et al. Wnt activation promotes neuronal differentiation of glioblastoma. Cell Death Dis. 2013, 4, e500. [Google Scholar] [CrossRef] [PubMed]
| Genes | GBM2 | G144 | Genes | GBM2 | G144 | Genes | GBM2 | G144 |
|---|---|---|---|---|---|---|---|---|
| AES | ↓ | ↓ | FRAT1 | = | = | SFRP1 | ↑ | ↓ |
| APC | = | = | FRZB | = | ↑ | SFRP4 | ↑ | ↑ |
| AXIN1 | ↓ | ↓ | FSHB | ↑ | ↑ | FBXW4 | ↓ | ↓ |
| BCL9 | = | = | FZD1 | ↑ | ↓ | SLC9A3R1 | ↓ | ↓ |
| BTRC | = | ↓ | FZD2 | = | ↓ | SOX17 | ↑ | ↑ |
| CCND1 | ↑ | ↓ | FZD3 | = | ↓ | T | ↑ | ↑ |
| CCND2 | ↑ | ↓ | FZD4 | ↑ | ↑ | TCF7 | ↑ | ↑ |
| CCND3 | ↑ | ↓ | FZD5 | = | = | TCF7L1 | = | ↑ |
| CSNK1A1 | = | ↓ | FZD6 | = | ↓ | TLE1 | ↑ | ↑ |
| CSNK1D | = | ↓ | FZD7 | = | ↓ | TLE2 | = | ↓ |
| CSNK1G1 | = | ↑ | FZD8 | ↑ | ↓ | WIF1 | ↑ | ↑ |
| CSNK2A1 | = | ↓ | GSK3A | ↓ | ↓ | WISP1 | = | ↑ |
| CTBP1 | = | ↓ | GSK3B | ↑ | = | WNT1 | ↑ | ↑ |
| CTBP2 | = | = | JUN | = | ↓ | WNT10A | ↑ | ↑ |
| CTNNB1 | = | = | KREMEN1 | = | ↓ | WNT11 | ↓ | ↑ |
| CTNNBIP1 | = | ↑ | LEF1 | = | ↓ | WNT16 | ↓ | ↑ |
| CXXC4 | ↑ | ↑ | LRP5 | ↓ | ↓ | WNT2 | ↑ | ↑ |
| DAAM1 | ↑ | = | LRP6 | = | ↓ | WNT2B | = | = |
| DIXDC1 | ↑ | ↑ | MYC | ↑ | ↓ | WNT3 | ↓ | ↓ |
| DKK1 | ↑ | ↑ | NKD1 | ↓ | ↑ | WNT3A | ↑ | ↑ |
| DVL1 | ↑ | = | NLK | ↑ | ↓ | WNT4 | ↑ | = |
| DVL2 | = | ↑ | PITX2 | = | ↓ | WNT5A | ↑ | ↓ |
| EP300 | = | = | PORCN | = | ↓ | WNT5B | ↑ | = |
| FBXW11 | = | = | PPP2CA | ↑ | ↓ | WNT6 | ↑ | ↑ |
| FBXW2 | ↑ | ↑ | PPP2R1A | = | ↓ | WNT7A | ↑ | ↑ |
| FGF4 | ↑ | ↑ | PYGO1 | = | ↑ | WNT7B | ↑ | ↓ |
| FOSL1 | ↑ | = | RHOU | ↑ | = | WNT8A | ↑ | ↑ |
| FOXN1 | = | ↑ | SENP2 | = | ↓ | WNT9A | ↓ | ↑ |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riva, G.; Cilibrasi, C.; Bazzoni, R.; Cadamuro, M.; Negroni, C.; Butta, V.; Strazzabosco, M.; Dalprà, L.; Lavitrano, M.; Bentivegna, A. Valproic Acid Inhibits Proliferation and Reduces Invasiveness in Glioma Stem Cells Through Wnt/β Catenin Signalling Activation. Genes 2018, 9, 522. https://doi.org/10.3390/genes9110522
Riva G, Cilibrasi C, Bazzoni R, Cadamuro M, Negroni C, Butta V, Strazzabosco M, Dalprà L, Lavitrano M, Bentivegna A. Valproic Acid Inhibits Proliferation and Reduces Invasiveness in Glioma Stem Cells Through Wnt/β Catenin Signalling Activation. Genes. 2018; 9(11):522. https://doi.org/10.3390/genes9110522
Chicago/Turabian StyleRiva, Gabriele, Chiara Cilibrasi, Riccardo Bazzoni, Massimiliano Cadamuro, Caterina Negroni, Valentina Butta, Mario Strazzabosco, Leda Dalprà, Marialuisa Lavitrano, and Angela Bentivegna. 2018. "Valproic Acid Inhibits Proliferation and Reduces Invasiveness in Glioma Stem Cells Through Wnt/β Catenin Signalling Activation" Genes 9, no. 11: 522. https://doi.org/10.3390/genes9110522
APA StyleRiva, G., Cilibrasi, C., Bazzoni, R., Cadamuro, M., Negroni, C., Butta, V., Strazzabosco, M., Dalprà, L., Lavitrano, M., & Bentivegna, A. (2018). Valproic Acid Inhibits Proliferation and Reduces Invasiveness in Glioma Stem Cells Through Wnt/β Catenin Signalling Activation. Genes, 9(11), 522. https://doi.org/10.3390/genes9110522






