Unfolded Protein Response Suppression in Yeast by Loss of tRNA Modifications
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids and Cultivation
2.2. Liquid Growth Inhibition Assays
2.3. Isolation of Total RNA
2.4. Quantification of mRNA Levels by qRT-PCR
2.5. RT-PCR of HAC1 mRNA
3. Results and Discussion
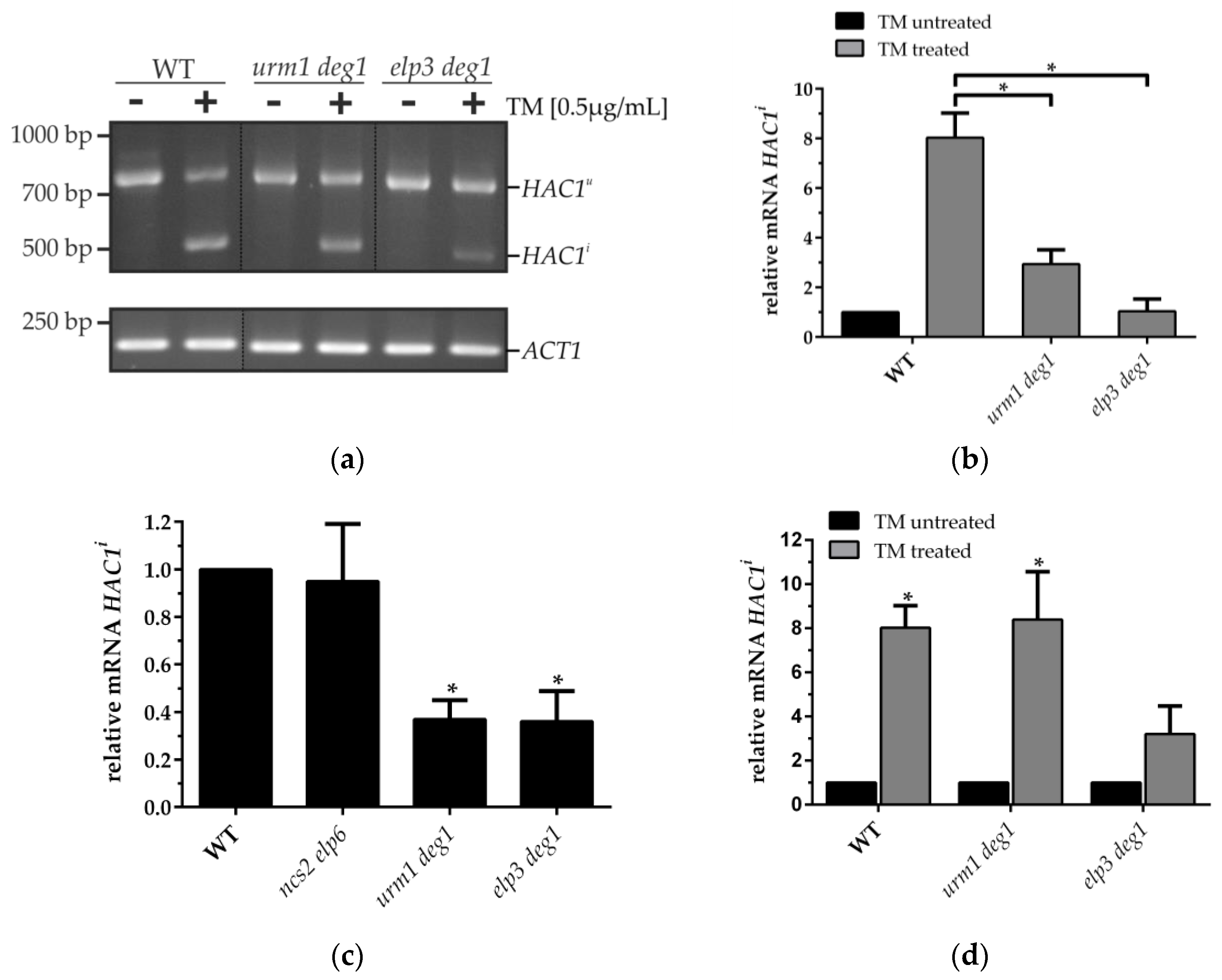
3.1. Dysfunction of the UPR System in tRNA Modification Mutants
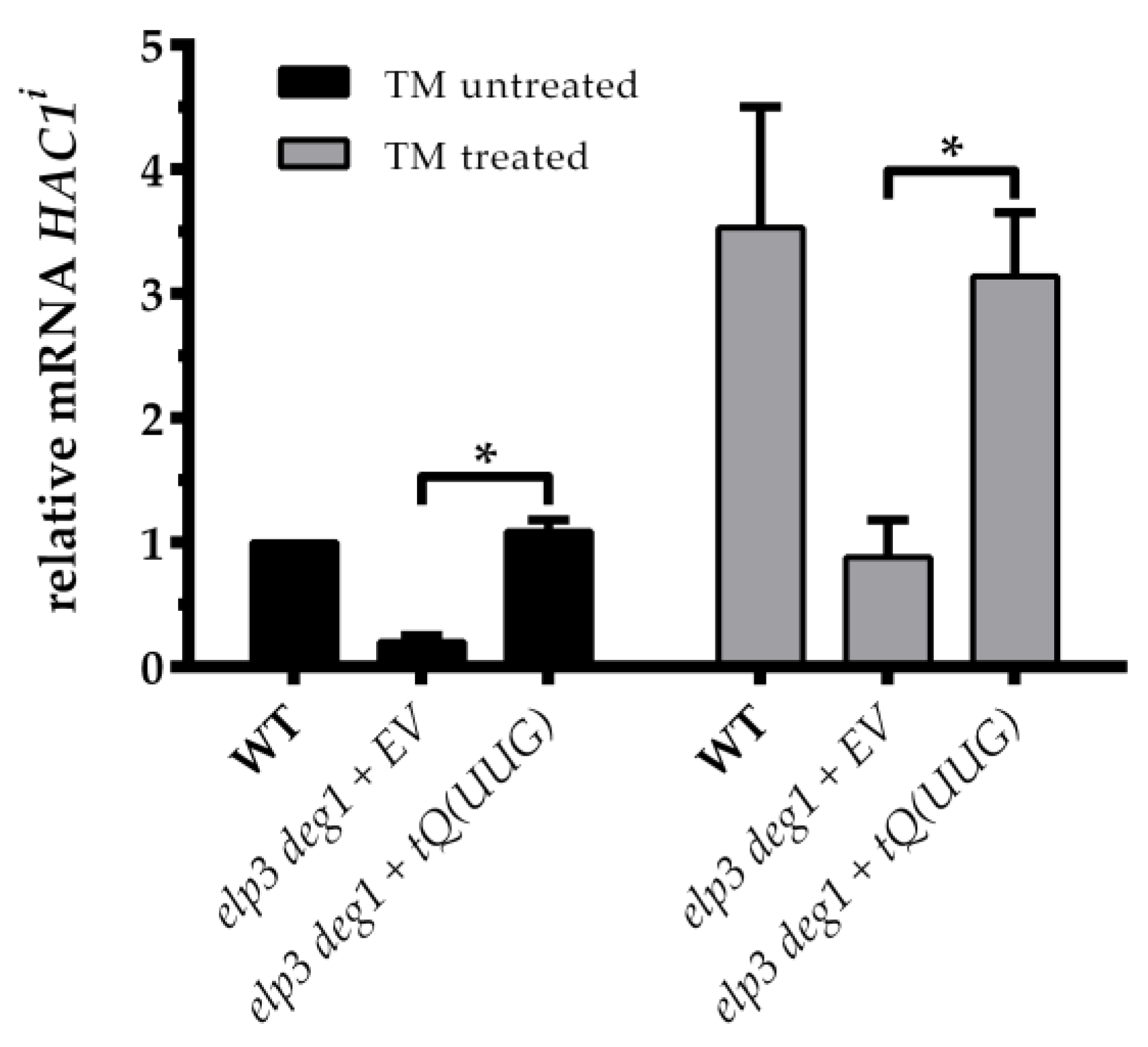
3.2. Rescue of UPR Suppression by tRNA Overexpression
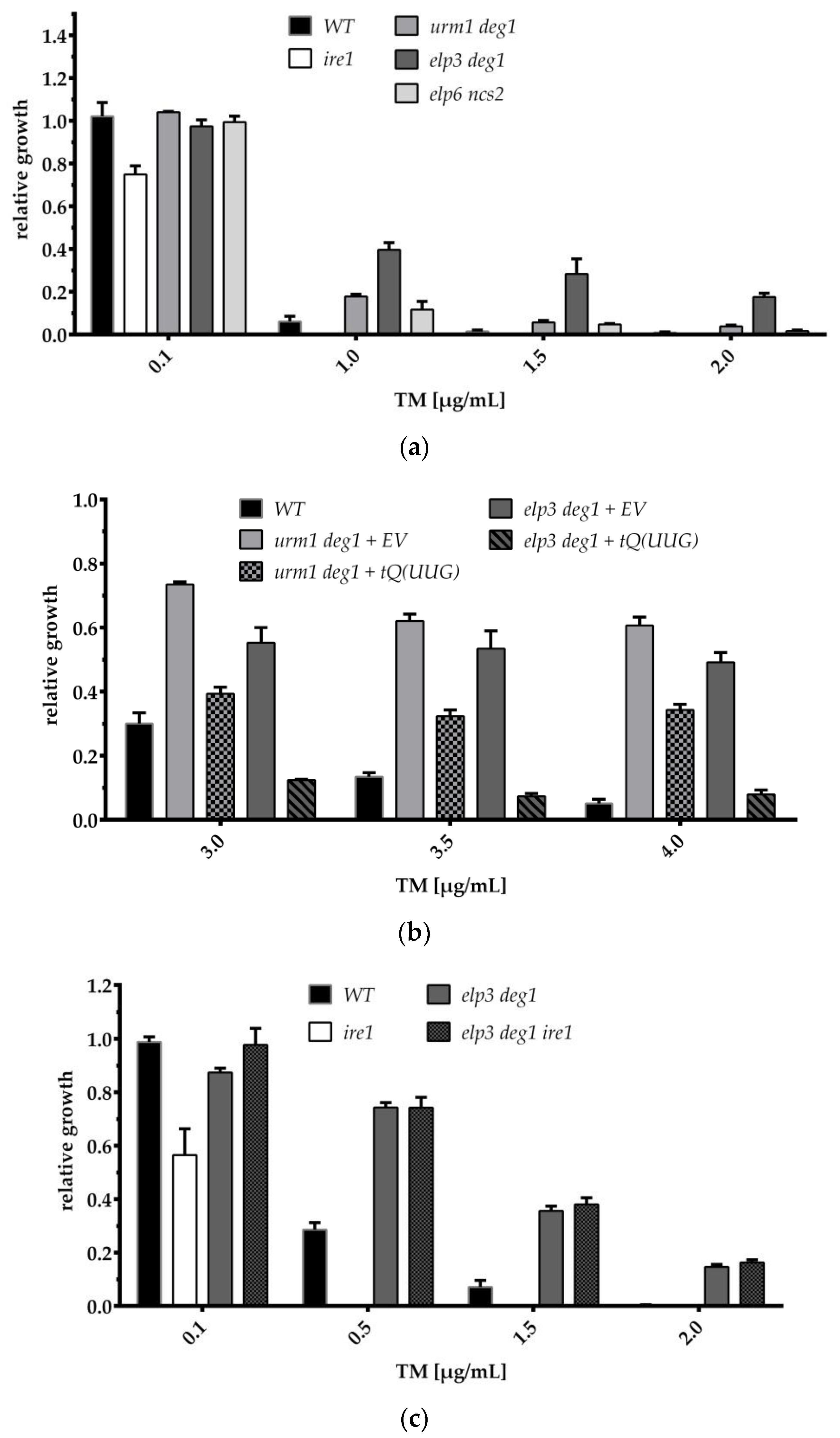
3.3. TM Phenotypes of tRNA Modification Mutants and Their Response to tRNAGln(UUG) Overexpression
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chou, H.-J.; Donnard, E.; Gustafsson, H.T.; Garber, M.; Rando, O.J. Transcriptome-wide analysis of roles for tRNA modifications in translational regulation. Mol. Cell 2017, 68, 978–992. [Google Scholar] [CrossRef] [PubMed]
- Nedialkova, D.D.; Leidel, S.A. Optimization of Codon Translation rates via tRNA modifications maintains proteome integrity. Cell 2015, 161, 1606–1618. [Google Scholar] [CrossRef] [PubMed]
- Esberg, A.; Huang, B.; Johansson, M.J.O.; Byström, A.S. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell 2006, 24, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Schaffrath, R.; Leidel, S.A. Wobble uridine modifications—A reason to live, a reason to die? RNA Biol. 2017, 14, 1209–1222. [Google Scholar] [CrossRef] [PubMed]
- Studte, P.; Zink, S.; Jablonowski, D.; Bär, C.; von der Haar, T.; Tuite, M.F.; Schaffrath, R. tRNA and protein methylase complexes mediate zymocin toxicity in yeast. Mol. Microbiol. 2008, 69, 1266–1277. [Google Scholar] [CrossRef] [PubMed]
- Kalhor, H.R.; Clarke, S. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol. Cell. Biol. 2003, 23, 9283–9292. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Lu, J.; Byström, A.S. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA 2008, 14, 2183–2194. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Johansson, M.J.O.; Byström, A.S. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 2005, 11, 424–436. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Huang, B.; Anderson, J.T.; Byström, A.S. Unexpected accumulation of ncm5U and ncm5S2U in a trm9 mutant suggests an additional step in the synthesis of mcm5U and mcm5S2U. PLoS ONE 2011, 6, e20783. [Google Scholar] [CrossRef] [PubMed]
- Leidel, S.; Pedrioli, P.G.A.; Bucher, T.; Brost, R.; Costanzo, M.; Schmidt, A.; Aebersold, R.; Boone, C.; Hofmann, K.; Peter, M. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature 2009, 458, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Noma, A.; Sakaguchi, Y.; Suzuki, T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 2009, 37, 1335–1352. [Google Scholar] [CrossRef] [PubMed]
- Nakai, Y.; Nakai, M.; Hayashi, H. Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J. Biol. Chem. 2008, 283, 27469–27476. [Google Scholar] [CrossRef] [PubMed]
- Klassen, R.; Grunewald, P.; Thüring, K.L.; Eichler, C.; Helm, M.; Schaffrath, R. Loss of anticodon wobble uridine modifications affects tRNALys function and protein levels in Saccharomyces cerevisiae. PLoS ONE 2015, 10, e0119261. [Google Scholar] [CrossRef] [PubMed]
- Klassen, R.; Bruch, A.; Schaffrath, R. Independent suppression of ribosomal +1 frameshifts by different tRNA anticodon loop modifications. RNA Biol. 2017, 14, 1252–1259. [Google Scholar] [CrossRef] [PubMed]
- Björk, G.R.; Huang, B.; Persson, O.P.; Byström, A.S. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA 2007, 13, 1245–1255. [Google Scholar] [CrossRef] [PubMed]
- Klassen, R.; Ciftci, A.; Funk, J.; Bruch, A.; Butter, F.; Schaffrath, R. tRNA anticodon loop modifications ensure protein homeostasis and cell morphogenesis in yeast. Nucleic Acids Res. 2016, 44, 10946–10959. [Google Scholar] [CrossRef] [PubMed]
- Laguesse, S.; Creppe, C.; Nedialkova, D.D.; Prévot, P.-P.; Borgs, L.; Huysseune, S.; Franco, B.; Duysens, G.; Krusy, N.; Lee, G.; et al. A dynamic unfolded protein response contributes to the control of cortical neurogenesis. Dev. Cell 2015, 35, 553–567. [Google Scholar] [CrossRef] [PubMed]
- Kojic, M.; Wainwright, B. The many faces of elongator in neurodevelopment and disease. Front. Mol. Neurosci. 2016, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sokołowski, M.; Klassen, R.; Bruch, A.; Schaffrath, R.; Glatt, S. Cooperativity between different tRNA modifications and their modification pathways. Biochim. Biophys. Acta Gene Regul. Mech. 2017. [Google Scholar] [CrossRef]
- Warren, J.D.; Rohrer, J.D.; Schott, J.M.; Fox, N.C.; Hardy, J.; Rossor, M.N. Molecular nexopathies: A new paradigm of neurodegenerative disease. Trends Neurosci. 2013, 36, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, R.; Han, L.; Faqeih, E.; Ewida, N.; Alobeid, E.; Phizicky, E.M.; Alkuraya, F.S. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum. Genet. 2016, 135, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.S.; Srivastava, S.; Farwell, K.D.; Lu, H.M.; Zeng, W.; Lu, H.; Chao, E.C.; Fatemi, A. ELP2 is a novel gene implicated in neurodevelopmental disabilities. Am. J. Med. Genet. Part A 2015, 167, 1391–1395. [Google Scholar] [CrossRef] [PubMed]
- Klassen, R.; Schaffrath, R. Role of pseudouridine formation by Deg1 for functionality of two glutamine isoacceptor tRNAs. Biomolecules 2017, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Scheidt, V.; Jüdes, A.; Bär, C.; Klassen, R.; Schaffrath, R. Loss of wobble uridine modification in tRNA anticodons interferes with TOR pathway signaling. Microb. Cell 2014, 1, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Kon, Y.; Phizicky, E.M. Functional importance of Ψ38 and Ψ39 in distinct tRNAs, amplified for tRNAGln(UUG) by unexpected temperature sensitivity of the s2U modification in yeast. RNA 2015, 21, 188–201. [Google Scholar] [CrossRef] [PubMed]
- Sidrauski, C.; Walter, P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 1997, 90, 1031–1039. [Google Scholar] [CrossRef]
- Mori, K.; Ma, W.; Gething, M.J.; Sambrook, J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 1993, 74, 743–756. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.S.; Walter, P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 1996, 87, 391–404. [Google Scholar] [CrossRef]
- Mori, K.; Kawahara, T.; Yoshida, H.; Yanagi, H.; Yura, T. Signalling from endoplasmic reticulum to nucleus: Transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells 1996, 1, 803–817. [Google Scholar] [CrossRef] [PubMed]
- Gueldener, U.; Heinisch, J.; Koehler, G.J.; Voss, D.; Hegemann, J.H. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002, 30, e23. [Google Scholar] [CrossRef] [PubMed]
- Sherman, F. Getting started with yeast. Methods Enzymol. 2002, 350, 3–41. [Google Scholar] [PubMed]
- Chen, D.C.; Yang, B.C.; Kuo, T.T. One-step transformation of yeast in stationary phase. Curr. Genet. 1992, 21, 83–84. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- De-Souza, E.A.; Pimentel, F.S.A.; Machado, C.M.; Martins, L.S.; da-Silva, W.S.; Montero-Lomeli, M.; Masuda, C.A. The unfolded protein response has a protective role in yeast models of classic galactosemia. Dis. Model. Mech. 2014, 7, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; Ogasawara, C.; Inada, T.; Englert, M.; Beier, H.; Takezawa, M.; Endo, T.; Yoshihisa, T. Dual functions of yeast tRNA ligase in the unfolded protein response: Unconventional cytoplasmic splicing of HAC1 pre-mRNA is not sufficient to release translational attenuation. Mol. Biol. Cell 2010, 21, 3722–3734. [Google Scholar] [CrossRef] [PubMed]
- Thiaville, P.; Legendre, R.; Rojas-Benitez, D.; Baudin-Baillieu, A.; Hatin, I.; Chalancon, G.; Glavic, A.; Namy, O.; de Crecy-Lagard, V. Global translational impacts of the loss of the tRNA modification t6A in yeast. Microb. Cell 2016, 3, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Nojima, H.; Leem, S.H.; Araki, H.; Sakai, A.; Nakashima, N.; Kanaoka, Y.; Ono, Y. Hac1: A novel yeast bZIP protein binding to the CRE motif is a multicopy suppressor for cdc10 mutant of Schizosaccharomyces pombe. Nucleic Acids Res. 1994, 22, 5279–5288. [Google Scholar] [CrossRef] [PubMed]
- Elbein, A.D. Inhibitors of the biosynthesis and processing of N-linked oligosaccharide chains. Annu. Rev. Biochem. 1987, 56, 497–534. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, A.; Chernyakov, I.; Gu, W.; Hiley, S.L.; Hughes, T.R.; Grayhack, E.J.; Phizicky, E.M. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 2006, 21, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Guy, M.P.; Kon, Y.; Phizicky, E.M. Lack of 2’-O-methylation in the tRNA anticodon loop of two phylogenetically distant yeast species activates the general amino acid control pathway. PLoS Genet. 2018, 14, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Halbleib, K.; Pesek, K.; Covino, R.; Hofbauer, H.F.; Wunnicke, D.; Hänelt, I.; Hummer, G.; Ernst, R. Activation of the unfolded protein response by lipid bilayer stress. Mol. Cell 2017, 67, 673–684. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, D.; Paira, S.; Das, B. Nuclear mRNA degradation tunes the gain of the unfolded protein response in Saccharomyces cerevisiae. Nucleic Acids Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T.; Masuda, Y.; Irie, K. The Saccharomyces cerevisiae AMPK, Snf1, negatively regulates the Hog1 MAPK pathway in ER stress response. PLoS Genet. 2015, 11, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Damon, J.R.; Pincus, D.; Ploegh, H.L. tRNA thiolation links translation to stress responses in Saccharomyces cerevisiae. Mol. Biol. Cell 2014, 26, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Steffen, K.K.; McCormick, M.A.; Pham, K.M.; Mackay, V.L.; Delaney, J.R.; Murakami, C.J.; Kaeberlein, M.; Kennedy, B.K. Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics 2012, 191, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, U.; Cullen, P.J. The tRNA modification complex elongator regulates the Cdc42-dependent mitogen-activated protein kinase pathway that controls filamentous growth in yeast. Eukaryot. Cell 2009, 8, 1362–1372. [Google Scholar] [CrossRef] [PubMed]
- Harris, M.A.; Deegan, J.I.; Ireland, A.; Lomax, J.; Ashburner, M.; Tweedie, S.; Carbon, S.; Lewis, S.; Mungall, C.; Day-Richter, J.; et al. The Gene Ontology project in 2008. Nucleic Acids Res. 2008, 36, 440–444. [Google Scholar] [CrossRef]
- Skrzypek, M.S.; Hirschman, J. Using the Saccharomyces Genome Database (SGD) for analysis of genomic information. Curr. Protoc. Bioinform. Chapter 2011, 1. [Google Scholar] [CrossRef]
| Strain | Genotype | References/Sources |
|---|---|---|
| BY4741 | MATa, his3Δ, leu2Δ, met15Δ, ura3Δ | Euroscarf, Frankfurt |
| AB43 | BY4741 ire1Δ::SpHIS5 | This study |
| RK520 | BY4741 ncs2Δ::SpHIS5 elp6Δ::KanMX4 | This study |
| RK206 | BY4741 urm1Δ::KanMX4 deg1Δ::SpHIS5 | [16] |
| RK220 | BY4741 elp3Δ::KanMX4 deg1Δ::SpHIS5 | [16] |
| AB97 | BY4741 elp3Δ::KanMX4 deg1Δ::SpHIS5 ire1Δ::KlURA3 | This study |
| Oligonucleotide | Sequence (5′–3′) | Target | References/Sources |
|---|---|---|---|
| Ire1_KO_Fwd | CATTAAAAAAACAGCATATCTGAGGAATTAATATTTTAGCACTTTGAAAACAGCTGAAGCTTCGTACGC | pUG27, pUG72 /IRE1 | This study |
| Ire1_KO_Rev | TAACATTAATGCAATAATCAACCAAGAAGAAGCAGAGGGGCATGAACATGGCATAGGCCACTAGTGGATCTG | pUG27, pUG72 /IRE1 | This study |
| Ire1_KO+_Fwd | CTTCGGGCAATACCTTCGACT | IRE1 | This study |
| Ire1_KO+_Rev | CAACCAAGAAGAAGCAGAGGG | IRE1 | This study |
| KO_NCS2_FW | TGCTATTGTCCATCCCTATCCTAGTTTTAAAAATATAATTCTATCAAGTTCAGCTGAAGCTTCGTACGC | pUG27 /NCS2 | This study |
| KO_NCS2_RV | TAAATAAATAAATACATAACCATTGGAATAGCGAAGCCTTTGACATTTCAGCATAGGCCACTAGTGGATCTG | pUG27 /NCS2 | This study |
| N_NCS2_FW | ACCGATGAGATGAGTGAGAC | NCS2 | This study |
| pUG27/SpHIS rev | GTCCAAAGCGATGGCAACGC | SpHIS5 | This study |
| HAC1_qPCR_Fwd | GACGACGCTACCTGCCG | HAC1i | This study |
| HAC1_qPCR_Rev | ACTGCGCTTCTGGATTACG | HAC1i | This study |
| qPCR_ACT1_FW | TTCCAGCCTTCTACGTTTCC | ACT1 | [16] |
| qPCR_ACT1_RV | AATCTCTACCGGCCAAATCG | ACT1 | [16] |
| HAC1F | CTGGCTGACCACGAAGACGC | HAC1u/i | [34] |
| HAC1R | TTGTCTTCATGAAGTGATGA | HAC1u/i | [34] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bruch, A.; Klassen, R.; Schaffrath, R. Unfolded Protein Response Suppression in Yeast by Loss of tRNA Modifications. Genes 2018, 9, 516. https://doi.org/10.3390/genes9110516
Bruch A, Klassen R, Schaffrath R. Unfolded Protein Response Suppression in Yeast by Loss of tRNA Modifications. Genes. 2018; 9(11):516. https://doi.org/10.3390/genes9110516
Chicago/Turabian StyleBruch, Alexander, Roland Klassen, and Raffael Schaffrath. 2018. "Unfolded Protein Response Suppression in Yeast by Loss of tRNA Modifications" Genes 9, no. 11: 516. https://doi.org/10.3390/genes9110516
APA StyleBruch, A., Klassen, R., & Schaffrath, R. (2018). Unfolded Protein Response Suppression in Yeast by Loss of tRNA Modifications. Genes, 9(11), 516. https://doi.org/10.3390/genes9110516





