The Obscure World of Integrative and Mobilizable Elements, Highly Widespread Elements that Pirate Bacterial Conjugative Systems
Abstract
1. Introduction
2. Maintenance of IMEs
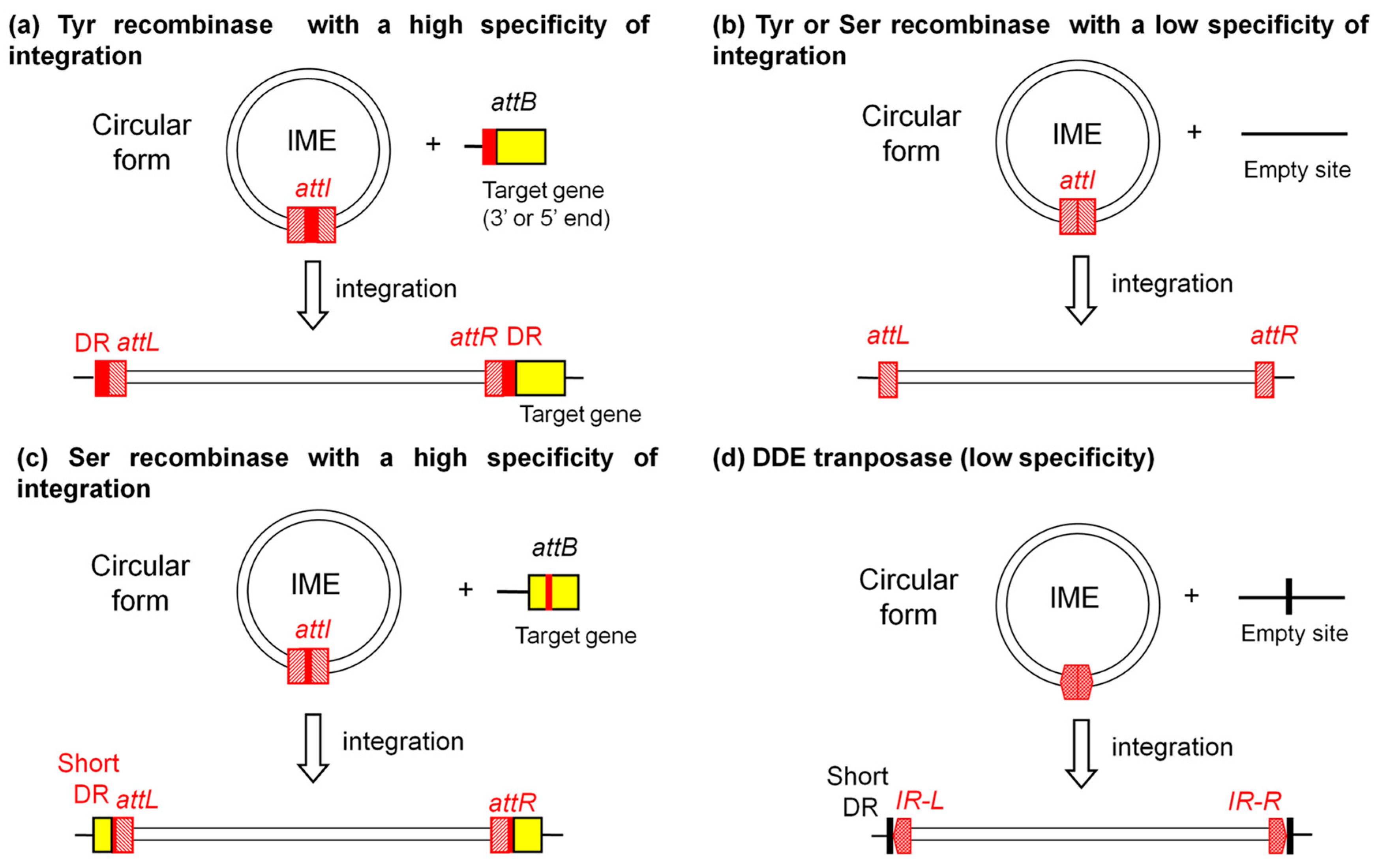
2.1. Integration and Excision
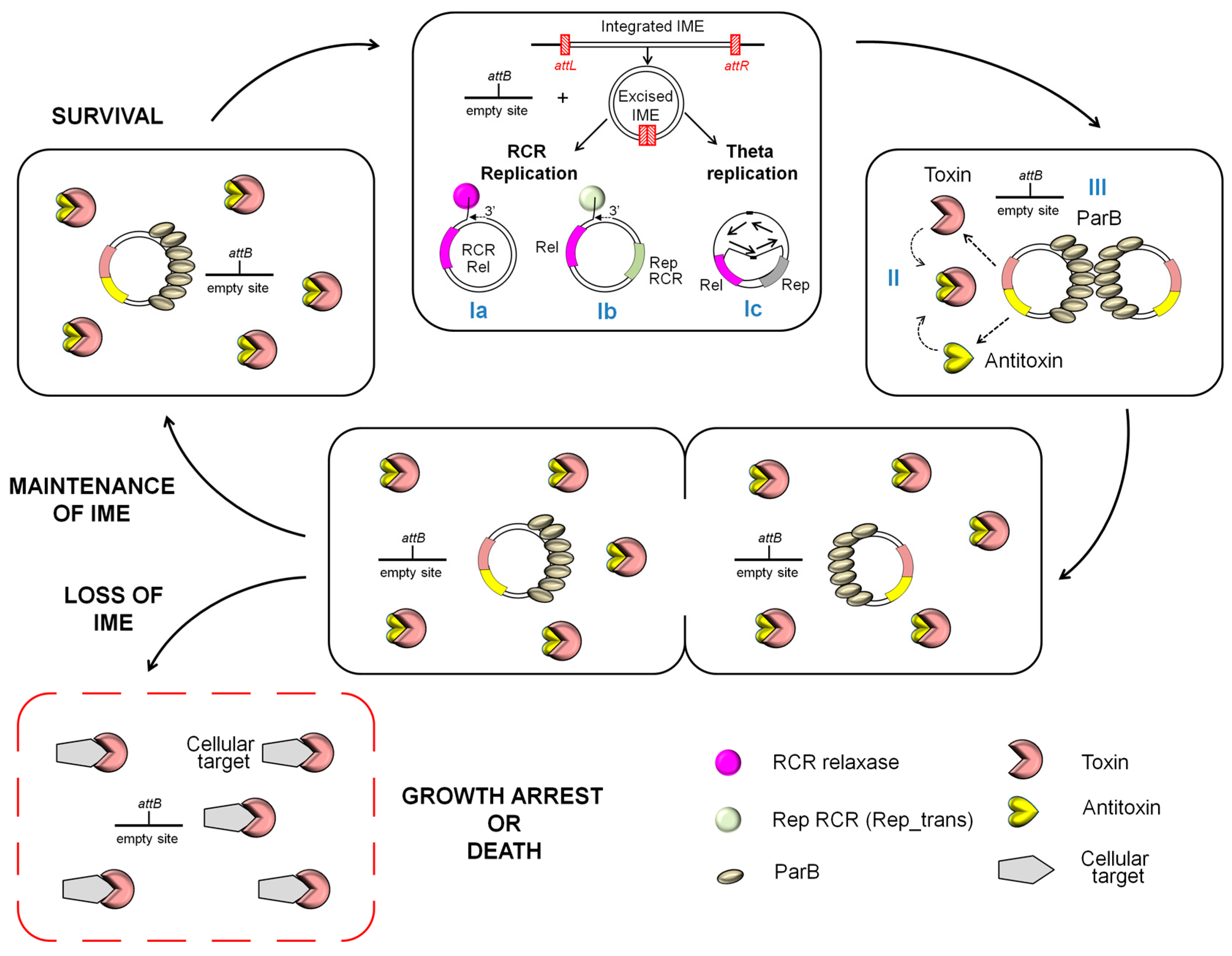
2.2. Maintenance of Excised IMEs: An Unexplored World
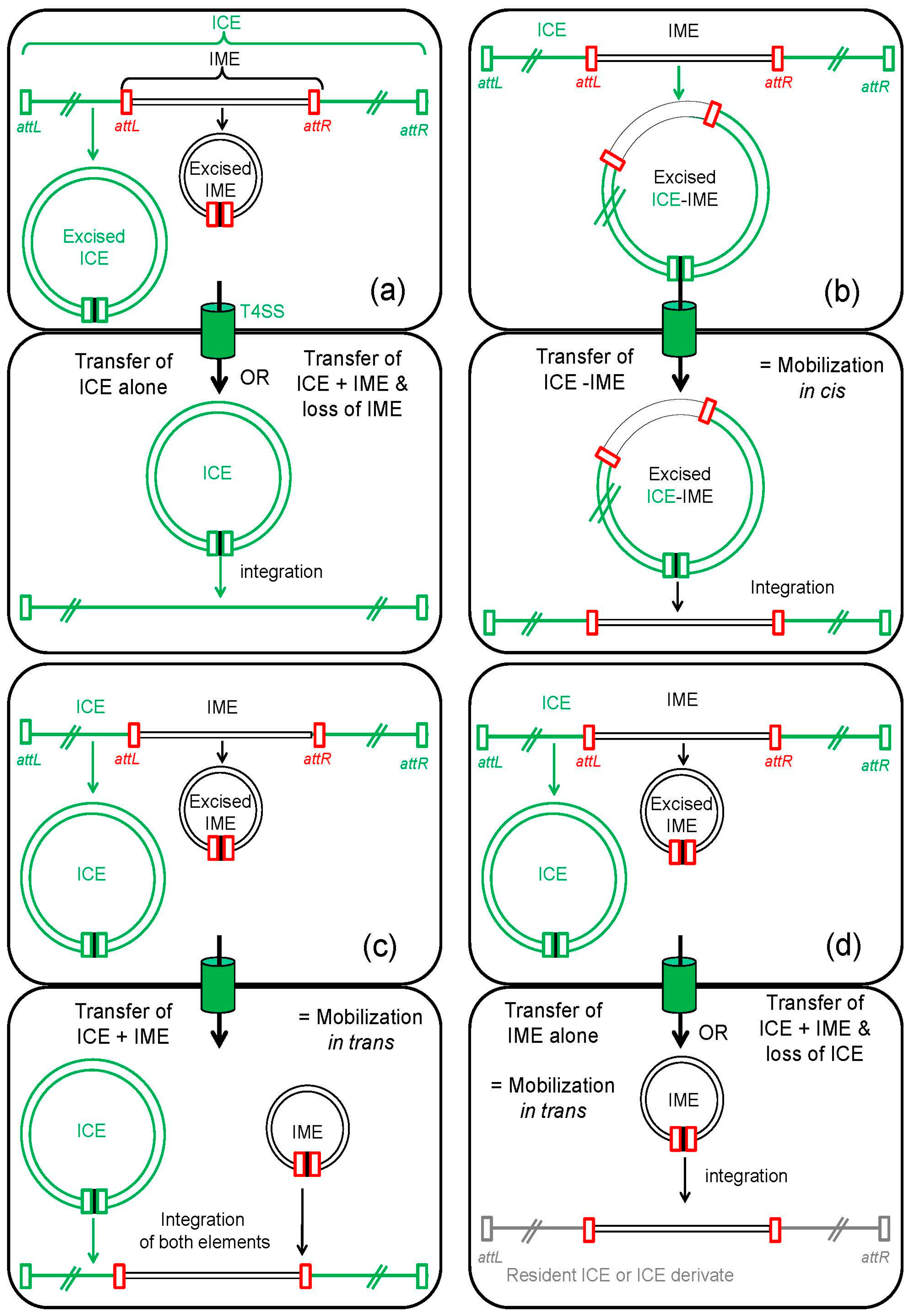
2.3. Impact of Other Mobile Genetic Elements on IME Maintenance
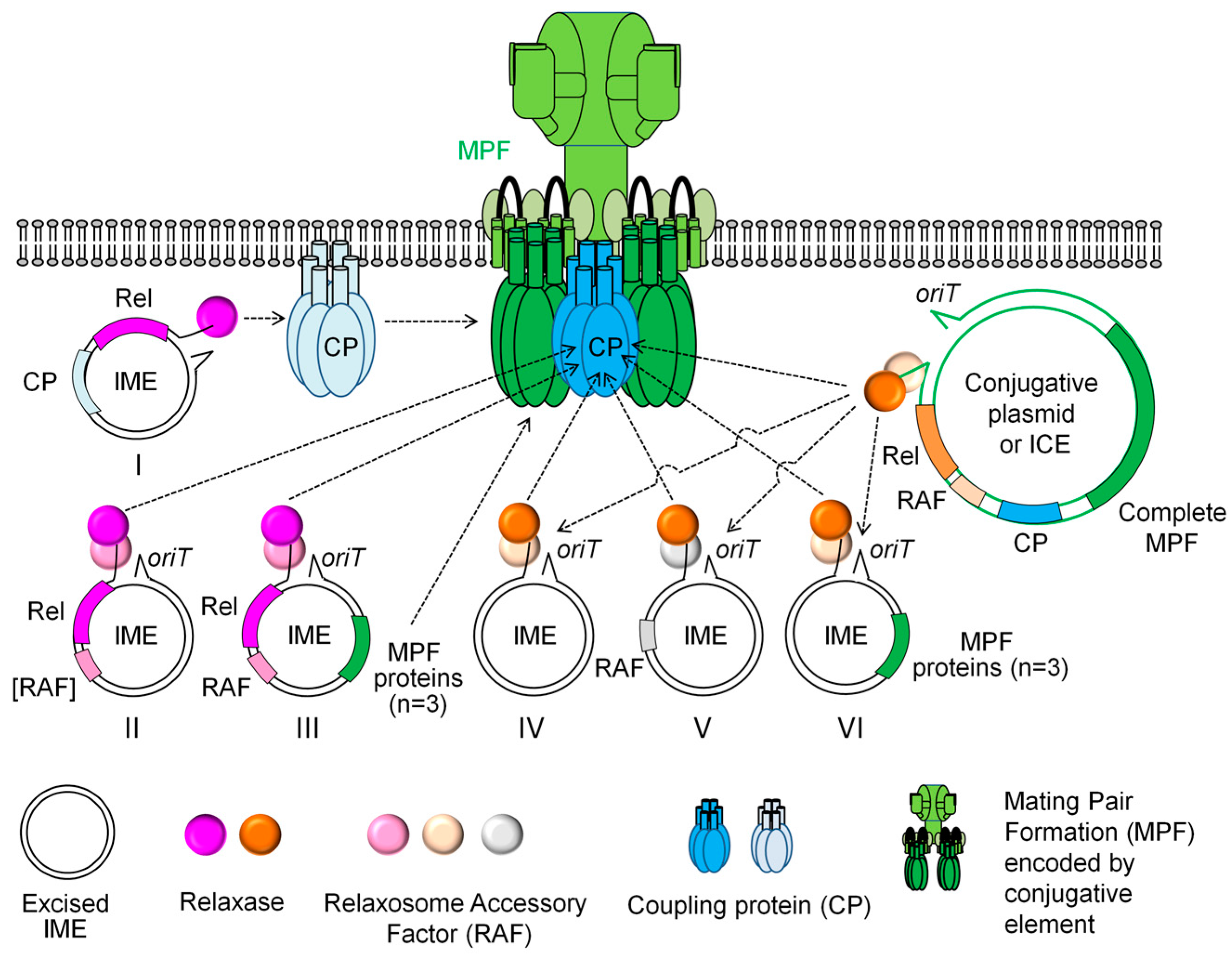
3. IMEs: Mobile Elements That Hijack the Conjugative Apparatus of Self-Transmissible Elements
3.1. Canonical IMEs Encoding Their Own Relaxases but no T4SS Protein
3.2. Non-Canonical IMEs Devoid of Relaxases
3.3. Non-Canonical IMEs Encoding Their Own Canonical Relaxase and Some Proteins of the T4SS
3.4. Non-Canonical IMEs Encoding a Non-Canonical Relaxase and/or CP
3.5. IMEs: Harmless Hitchhikers or Harmful Pirates of Conjugative Elements?
4. Moving with IMEs: Their Cargo Genes
4.1. IMEs: A Reservoir of Antibiotic Resistance Genes
4.2. Other Putative Functions Encoded by IMEs
5. Evolution of IMEs
6. IMEs: An Obscure World to Explore
7. Concluding Remarks
Conflicts of Interest
References
- Guglielmini, J.; Quintais, L.; Garcillan-Barcia, M.P.; de la Cruz, F.; Rocha, E.P. The repertoire of ICE in prokaryotes underscores the unity, diversity and ubiquity of conjugation. PLoS Genet. 2011, 7, e1002222. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Cabezon, E.; Ripoll-Rozada, J.; Pena, A.; de la Cruz, F.; Arechaga, I. Towards an integrated model of bacterial conjugation. FEMS Microbiol. Rev. 2015, 39, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Frost, L.S.; Leplae, R.; Summers, A.O.; Toussaint, A. Mobile genetic elements: The agents of open source evolution. Nat. Rev. Microbiol. 2005, 3, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Smillie, C.; Garcillan-Barcia, M.P.; Francia, M.V.; Rocha, E.P.; de la Cruz, F. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 2010, 74, 434–452. [Google Scholar] [CrossRef] [PubMed]
- Thoma, L.; Muth, G. The conjugative DNA-transfer apparatus of Streptomyces. Int. J. Med. Microbiol. 2015, 305, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Garcillan-Barcia, M.P.; Francia, M.V.; de la Cruz, F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 2009, 33, 657–687. [Google Scholar] [CrossRef] [PubMed]
- Ilangovan, A.; Connery, S.; Waksman, G. Structural biology of the gram-negative bacterial conjugation systems. Trends Microbiol. 2015, 23, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Goessweiner-Mohr, N.; Arends, K.; Keller, W.; Grohmann, E. Conjugative type IV secretion systems in gram-positive bacteria. Plasmid 2013, 70, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Guglielmini, J.; Neron, B.; Abby, S.S.; Garcillan-Barcia, M.P.; de la Cruz, F.; Rocha, E.P. Key components of the eight classes of type IV secretion systems involved in bacterial conjugation or protein secretion. Nucleic Acids Res. 2014, 42, 5715–5727. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, J.P.; Kwong, S.M.; Murphy, R.J.; Yui Eto, K.; Price, K.J.; Nguyen, Q.T.; O’Brien, F.G.; Grubb, W.B.; Coombs, G.W.; Firth, N. An updated view of plasmid conjugation and mobilization in Staphylococcus. Mob. Genet. Elements 2016, 6, e1208317. [Google Scholar] [CrossRef] [PubMed]
- Bellanger, X.; Payot, S.; Leblond-Bourget, N.; Guedon, G. Conjugative and mobilizable genomic islands in bacteria: Evolution and diversity. FEMS Microbiol. Rev. 2014, 38, 720–760. [Google Scholar] [CrossRef] [PubMed]
- Burrus, V.; Pavlovic, G.; Decaris, B.; Guedon, G. Conjugative transposons: The tip of the iceberg. Mol. Microbiol. 2002, 46, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Ambroset, C.; Coluzzi, C.; Guedon, G.; Devignes, M.D.; Loux, V.; Lacroix, T.; Payot, S.; Leblond-Bourget, N. New insights into the classification and integration specificity of Streptococcus integrative conjugative elements through extensive genome exploration. Front. Microbiol. 2016, 6, 1483. [Google Scholar] [CrossRef] [PubMed]
- Ghinet, M.G.; Bordeleau, E.; Beaudin, J.; Brzezinski, R.; Roy, S.; Burrus, V. Uncovering the prevalence and diversity of integrating conjugative elements in actinobacteria. PLoS ONE 2011, 6, e27846. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.M.; Grossman, A.D. Integrative and conjugative elements (ICEs): What they do and how they work. Annu. Rev. Genet. 2015, 49, 577–601. [Google Scholar] [CrossRef] [PubMed]
- Guglielmini, J.; de la Cruz, F.; Rocha, E.P. Evolution of conjugation and type IV secretion systems. Mol. Biol. Evol. 2013, 30, 315–331. [Google Scholar] [CrossRef] [PubMed]
- Parsons, J.A.; Bannam, T.L.; Devenish, R.J.; Rood, J.I. TcpA, an FtsK/SpoIIIE homolog, is essential for transfer of the conjugative plasmid pCW3 in Clostridium perfringens. J. Bacteriol. 2007, 189, 7782–7790. [Google Scholar] [CrossRef] [PubMed]
- Steen, J.A.; Bannam, T.L.; Teng, W.L.; Devenish, R.J.; Rood, J.I. The putative coupling protein TcpA interacts with other pCW3-encoded proteins to form an essential part of the conjugation complex. J. Bacteriol. 2009, 191, 2926–2933. [Google Scholar] [CrossRef] [PubMed]
- Burrus, V. Mechanisms of stabilization of integrative and conjugative elements. Curr. Opin. Microbiol. 2017, 38, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Carraro, N.; Burrus, V. The dualistic nature of integrative and conjugative elements. Mob. Genet. Elements 2015, 5, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Toussaint, A.; Merlin, C. Mobile elements as a combination of functional modules. Plasmid 2002, 47, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Burrus, V.; Pavlovic, G.; Decaris, B.; Guédon, G. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid 2002, 48, 77–97. [Google Scholar] [CrossRef]
- Ramsay, J.P.; Firth, N. Diverse mobilization strategies facilitate transfer of non-conjugative mobile genetic elements. Curr. Opin. Microbiol. 2017, 38, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Carraro, N.; Rivard, N.; Burrus, V.; Ceccarelli, D. Mobilizable genomic islands, different strategies for the dissemination of multidrug resistance and other adaptive traits. Mob. Genet. Elements 2017, 7, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Coluzzi, C.; Guedon, G.; Devignes, M.D.; Ambroset, C.; Loux, V.; Lacroix, T.; Payot, S.; Leblond-Bourget, N. A glimpse into the world of integrative and mobilizable elements in streptococci reveals an unexpected diversity and novel families of mobilization proteins. Front. Microbiol. 2017, 8, 443. [Google Scholar] [CrossRef] [PubMed]
- Daccord, A.; Ceccarelli, D.; Burrus, V. Integrating conjugative elements of the SXT/R391 family trigger the excision and drive the mobilization of a new class of Vibrio genomic islands. Mol. Microbiol. 2010, 78, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Achard, A.; Leclercq, R. Characterization of a small mobilizable transposon, MTnSag1, in Streptococcus agalactiae. J. Bacteriol. 2007, 189, 4328–4331. [Google Scholar] [CrossRef] [PubMed]
- Lyras, D.; Adams, V.; Ballard, S.A.; Teng, W.L.; Howarth, P.M.; Crellin, P.K.; Bannam, T.L.; Songer, J.G.; Rood, J.I. tISCpe8, an IS1595-family lincomycin resistance element located on a conjugative plasmid in Clostridium perfringens. J. Bacteriol. 2009, 191, 6345–6351. [Google Scholar] [CrossRef] [PubMed]
- Giovanetti, E.; Brenciani, A.; Tiberi, E.; Bacciaglia, A.; Varaldo, P.E. ICESp2905, the erm(TR)-tet(O) element of Streptococcus pyogenes, is formed by two independent integrative and conjugative elements. Antimicrob. Agents Chemother. 2012, 56, 591–594. [Google Scholar] [CrossRef] [PubMed]
- Brenciani, A.; Tiberi, E.; Bacciaglia, A.; Petrelli, D.; Varaldo, P.E.; Giovanetti, E. Two distinct genetic elements are responsible for erm(TR)-mediated erythromycin resistance in tetracycline-susceptible and tetracycline-resistant strains of Streptococcus pyogenes. Antimicrob. Agents Chemother. 2011, 55, 2106–2112. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, M.S.; Warburton, P.J.; Roberts, A.P.; Mullany, P.; Allan, E. Genetic organisation, mobility and predicted functions of genes on integrated, mobile genetic elements in sequenced strains of Clostridium difficile. PLoS ONE 2011, 6, e23014. [Google Scholar] [CrossRef] [PubMed]
- Adams, V.; Lyras, D.; Farrow, K.A.; Rood, J.I. The clostridial mobilisable transposons. Cell. Mol. Life Sci. 2002, 59, 2033–2043. [Google Scholar] [CrossRef] [PubMed]
- Billington, S.J.; Songer, J.G.; Jost, B.H. Widespread distribution of a Tet W determinant among tetracycline-resistant isolates of the animal pathogen Arcanobacterium pyogenes. Antimicrob. Agents Chemother. 2002, 46, 1281–1287. [Google Scholar] [CrossRef] [PubMed]
- Lavigne, J.P.; Vergunst, A.C.; Bourg, G.; O’Callaghan, D. The IncP island in the genome of Brucella suis 1330 was acquired by site-specific integration. Infect. Immun. 2005, 73, 7779–7783. [Google Scholar] [CrossRef] [PubMed]
- Harmer, C.J.; Hamidian, M.; Hall, R.M. pIP40a, a type 1 IncC plasmid from 1969 carries the integrative element GIsul2 and a novel class II mercury resistance transposon. Plasmid 2017, 92, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo-Diaz, F.; Fernandez-Lopez, C.; Douarre, P.E.; Baez-Ortega, A.; Flores, C.; Glaser, P.; Espinosa, M. Streptococcal group B integrative and mobilizable element IMESag-rpsI encodes a functional relaxase involved in its transfer. Open Biol. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Doublet, B.; Boyd, D.; Mulvey, M.R.; Cloeckaert, A. The Salmonella genomic island 1 is an integrative mobilizable element. Mol. Microbiol. 2005, 55, 1911–1924. [Google Scholar] [CrossRef] [PubMed]
- Carraro, N.; Rivard, N.; Ceccarelli, D.; Colwell, R.R.; Burrus, V. IncA/C conjugative plasmids mobilize a new family of multidrug resistance islands in clinical Vibrio cholerae non-O1/non-O139 isolates from Haiti. MBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Daccord, A.; Ceccarelli, D.; Rodrigue, S.; Burrus, V. Comparative analysis of mobilizable genomic islands. J. Bacteriol. 2013, 195, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Carraro, N.; Matteau, D.; Luo, P.; Rodrigue, S.; Burrus, V. The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genet. 2014, 10, e1004714. [Google Scholar] [CrossRef] [PubMed]
- Holden, M.T.; Seth-Smith, H.M.; Crossman, L.C.; Sebaihia, M.; Bentley, S.D.; Cerdeno-Tarraga, A.M.; Thomson, N.R.; Bason, N.; Quail, M.A.; Sharp, S.; et al. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J. Bacteriol. 2009, 191, 261–277. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, N.B.; Wang, G.R.; Stevens, A.M.; Salyers, A.A. Excision, transfer and integration of NBU1, a mobilizable site-selective insertion element. J. Bacteriol. 1993, 175, 6578–6587. [Google Scholar] [CrossRef] [PubMed]
- Naito, M.; Ogura, Y.; Itoh, T.; Shoji, M.; Okamoto, M.; Hayashi, T.; Nakayama, K. The complete genome sequencing of Prevotella intermedia strain OMA14 and a subsequent fine-scale, intra-species genomic comparison reveal an unusual amplification of conjugative and mobile transposons and identify a novel prevotella-lineage-specific repeat. DNA Res. 2016, 23, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Puymege, A.; Bertin, S.; Chuzeville, S.; Guedon, G.; Payot, S. Conjugative transfer and cis-mobilization of a genomic island by an integrative and conjugative element of Streptococcus agalactiae. J. Bacteriol. 2013. [CrossRef] [PubMed]
- Ghosh, S.; Sadowsky, M.J.; Roberts, M.C.; Gralnick, J.A.; LaPara, T.M. Sphingobacterium sp. Strain PM2-P1–29 harbours a functional tet(X) gene encoding for the degradation of tetracycline. J. Appl. Microbiol. 2009, 106, 1336–1342. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shoemaker, N.B.; Wang, G.R.; Salyers, A.A. Characterization of a Bacteroides mobilizable transposon, NBU2, which carries a functional lincomycin resistance gene. J. Bacteriol. 2000, 182, 3559–3571. [Google Scholar] [CrossRef] [PubMed]
- Puymege, A.; Bertin, S.; Guedon, G.; Payot, S. Analysis of Streptococcus agalactiae pan-genome for prevalence, diversity and functionality of integrative and conjugative or mobilizable elements integrated in the tRNA(Lys CTT) gene. Mol. Genet. Genom. 2015, 290, 1727–1740. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.J.; Parker, A.C. Identification of a circular intermediate in the transfer and transposition of Tn4555, a mobilizable transposon from Bacteroides spp. J. Bacteriol. 1993, 175, 2682–2691. [Google Scholar] [CrossRef] [PubMed]
- Vedantam, G.; Novicki, T.J.; Hecht, D.W. Bacteroides fragilis transfer factor Tn5520: The smallest bacterial mobilizable transposon containing single integrase and mobilization genes that function in Escherichia coli. J. Bacteriol. 1999, 181, 2564–2571. [Google Scholar] [PubMed]
- Dingle, K.E.; Elliott, B.; Robinson, E.; Griffiths, D.; Eyre, D.W.; Stoesser, N.; Vaughan, A.; Golubchik, T.; Fawley, W.N.; Wilcox, M.H.; et al. Evolutionary history of the Clostridium difficile pathogenicity locus. Genome Biol. Evol. 2014, 6, 36–52. [Google Scholar] [CrossRef] [PubMed]
- Bass, K.A.; Hecht, D.W. Isolation and characterization of cLV25, a Bacteroides fragilis chromosomal transfer factor resembling multiple Bacteroides sp. mobilizable transposons. J. Bacteriol. 2002, 184, 1895–1904. [Google Scholar] [CrossRef] [PubMed]
- Hecht, D.W.; Malamy, M.H. Tn4399, a conjugal mobilizing transposon of Bacteroides fragilis. J. Bacteriol. 1989, 171, 3603–3608. [Google Scholar] [CrossRef] [PubMed]
- Bannam, T.L.; Crellin, P.K.; Rood, J.I. Molecular genetics of the chloramphenicol-resistance transposon Tn4451 from Clostridium perfringens: The TnpX site-specific recombinase excises a circular transposon molecule. Mol. Microbiol. 1995, 16, 535–551. [Google Scholar] [CrossRef] [PubMed]
- Lyras, D.; Adams, V.; Lucet, I.; Rood, J.I. The large resolvase TnpX is the only transposon-encoded protein required for transposition of the Tn4451/3 family of integrative mobilizable elements. Mol. Microbiol. 2004, 51, 1787–1800. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Gourbeyre, E.; Varani, A.; Ton-Hoang, B.; Chandler, M. Everyman’s guide to bacterial insertion sequences. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Groth, A.C.; Calos, M.P. Phage integrases: Biology and applications. J. Mol. Biol. 2004, 335, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Daccord, A.; Mursell, M.; Poulin-Laprade, D.; Burrus, V. Dynamics of the SetCD-regulated integration and excision of genomic islands mobilized by integrating conjugative elements of the SXT/R391 family. J. Bacteriol. 2012, 194, 5794–5802. [Google Scholar] [CrossRef] [PubMed]
- Parker, A.C.; Smith, C.J. A multicomponent system is required for tetracycline-induced excision of Tn4555. J. Bacteriol. 2004, 186, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Wood, M.M.; Rajeev, L.; Gardner, J.F. Interactions of NBU1 IntN1 and Orf2x proteins with attachment site DNA. J. Bacteriol. 2013, 195, 5516–5525. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, N.B.; Wang, G.R.; Salyers, A.A. Multiple gene products and sequences required for excision of the mobilizable integrated bacteroides element NBU1. J. Bacteriol. 2000, 182, 928–936. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, K.; Van Duyne, G.D. The ins and outs of serine integrase site-specific recombination. Curr. Opin. Struct. Biol. 2014, 24, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Douarre, P.E.; Sauvage, E.; Poyart, C.; Glaser, P. Host specificity in the diversity and transfer of lsa resistance genes in group B Streptococcus. J. Antimicrob. Chemother. 2015, 70, 3205–3213. [Google Scholar] [CrossRef] [PubMed]
- Persaud, C.; Lu, Y.; Vila-Sanjurjo, A.; Campbell, J.L.; Finley, J.; O’Connor, M. Mutagenesis of the modified bases, m(5)U1939 and psi2504, in Escherichia coli 23S rRNA. Biochem. Biophys. Res. Commun. 2010, 392, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Doublet, B.; Golding, G.R.; Mulvey, M.R.; Cloeckaert, A. Secondary chromosomal attachment site and tandem integration of the mobilizable Salmonella genomic island 1. PLoS ONE 2008, 3, e2060. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, N.B.; Wang, G.R.; Salyers, A.A. NBU1, a mobilizable site-specific integrated element from Bacteroides spp., can integrate nonspecifically in Escherichia coli. J. Bacteriol. 1996, 178, 3601–3607. [Google Scholar] [CrossRef] [PubMed]
- Tribble, G.D.; Parker, A.C.; Smith, C.J. The Bacteroides mobilizable transposon Tn4555 integrates by a site-specific recombination mechanism similar to that of the gram-positive bacterial element Tn916. J. Bacteriol. 1997, 179, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Tribble, G.D.; Parker, A.C.; Smith, C.J. Transposition genes of the Bacteroides mobilizable transposon Tn4555: Role of a novel targeting gene. Mol. Microbiol. 1999, 34, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Lyras, D.; Storie, C.; Huggins, A.S.; Crellin, P.K.; Bannam, T.L.; Rood, J.I. Chloramphenicol resistance in Clostridium difficile is encoded on Tn4453 transposons that are closely related to Tn4451 from Clostridium perfringens. Antimicrob. Agents Chemother. 1998, 42, 1563–1567. [Google Scholar] [PubMed]
- Salyers, A.A.; Shoemaker, N.B.; Li, L.Y. In the driver’s seat: The bacteroides conjugative transposons and the elements they mobilize. J. Bacteriol. 1995, 177, 5727–5731. [Google Scholar] [CrossRef] [PubMed]
- Guerillot, R.; Da Cunha, V.; Sauvage, E.; Bouchier, C.; Glaser, P. Modular evolution of TnGBSs, a new family of integrative and conjugative elements associating insertion sequence transposition, plasmid replication and conjugation for their spreading. J. Bacteriol. 2013, 195, 1979–1990. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.A.; Babic, A.; Grossman, A.D. Autonomous plasmid-like replication of a conjugative transposon. Mol. Microbiol. 2010, 75, 268–279. [Google Scholar] [CrossRef] [PubMed]
- Wright, L.D.; Grossman, A.D. Autonomous replication of the conjugative transposon Tn916. J. Bacteriol. 2016, 198, 3355–3366. [Google Scholar] [CrossRef] [PubMed]
- Carraro, N.; Poulin, D.; Burrus, V. Replication and active partition of integrative and conjugative elements (ICEs) of the SXT/R391 family: The line between ICEs and conjugative plasmids is getting thinner. PLoS Genet. 2015, 11, e1005298. [Google Scholar] [CrossRef] [PubMed]
- Burrus, V.; Bontemps, C.; Decaris, B.; Guedon, G. Characterization of a novel type II restriction-modification system, Sth368I, encoded by the integrative element ICESt1 of Streptococcus thermophilus CNRZ368. Appl. Environ. Microbiol. 2001, 67, 1522–1528. [Google Scholar] [CrossRef] [PubMed]
- Mruk, I.; Kobayashi, I. To be or not to be: Regulation of restriction-modification systems and other toxin-antitoxin systems. Nucleic Acids Res. 2014, 42, 70–86. [Google Scholar] [CrossRef] [PubMed]
- Kiss, J.; Nagy, B.; Olasz, F. Stability, entrapment and variant formation of Salmonella genomic island 1. PLoS ONE 2012, 7, e32497. [Google Scholar] [CrossRef] [PubMed]
- Huguet, K.T.; Gonnet, M.; Doublet, B.; Cloeckaert, A. A toxin antitoxin system promotes the maintenance of the IncA/C-mobilizable Salmonella genomic island 1. Sci. Rep. 2016, 6, 32285. [Google Scholar] [CrossRef] [PubMed]
- Pavlovic, G.; Burrus, V.; Gintz, B.; Decaris, B.; Guedon, G. Evolution of genomic islands by deletion and tandem accretion by site-specific recombination: ICESt1-related elements from Streptococcus thermophilus. Microbiology 2004, 150, 759–774. [Google Scholar] [CrossRef] [PubMed]
- Bellanger, X.; Morel, C.; Gonot, F.; Puymege, A.; Decaris, B.; Guedon, G. Site-specific accretion of an integrative conjugative element together with a related genomic island leads to cis mobilization and gene capture. Mol. Microbiol. 2011, 81, 912–925. [Google Scholar] [CrossRef] [PubMed]
- Carraro, N.; Libante, V.; Morel, C.; Charron-Bourgoin, F.; Leblond, P.; Guedon, G. Plasmid-like replication of a minimal streptococcal integrative and conjugative element. Microbiology 2016. [Google Scholar] [CrossRef] [PubMed]
- Lautner, M.; Schunder, E.; Herrmann, V.; Heuner, K. Regulation, integrase-dependent excision and horizontal transfer of genomic islands in Legionella pneumophila. J. Bacteriol. 2013, 195, 1583–1597. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Loria, R. Emergence of novel pathogenic Streptomyces species by site-specific accretion and cis-mobilization of pathogenicity islands. Mol. Plant Microbe Interact. 2017, 30, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Brochet, M.; Couve, E.; Glaser, P.; Guedon, G.; Payot, S. Integrative conjugative elements and related elements are major contributors to the genome diversity of Streptococcus agalactiae. J. Bacteriol. 2008, 190, 6913–6917. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.G.; Malamy, M.H. Requirements for strand- and site-specific cleavage within the oriT region of Tn4399, a mobilizing transposon from Bacteroides fragilis. J. Bacteriol. 1995, 177, 3158–3165. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.J.; Parker, A.C. The transfer origin for Bacteroides mobilizable transposon Tn4555 is related to a plasmid family from gram-positive bacteria. J. Bacteriol. 1998, 180, 435–439. [Google Scholar] [PubMed]
- Marcoleta, A.E.; Berrios-Pasten, C.; Nunez, G.; Monasterio, O.; Lagos, R. Klebsiella pneumoniae asparagine tDNAs are integration hotspots for different genomic islands encoding microcin e492 production determinants and other putative virulence factors present in hypervirulent strains. Front. Microbiol. 2016, 7, 849. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.G.; Malamy, M.H. Characterization of a “mobilization cassette” in transposon Tn4399 from Bacteroides fragilis. J. Bacteriol. 1993, 175, 5814–5823. [Google Scholar] [CrossRef] [PubMed]
- Yoshii, A.; Moriyama, H.; Fukuhara, T. The novel kasugamycin 2′-N-acetyltransferase gene aac(2′)-IIa, carried by the IncP island, confers kasugamycin resistance to rice-pathogenic bacteria. Appl. Environ. Microbiol. 2012, 78, 5555–5564. [Google Scholar] [CrossRef] [PubMed]
- Goh, S.; Hussain, H.; Chang, B.J.; Emmett, W.; Riley, T.V.; Mullany, P. Phage phiC2 mediates transduction of Tn6215, encoding erythromycin resistance, between Clostridium difficile strains. MBio 2013, 4, e00840-13. [Google Scholar] [CrossRef] [PubMed]
- Crellin, P.K.; Rood, J.I. Tn4451 from Clostridium perfringens is a mobilizable transposon that encodes the functional Mob protein, TnpZ. Mol. Microbiol. 1998, 27, 631–642. [Google Scholar] [CrossRef] [PubMed]
- Douard, G.; Praud, K.; Cloeckaert, A.; Doublet, B. The Salmonella genomic island 1 is specifically mobilized in trans by the IncA/C multidrug resistance plasmid family. PLoS ONE 2010, 5, e15302. [Google Scholar] [CrossRef] [PubMed]
- Boyd, D.; Peters, G.A.; Cloeckaert, A.; Boumedine, K.S.; Chaslus-Dancla, E.; Imberechts, H.; Mulvey, M.R. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 2001, 183, 5725–5732. [Google Scholar] [CrossRef] [PubMed]
- Siebor, E.; de Curraize, C.; Amoureux, L.; Neuwirth, C. Mobilization of the Salmonella genomic island SGI1 and the Proteus genomic island PGI1 by the A/C2 plasmid carrying blaTEM-24 harboured by various clinical species of Enterobacteriaceae. J. Antimicrob. Chemother. 2016, 71, 2167–2170. [Google Scholar] [CrossRef] [PubMed]
- Haase, J.; Kalkum, M.; Lanka, E. TrbK, a small cytoplasmic membrane lipoprotein, functions in entry exclusion of the IncP alpha plasmid RP4. J. Bacteriol. 1996, 178, 6720–6729. [Google Scholar] [CrossRef] [PubMed]
- Haase, J.; Lurz, R.; Grahn, A.M.; Bamford, D.H.; Lanka, E. Bacterial conjugation mediated by plasmid RP4: RSF1010 mobilization, donor-specific phage propagation and pilus production require the same Tra2 core components of a proposed DNA transport complex. J. Bacteriol. 1995, 177, 4779–4791. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, J.A.; Traore, D.A.; Bannam, T.L.; Lyras, D.; Whisstock, J.C.; Rood, J.I. TcpM: A novel relaxase that mediates transfer of large conjugative plasmids from Clostridium perfringens. Mol. Microbiol. 2016, 99, 884–896. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.A.; Thomas, J.; Grossman, A.D. The Bacillus subtilis conjugative transposon ICEBs1 mobilizes plasmids lacking dedicated mobilization functions. J. Bacteriol. 2012, 194, 3165–3172. [Google Scholar] [CrossRef] [PubMed]
- Naglich, J.G.; Andrews, R.E., Jr. Tn916-dependent conjugal transfer of pC194 and pUB110 from Bacillus subtilis into Bacillus thuringiensis subsp. israelensis. Plasmid 1988, 20, 113–126. [Google Scholar] [CrossRef]
- Showsh, S.A.; Andrews, R.E., Jr. Analysis of the requirement for a pUB110 mob region during Tn916-dependent mobilization. Plasmid 1999, 41, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Mingoia, M.; Morici, E.; Marini, E.; Brenciani, A.; Giovanetti, E.; Varaldo, P.E. Macrolide resistance gene erm(TR) and erm(TR)-carrying genetic elements in Streptococcus agalactiae: Characterization of ICESagTR7, a new composite element containing IMESp2907. J. Antimicrob. Chemother. 2016, 71, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Morici, E.; Simoni, S.; Brenciani, A.; Giovanetti, E.; Varaldo, P.E.; Mingoia, M. A new mosaic integrative and conjugative element from Streptococcus agalactiae carrying resistance genes for chloramphenicol (catQ) and macrolides [mef(I) and erm(TR)]. J. Antimicrob. Chemother. 2016, 72, 64–67. [Google Scholar] [CrossRef] [PubMed]
- Harmer, C.J.; Hamidian, M.; Ambrose, S.J.; Hall, R.M. Destabilization of IncA and IncC plasmids by SGI1 and SGI2 type Salmonella genomic islands. Plasmid 2016, 87–88, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Carraro, N.; Durand, R.; Rivard, N.; Anquetil, C.; Barrette, C.; Humbert, M.; Burrus, V. Salmonella genomic island 1 (SGI1) reshapes the mating apparatus of IncC conjugative plasmids to promote self-propagation. PLoS Genet. 2017, 13, e1006705. [Google Scholar] [CrossRef] [PubMed]
- Billington, S.J.; Jost, B.H. Multiple genetic elements carry the tetracycline resistance gene tet(W) in the animal pathogen Arcanobacterium pyogenes. Antimicrob. Agents Chemother. 2006, 50, 3580–3587. [Google Scholar] [CrossRef] [PubMed]
- Whittle, G.; Hund, B.D.; Shoemaker, N.B.; Salyers, A.A. Characterization of the 13-kilobase ermF region of the Bacteroides conjugative transposon cTnDOT. Appl. Environ. Microbiol. 2001, 67, 3488–3495. [Google Scholar] [CrossRef] [PubMed]
- Siebor, E.; Neuwirth, C. The new variant of Salmonella genomic island 1 (SGI1-V) from a Proteus mirabilis French clinical isolate harbours blaVEB-6 and qnrA1 in the multiple antibiotic resistance region. J. Antimicrob. Chemother. 2011, 66, 2513–2520. [Google Scholar] [CrossRef] [PubMed]
- Schultz, E.; Barraud, O.; Madec, J.Y.; Haenni, M.; Cloeckaert, A.; Ploy, M.C.; Doublet, B. Multidrug resistance Salmonella genomic island 1 in a Morganella morganii subsp. Morganii human clinical isolate from france. mSphere 2017, 2. [Google Scholar] [CrossRef]
- Ferreira, L.Q.; Avelar, K.E.; Vieira, J.M.; de Paula, G.R.; Colombo, A.P.; Domingues, R.M.; Ferreira, M.C. Association between the cfxA gene and transposon Tn4555 in Bacteroides distasonis strains and other bacteroides species. Curr. Microbiol. 2007, 54, 348–353. [Google Scholar] [CrossRef] [PubMed]
- Soliman, A.M.; Ahmed, A.M.; Shimamoto, T.; El-Domany, R.A.; Nariya, H. First report in Africa of two clinical isolates of Proteus mirabilis carrying Salmonella genomic island (SGI1) variants, SGI1-PmABB and SGI1-W. Infect. Genet. Evol. 2017, 51, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Schultz, E.; Haenni, M.; Mereghetti, L.; Siebor, E.; Neuwirth, C.; Madec, J.Y.; Cloeckaert, A.; Doublet, B. Survey of multidrug resistance integrative mobilizable elements SGI1 and PGI1 in Proteus mirabilis in humans and dogs in france, 2010–2013. J. Antimicrob. Chemother. 2015, 70, 2543–2546. [Google Scholar] [CrossRef] [PubMed]
- Yoshii, A.; Omatsu, T.; Katayama, Y.; Koyama, S.; Mizutani, T.; Moriyama, H.; Fukuhara, T. Two types of genetic carrier, the IncP genomic island and the novel IncP-1beta plasmid, for the aac(2′)-IIa gene that confers kasugamycin resistance in Acidovorax avenae ssp. avenae. Mol. Plant Pathol. 2015, 16, 288–300. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, N.B.; Barber, R.D.; Salyers, A.A. Cloning and characterization of a Bacteroides conjugal tetracycline-erythromycin resistance element by using a shuttle cosmid vector. J. Bacteriol. 1989, 171, 1294–1302. [Google Scholar] [CrossRef] [PubMed]
- Magot, M. Physical characterization of the Clostridium perfringens tetracycline-chloramphenicol resistance plasmid pIP401. Ann. Microbiol. (Paris) 1984, 135, 269–282. [Google Scholar] [CrossRef]
- Balado, M.; Lemos, M.L.; Osorio, C.R. Integrating conjugative elements of the SXT/R391 family from fish-isolated Vibrios encode restriction-modification systems that confer resistance to bacteriophages. FEMS Microbiol. Ecol. 2013, 83, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Ershova, A.S.; Rusinov, I.S.; Spirin, S.A.; Karyagina, A.S.; Alexeevski, A.V. Role of restriction-modification systems in prokaryotic evolution and ecology. Biochemistry (Mosc.) 2015, 80, 1373–1386. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Dietrich, C.; Hongoh, Y.; Brune, A. Restriction-modification systems as mobile genetic elements in the evolution of an intracellular symbiont. Mol. Biol. Evol. 2016, 33, 721–725. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K.; Christensen, S.K.; Lobner-Olesen, A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 2005, 3, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Awano, N.; Masuda, H.; Park, J.H.; Inouye, M. Transcriptional repressor HipB regulates the multiple promoters in Escherichia coli. J. Mol. Microbiol. Biotechnol. 2013, 23, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, M.A.; Balani, P.; Min, J.; Chinnam, N.B.; Hansen, S.; Vulic, M.; Lewis, K.; Brennan, R.G. HipBA-promoter structures reveal the basis of heritable multidrug tolerance. Nature 2015, 524, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, M.A.; Piro, K.M.; Xu, W.; Hansen, S.; Lewis, K.; Brennan, R.G. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 2009, 323, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Cury, J.; Touchon, M.; Rocha, E.P.C. Integrative and conjugative elements and their hosts: Composition, distribution and organization. Nucleic Acids Res. 2017, 45, 8943–8956. [Google Scholar] [CrossRef] [PubMed]
- Trudel, M.V.; Vincent, A.T.; Attere, S.A.; Labbe, M.; Derome, N.; Culley, A.I.; Charette, S.J. Diversity of antibiotic-resistance genes in Canadian isolates of Aeromonas salmonicida subsp. salmonicida: Dominance of pSN254b and discovery of pAsa8. Sci. Rep. 2016, 6, 35617. [Google Scholar] [CrossRef] [PubMed]
- Adams, V.; Johanesen, P.A.; Rood, J.I.; Lyras, D. Mobilisable genetic elements from the Clostridia. In Bacterial Integrative Mobile Genetic Elements; Roberts, A., Mullany, P., Eds.; Landes Bioscience: Austin, TX, USA, 2013; pp. 120–134. [Google Scholar]
- Mulvey, M.R.; Boyd, D.A.; Olson, A.B.; Doublet, B.; Cloeckaert, A. The genetics of Salmonella genomic island 1. Microbes Infect. 2006, 8, 1915–1922. [Google Scholar] [CrossRef] [PubMed]
- Bardaji, L.; Echeverria, M.; Rodriguez-Palenzuela, P.; Martinez-Garcia, P.M.; Murillo, J. Four genes essential for recombination define GInts, a new type of mobile genomic island widespread in bacteria. Sci. Rep. 2017, 7, 46254. [Google Scholar] [CrossRef] [PubMed]
- Boyd, E.F.; Almagro-Moreno, S.; Parent, M.A. Genomic islands are dynamic, ancient integrative elements in bacterial evolution. Trends Microbiol. 2009, 17, 47–53. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Hug, L.A.; Edwards, E.A.; Holmes, S.; Spormann, A.M. Site-specific mobilization of vinyl chloride respiration islands by a mechanism common in Dehalococcoides. BMC Genom. 2011, 12, 287. [Google Scholar] [CrossRef] [PubMed]
- Christie, G.E.; Dokland, T. Pirates of the Caudovirales. Virology 2012, 434, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Gagnevin, L.; Chandler, M. The new IS1595 family, its relation to IS1 and the frontier between insertion sequences and transposons. Res. Microbiol. 2009, 160, 232–241. [Google Scholar] [CrossRef] [PubMed]
| IME a | Species (Division) b | Int c | Integration Site d | Putative Maintenance Genes e | Reference |
|---|---|---|---|---|---|
| MTnSag1 | Streptococcus agalactiae (fir.) | DDE | AT-rich regions | None | [28] |
| tISCpe8 | Clostridium perfringens (fir.) | DDE | AT-rich regions | None | [29] |
| IME_SpnAP200_rumA | Streptococcus pneumoniae (fir.) | Ser | Internal site of rumA (23S rRNA methyltransferase) | Replisome organizer | [26] |
| IME_ScoC232_maff2_site1 f | Streptococcus constellatus (fir.) | Ser | Internal site S1 of a gene (Maff2-related) from Tn5252-related ICEs | Replisome organizer, DnaC | [26] |
| IMESp2907 f | Streptococcus pyogenes (fir.) | Ser | Internal site S2 of a gene (Maff2-related) from Tn5252-related ICEs | Replisome organizer | [30] |
| tet(O) fragment | Streptococcus pyogenes (fir.) | Ser | Internal site of a gene (SNF2 helicase) from Tn5252-related ICEs | RepA, ParB | [31] |
| Tn6104 | Clostridioides difficile (fir.) | Ser | Internal site of traG (VirD4 CP) from Tn5252 related ICEs | Replisome organizer, DnaC, TA | [32] |
| IME_Sco1050_traG_site1 f | Streptococcus constellatus C1050 (fir.) | Ser | Internal site S1 of traG (VirD4 CP) from Tn5252-related ICEs | Replisome organizer | [26] |
| IME_Sco1050_traG_site2 f | Streptococcus constellatus C1050 (fir.) | Ser | Internal site S2 of traG (VirD4 CP) from Tn5252-related ICEs | Replisome organizer | [26] |
| Tn4451 f | Clostridium perfringens (fir.) | Ser | Numerous sites (GA) | [33] | |
| ATE-1 | Trueperella pyogenes (act.) | Tyr | 3′ end of guaA (GMP synthase) | TA | [34] |
| IncP island g | Brucella suis (α) | Tyr | 3′ end of guaA (GMP synthase) | RepA, antitoxin | [35] |
| Gisul2 | Pseudomonas aeruginosa (γ) h | Tyr | 3′ end of guaA (GMP synthase) | RepA, RepC | [36] |
| IME_SagNEM316_rplL f | Streptococcus agalactiae (fir.) | Tyr | 3′ end of rplL (L7/L12 ribosomal protein) | [26] | |
| IME_Sga2069_rpmE f | Streptococcus gallolyticus (fir.) | Tyr | 3′ end of rpmE (L31 ribosomal protein) | Rep_Trans | [26] |
| IME_SSalJIM777_rpmG f | Streptococcus salivarius (fir.) | Tyr | 3′ end of rpmG (L33 ribosomal protein) | [26] | |
| IMESag-rpsI g | Streptococcus agalactiae HRC (fir.) | Tyr | 3′ end of rpsI (S9 ribosomal protein) | Rep_Trans, TA | [37] |
| SGI1 g | Salmonella enterica DT104 (γ) | Tyr | 3′ end of trmE (tRNA modification GTPase) | Rep_3, TA | [38] |
| MGIVchHai6 | Vibrio cholerae (γ) | Tyr | 3′ end of trmE (tRNA modification GTPase) | [39] | |
| MGIVchMoz6 | Vibrio cholerae (γ) | Tyr | 3′ end of yicC (unknown) | RM II | [40] |
| MGIVchUSA1 f | Vibrio cholerae (γ) | Tyr | 3′ end of yicC (unknown) | 2 TAs | [27] |
| MGIVflInd1 f | Vibrio fluvialis (γ) | Tyr | 3′ end of yicC (unknown) | [27] | |
| MGIVmi1 | Vibrio mimicus (γ) | Tyr | 3′ end of yicC (unknown) | [41] | |
| BcenGI2 f | Burkholderia cenocepacia (β) | Tyr | 3′ end of tRNAala gene | Rep_3, TA | [42] |
| IME_SsuTL13-tRNAasn | Streptococcus suis (fir.) | Tyr | 3′ end of tRNAarg gene | [26] | |
| IME_SanC1051_tRNAarg | Streptococcus anginosus (fir.) | Tyr | 3′ end of tRNAasn gene | [26] | |
| NBU1 | Bacteroides uniformis (bac.) | Tyr | 3′ end of tRNAleu gene | TA | [43] |
| MTnPi4 | Prevotella intermedia (bac.) | Tyr | 3′ end of tRNAleu gene | [44] | |
| IME_Sag2603_tRNAlys f | Streptococcus agalactiae (fir.) | Tyr | 3′ end of tRNAlys gene | [45] | |
| Tn6031 | Sphingobacterium sp. (bac.) | Tyr | 3′ end of tRNApro gene | [46] | |
| NBU2 | Bacteroides fragilis (bac.) | Tyr | 3′ end of tRNAser gene | [47] | |
| IME_SdyRE378_ebfC f | Streptococcus dysgalactiae (fir.) | Tyr | 5′ end of ebfC (nucleoid associated protein) | [26] | |
| IME_SanC238_tatD | Streptococcus anginosus (fir.) | Tyr | 5′ end of tatD (DNAse) | [26] | |
| IME_oriT g | Streptococcus agalactiae (fir.) | Tyr | oriT from Tn916 and ICESt3-related ICEs | [48] | |
| Tn4555 | Bacteroides vulgatus (bac.) | Tyr | Two preferred sites | [49] | |
| Tn5520 | Bacteroides fragilis (bac.) | Tyr | AT-rich regions | None | [50] |
| Tn6218 g | Clostridioides difficile (fir.) | Tyr | AT-rich regions | [51] | |
| cLV25 | Bacteroides fragilis (bac.) | Tyr | ND | [52] | |
| MTnPi1 | Prevotella intermedia (bac.) | Tyr duo | TTAC NNNNN AA | [44] | |
| MTnPi2 | Prevotella intermedia (bac.) | Tyr duo | TTGC NNNNN AA | [44] | |
| MTnPi3 | Prevotella intermedia (bac.) | Tyr duo | TTAC NNNNN A/G A/G | [44] | |
| Tn4399 | Bacteroides fragilis (bac.) | ND | Numerous sites | [53] |
| IME a | Species (Division) b | Mobilization Proteins Encoded by the IME c | Mobilizing Element (CP, MPF) d | Reference | |
|---|---|---|---|---|---|
| Relaxase | Others | ||||
| MTnSag1 | Streptococcus agalactiae (fir.) | None | None | Tn916 (TcpA, FA) | [28] |
| tISCpe8 | Clostridium perfringens (fir.) | None | None | Tn916 (TcpA, FA) | [29] |
| GIE492 e | Klebsiella pneumoniae (γ) | None | Proposed: ICEKp1 (VirD4, T) | [87] | |
| MGIVflInd1 e | Vibrio fluvialis (γ) | None | ICEVflInd1 and SXT (VirD4, F) | [27] | |
| MGIVchHai6 e | Vibrio cholerae (γ) | None | 1 RAF | IncA/C plasmids (VirD4, F) | [39] |
| MGIVmi1 e | Vibrio mimicus (γ) | None | 1 RAF | IncA/C plasmids (VirD4, F) | [41] |
| SGI1 e,f | Salmonella enterica (γ) | None | TraG, TraH, TraN | IncA/C plasmids (VirD4, F) | [38] |
| Gisul2 g | Pseudomonas aeruginosa (γ) | None | TrbJ, TrbK, TrbL | Proposed: IncP plasmids (VirD4, T) | [36] |
| IME_SsalCCHSS3_ND | Streptococcus salivarius (fir.) | MobC | VirD4 | [26] | |
| NBU1 | Bacteroides uniformis (bac.) | MobP | CTnERL and CTnDOT (VirD4, B); IncP plasmids (VirD4, T) | [43] | |
| NBU2 | Bacteroides fragilis (bac.) | MobP | CTnERL (VirD4, B); IncP plasmids (VirD4, T) | [47] | |
| Tn4555 | Bacteroides vulgatus (bac.) | MobP | CTn341 (VirD4, B); IncP plasmids (VirD4, T) | [49] | |
| cLV25 | Bacteroides fragilis (bac.) | MobP | 1 RAF | IncP plasmids (VirD4, T) | [52] |
| Tn4399 | Bacteroides fragilis (bac.) | MobP | 1 RAF | CTnDOT (VirD4, B); IncP plasmids (VirD4, T) | [88] |
| IncP island f | Burkholderia glumae (β) | MobP | 2 RAFs, TrbJ, TrbK, TrbL | Proposed: IncP plasmids (VirD4, T) | [89] |
| IME_ScoC232_maff2_site 1 | Streptococcus constellatus (fir.) | MobP | [26] | ||
| IMESp2907 e | Streptococcus pyogenes (fir.) | MobQ | Proposed: Tn5252 superfamily (VirD4, FATA) | [30] | |
| ATE-1 | Trueperella pyogenes (act.) | MobV | IncP plasmids (VirD4, T) | [34] | |
| Tn5520 | Bacteroides fragilis (bac.) | MobV | None | IncP plasmids (VirD4, T) | [50] |
| Tn6215 | Clostridioides difficile (fir.) | MobV | ND | [90] | |
| Tn4451 e | Clostridium perfringens (fir.) | MobV | IncP plasmids (VirD4, T) | [91] | |
| IMESag-rpsI f,g | Streptococcus agalactiae HRC (fir.) | MobV | pAMβ1 plasmid (VirD4, FATA) | [37] | |
| tet(O) fragment | Streptococcus pyogenes (fir.) | MobV | Proposed: Tn5252 superfamily (VirD4, FATA) | [31] | |
| IME-oriT e,f,h | Streptococcus agalactiae (fir.) | MobT | Proposed: Tn916 and ICESt3 (TcpA, FA) | [63] | |
| IME_Sag2603_tRNAlys e | Streptococcus agalactiae (fir.) | MobT | Proposed: helpers with TcpA and FA | [26] | |
| IME_SsuBM407_tRNAleu e | Streptococcus suis (fir.) | MobT | TcpA | Proposed: helpers with TcpA and FA | [26] |
| IME_SdyRE378_ebfC e | Streptococcus dysgalactiae (fir.) | PF01719 | Proposed: helpers with TcpA and FA | [26] | |
| IME_SsalJIM8777_rpmG e | Streptococcus salivarius (fir.) | PF01719 | TcpA | Proposed: helpers with TcpA and FA | [26] |
| IME_SsuTL13_rpsI e | Streptococcus suis (fir.) | PF01719-helicase | Proposed: helpers with TcpA and FA | [26] | |
| IME_Seq35246_rpsI e | Streptococcus equi (fir.) | PF01719-helicase | TcpA | Proposed: helpers with TcpA and FA | [26] |
| IME_SanC238_tatD | Streptococcus anginosus (fir.) | PHA00330 | Proposed: helpers with TcpA and FA | [26] | |
| IME_SpnA45_tRNAleu e | Streptococcus pneumoniae (fir.) | PHA00330 | TcpA | Proposed: helpers with TcpA and FA | [26] |
| IME_SiniSF1_ebfC | Streptococcus iniae (fir.) | PF02407 | TcpA | Proposed: helpers with TcpA and FA | [26] |
| IME a | Species (Division) b | Size | Putative Cargo Genes | Reference | |
|---|---|---|---|---|---|
| Resistance Genes | Others | ||||
| ATE-1 | Trueperella pyogenes (act.) | 10.8 | tet(W) (tetracycline) | TA, 3 unknown | [105] |
| Tn6031 | Sphingobacterium sp. (bac.) | 13.0 | tet(X) (tetracycline), aadS (streptomycin) | 5 unknown | [46] |
| tet(O) fragment | Streptococcus pyogenes (fir.) | 13.4 | tet(O) (tetracycline) | RNA polymerase sigma factor sigma-70, 5 unknown | [31] |
| IMESp2907 | Streptococcus pyogenes (fir.) Streptococcus agalactiae (fir.) | 12.6 | erm(TR) (macrolide, lincosamide, streptogramin) | 11 unknown | [31] [101] |
| ermF region | Bacteroides thetaiotaomicron (bac.) | 13.0 | ermF (clindamycin, erythromycin) | 4 unknown | [106] |
| tISCpe8 | Clostridium perfringens (fir.) | 2.0 | lnu(P) (lincomycin) | [29] | |
| MTnSag1 (tISSag10) | Streptococcus agalactiae (fir.) | 1.7 | lnu(C) (lincomycin) | [28] | |
| IME_GB00957_oriT | Streptococcus agalactiae (fir.) | 5.2 | lsa(C) (lincosamide, streptogramin A and pleuromutilin) | 2 unknown | [48] |
| NBU2 | Bacteroides fragilis (bac.) | 11.1 | linAN2 (lincomycin, clindamycin), mefEN2 (erythromycin) | 2 unknown | [47] |
| SGI1 | Salmonella enterica (γ) | 42.4 | aadA2 (streptomycin, spectinomycin), floR (chloramphenicol, florfenicol), tet(G) (tetracycline), blaPSE-1 (ampicillin), sul1 (sulfonamides) | TA, 18 unknown | [38] |
| SGI1-V | Proteus mirabilis (γ) | 42.9 | aacA4 (kanamycin, tobramycin, netilmicin, amikacin), aadB (kanamycin, gentamicin, tobramycin); dhfrA1 (trimethoprim), blaVEB-6 (extended-spectrum cephalosporin), sul1 (sulfonamides), qnrA1 (quinolones) | 20 unknown | [107] |
| SGI1-L | Morganella morganii (γ) | 50.3 | tet(G) (tetracycline), floR (chloramphenicol, florfenicol), dhfrA15 (trimethoprim), blaPSE-1 (amoxicillin, clavulanate), sul1 (sulfonamides) | 11 unknown | [108] |
| MGIVchHai6 | Vibrio cholerae (γ) | 47.4 | aadA2 (streptomycin, spectinomycin), floR (chloramphenicol, florfenicol), tet(G) (tetracycline), blaPSE-1 (ampicillin), sul1 (sulfonamides), merEDAFPT (mercury) | RM I, 6 unknown | [39] |
| IncP island | Brucella suis (α) | 12.7 | antitoxin, 2 unknown | [35] | |
| IncP island c | Burkholderia glumae (β), Acidovorax avenae (β) | 14.1 | aac(2′)-IIa (kasugamycin) | antitoxin, 3 unknown | [89] |
| Tn4453 c | Clostridioides difficile | 6.3 | catD (chloramphenicol) | 2 unknown | [69] |
| Tn4555 | Bacteroides vulgatus | 12.2 | cfxA (cefoxitin) | 2 unknown | [67] |
| GIsul2 | Pseudomonas aeruginosa (γ) | 15.4 | sul2 (sulphonamide), arsBCHR (arsenate/arsenite) | TA, 4 unknown | [36] |
| IME_Sag2603_tRNAlys | Clostridioides difficile (fir.) | 10.5 | ABC transporter of the drug resistance transporter subfamily, 8 unknown | [48,84] | |
| IME_SagA909_tRNAlys | Clostridium perfringens (fir.) | 8.3 | Intracellular protease, 7 unknown | [48,84] | |
| IMESag-rpsI | Streptococcus agalactiae (fir.) | 9.1 | TA, 6 unknown | [37] | |
| IME_Sag2603_rpsI | Streptococcus agalactiae (fir.) | 9.0 | arsR (arsenate reductase) | TA, 5 unknown | [84] |
| IME_18RS21_oriT | Streptococcus agalactiae (fir.) | 6.4 | merA, merR (mercury) | 3 unknown | [48] |
| MTnPi2 | Prevotella intermedia (bac.) | 16.6 | LuxR family transcriptional regulator, 9 unknown | [44] | |
| MTnPi3 | Prevotella intermedia (bac.) | 18.4 | 2 ABC transporter components, 6 unknown | [44] | |
| MTnPi4 | Prevotella intermedia (bac.) | 12.4 | Subtilase-like protease, 7 unknown | [44] | |
| MGIVchUSA1 c | Vibrio cholerae (γ) | 22.0 | two TA, 10 unknown | [27] | |
| MGIVchMoz6 | Vibrio cholerae (γ) | 19.7 | RM II, 7 unknown | [40] | |
| MGIVmi1 | Vibrio mimicus (γ) | 16.5 | RM III, 10 unknown | [87] | |
| GIE492 | Klebsiella pneumoniae (γ) | 22.3 | MccE492 microcin (bacteriocin), 7 unknown | [32] | |
| Tn6104 | Clostridioides difficile (fir.) | 15.6 | Lantibiotic synthesis (bacteriocin), T/A, 7 unknown | [61] | |
| Tn5520 | Bacteroides fragilis (bac.) | 4.7 | [87] | ||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guédon, G.; Libante, V.; Coluzzi, C.; Payot, S.; Leblond-Bourget, N. The Obscure World of Integrative and Mobilizable Elements, Highly Widespread Elements that Pirate Bacterial Conjugative Systems. Genes 2017, 8, 337. https://doi.org/10.3390/genes8110337
Guédon G, Libante V, Coluzzi C, Payot S, Leblond-Bourget N. The Obscure World of Integrative and Mobilizable Elements, Highly Widespread Elements that Pirate Bacterial Conjugative Systems. Genes. 2017; 8(11):337. https://doi.org/10.3390/genes8110337
Chicago/Turabian StyleGuédon, Gérard, Virginie Libante, Charles Coluzzi, Sophie Payot, and Nathalie Leblond-Bourget. 2017. "The Obscure World of Integrative and Mobilizable Elements, Highly Widespread Elements that Pirate Bacterial Conjugative Systems" Genes 8, no. 11: 337. https://doi.org/10.3390/genes8110337
APA StyleGuédon, G., Libante, V., Coluzzi, C., Payot, S., & Leblond-Bourget, N. (2017). The Obscure World of Integrative and Mobilizable Elements, Highly Widespread Elements that Pirate Bacterial Conjugative Systems. Genes, 8(11), 337. https://doi.org/10.3390/genes8110337






