The Small and the Dead: A Review of Ancient DNA Studies Analysing Micromammal Species
Abstract
1. Introduction
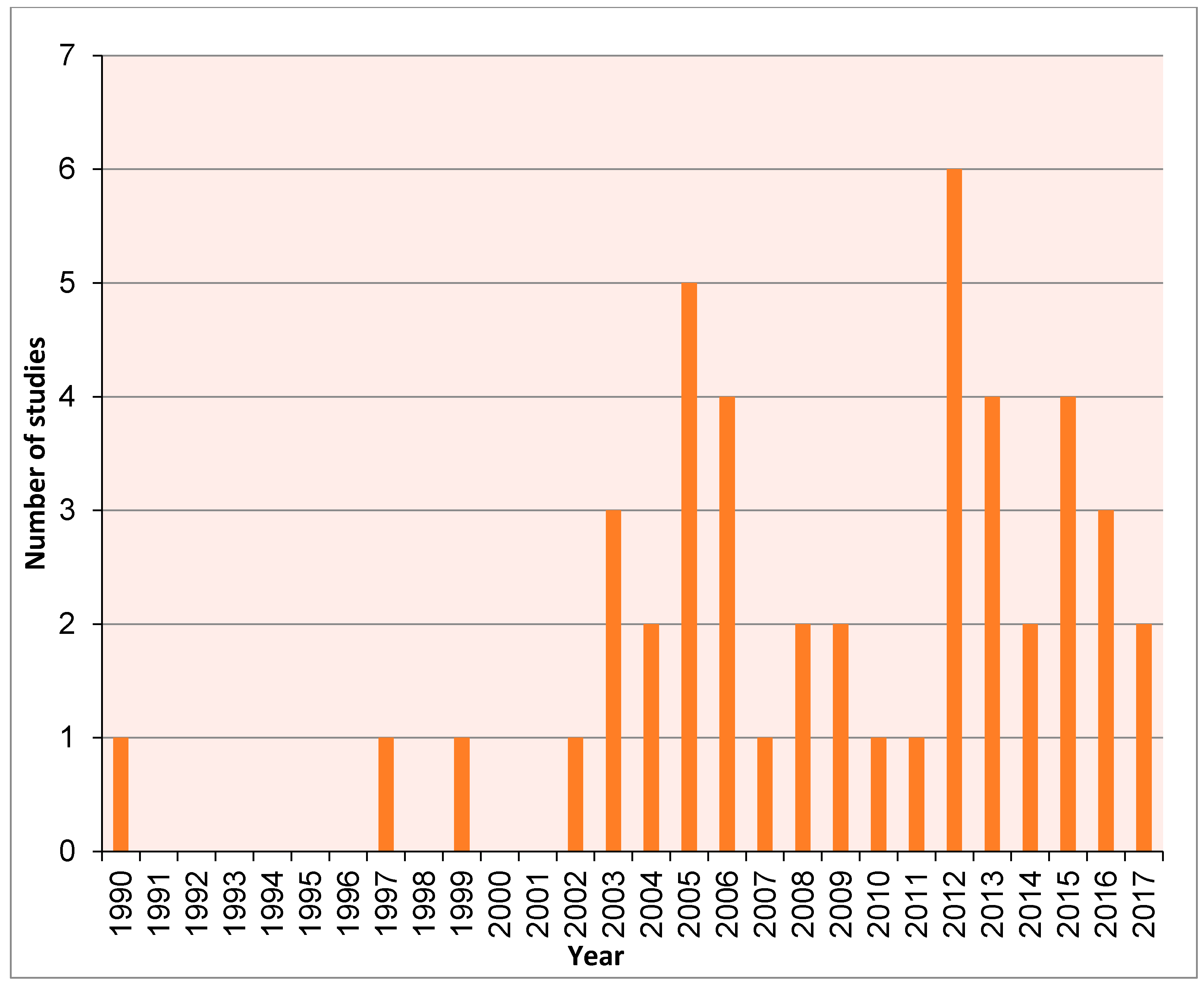
2. Methods
3. Results and Discussion
3.1. Next Generation Sequencing
3.2. DNA Metabarcoding
3.3. Cave Sites
3.4. Avian Pellets
3.5. Resolving the Phylogenetic Placement of Extinct Species
3.6. Species Identification
3.7. Biogeographic Hypotheses and Island Micromammals
3.8. Interactions with Humans
3.9. Population Genetics
3.10. Conservation
3.11. Climate Change
4. Conclusions
Conflicts of Interest
References
- Hagelberg, E.; Hofreiter, M.; Keyser, C. Introduction. Ancient DNA: The first three decades. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20130371. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, R.; Bowman, B.; Freiberger, M.; Ryder, O.A.; Wilson, A.C. DNA sequences from the quagga, an extinct member of the horse family. Nature 1984, 312, 282–284. [Google Scholar] [CrossRef] [PubMed]
- Bos, K.I.; Jäger, G.; Schuenemann, V.J.; Vågene, Å.J.; Spyrou, M.A.; Herbig, A.; Nieselt, K.; Krause, J. Parallel detection of ancient pathogens via array-based DNA capture. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20130375. [Google Scholar] [CrossRef] [PubMed]
- Roberts, C.; Ingham, S. Using ancient DNA analysis in palaeopathology: A critical analysis of published papers, with recommendations for future work. Int. J. Osteoarchaeol. 2008, 18, 600–613. [Google Scholar] [CrossRef]
- Hofreiter, M.; Barnes, I. Diversity Lost: Are All Holarctic Large Mammal Species Just Relict Populations? BMC Biol. 2010, 8, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Leonard, J.A. Ancient DNA applications for wildlife conservation. Mol. Ecol. 2008, 17, 4186–4196. [Google Scholar] [CrossRef] [PubMed]
- MacHugh, D.E.; Larson, G.; Orlando, L. Taming the Past: Ancient DNA and the study of animal domestication. Annu. Rev. Anim. Biosci. 2016, 5, 329–351. [Google Scholar] [CrossRef] [PubMed]
- Anderson-Carpenter, L.L.; McLachlan, J.S.; Jackson, S.T.; Kuch, M.; Lumibao, C.Y.; Poinar, H.N. Ancient DNA from lake sediments: Bridging the gap between paleoecology and genetics. BMC Evol. Biol. 2011, 11, 30. [Google Scholar] [CrossRef] [PubMed]
- Hofreiter, M.; Serre, D.; Poinar, H.N.; Kuch, M.; Pääbo, S. Ancient DNA. Nat. Rev. Genet. 2001, 2, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Pääbo, S.; Poinar, H.; Serre, D.; Jaenicke-Despres, V.; Hebler, J.; Rohland, N.; Kuch, M.; Krause, J.; Vigilant, L.; Hofreiter, M. Genetic analyses from ancient DNA. Annu. Rev. Genet. 2004, 38, 645–679. [Google Scholar] [CrossRef] [PubMed]
- Palkopoulou, E.; Mallick, S.; Skoglund, P.; Enk, J.; Rohland, N.; Li, H.; Omrak, A.; Vartanyan, S.; Poinar, H.; Götherström, A.; et al. Complete genomes reveal signatures of demographic and genetic declines in the woolly mammoth. Curr. Biol. 2015, 25, 1395–1400. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.; Knapp, M.; Enk, J.; Lippold, S.; Kircher, M.; Lister, A.; MacPhee, R.D.E.; Widga, C.; Czechowsk, P.; Sommer, R.; et al. The evolutionary and phylogeographic history of woolly mammoths: A comprehensive mitogenomic analysis. Sci. Rep. 2017, 7, 44585. [Google Scholar] [CrossRef] [PubMed]
- Dalton, R. Ancient DNA set to rewrite human history. Nature 2010, 465, 148–149. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Posth, C.; Hajdinjak, M.; Petr, M.; Mallick, S.; Fernandes, D.; Furtwängler, A.; Haak, W.; Meyer, M.; Mittnik, A.; et al. The genetic history of Ice Age Europe. Nature 2016, 534, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Llamas, B.; Willerslev, E.; Orlando, L. Human evolution: A tale from ancient genomes. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20150484. [Google Scholar] [CrossRef] [PubMed]
- Blanga-Kanfi, S.; Miranda, H.; Penn, O.; Pupko, T.; DeBry, R.W.; Huchon, D. Rodent phylogeny revised: Analysis of six nuclear genes from all major rodent clades. BMC Evol. Biol. 2009, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Reed, D.N. Taphonomic implications of roosting behavior and trophic habits in two species of African owl. J. Archaeol. Sci. 2005, 32, 1669–1676. [Google Scholar] [CrossRef]
- Martin, A.P.; Palumbi, S.R. Body size, metabolic rate, generation time, and the molecular clock. Proc. Natl. Acad. Sci. USA 1993, 90, 4087–4091. [Google Scholar] [CrossRef] [PubMed]
- Li, W.H.; Tanimura, M.; Sharp, P.M. An evaluation of the molecular clock hypothesis using mammalian DNA sequences. J. Mol. Evol. 1987, 25, 330–342. [Google Scholar] [CrossRef] [PubMed]
- Bromham, L.; Rambaut, A.; Harvey, P.H. Determinants of rate variation in mammalian DNA sequence evolution. J. Mol. Evol. 1996, 43, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Nabholz, B.; Glémin, S.; Galtier, N. Strong variations of mitochondrial mutation rate across mammals—The longevity hypothesis. Mol. Biol. Evol. 2008, 25, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Orr, H.A. The genetic theory of adaptation: A brief history. Nat. Rev. Genet. 2005, 6, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Bromham, L. Why do species vary in their rate of molecular clock evolution? Biol. Lett. 2009, 5, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gaines, M.S.; McClenaghan, L.R., Jr. Dispersal in Small Mammals. Annu. Rev. Ecol. Syst. 1980, 11, 163–196. [Google Scholar] [CrossRef]
- Lister, A.M. Mammalian fossils and quaternary biostratigraphy. Quat. Sci. Rev. 1992, 11, 329–344. [Google Scholar] [CrossRef]
- Schreve, D.C. Mammalian evidence from Middle Pleistocene fluvial sequences for complex environmental change at the oxygen isotope substage level. Quat. Int. 2001, 79, 65–74. [Google Scholar] [CrossRef]
- Currant, A.; Jacobi, R. A formal mammalian biostratigraphy for the Late Pleistocene of Britain. Quat. Sci. Rev. 2001, 20, 1707–1716. [Google Scholar] [CrossRef]
- Chaline, J.; Brunet-Lecomte, P.; Montuire, S.; Viriot, L. Anatomy of the arvicoline radiation (Rodentia): Palaeogeographical, palaeoecological history and evolutionary data. Ann. Zool. Fennici 1999, 36, 239–267. [Google Scholar]
- Abramson, N.I.; Lebedev, V.S.; Tesakov, A.S. Bannikova Supraspecies relationships in the subfamily Arvicolinae (Rodentia, Cricetidae): An unexpected result of nuclear gene analysis. Mol. Biol. 2009, 43, 834–846. [Google Scholar] [CrossRef]
- Knapp, M.; Hofreiter, M. Next generation sequencing of ancient DNA: Requirements, strategies and perspectives. Genes 2010, 1, 227–243. [Google Scholar] [CrossRef] [PubMed]
- Enk, J.M.; Devault, A.M.; Kuch, M.; Murgha, Y.E.; Rouillard, J.M.; Poinar, H.N. Ancient whole genome enrichment using baits built from modern DNA. Mol. Biol. Evolut. 2014, 31, 1292–1294. [Google Scholar] [CrossRef] [PubMed]
- Burbano, H.A.; Hodges, E.; Green, R.E.; Briggs, A.W.; Krause, J.; Meyer, M.; Good, J.M.; Maricic, T.; Johnson, P.L.; Xuan, Z.; et al. Targeted investigation of the Neandertal genome by array-based sequence capture. Science 2010, 328, 723–725. [Google Scholar] [CrossRef] [PubMed]
- Westbury, M.; Baleka, S.; Barlow, A.; Hartmann, S.; Paijmans, J.L.; Kramarz, A.; Forasiepi, A.M.; Bond, M.; Gelfo, J.N.; Reguero, M.A.; et al. A mitogenomic timetree for Darwin’s enigmatic South American mammal Macrauchenia patachonica. Nat. Commun. 2017, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, B.; Hofreiter, M. A paleogenomic perspective on evolution and gene function: New insights from ancient DNA. Science 2014, 343, 1236573. [Google Scholar] [CrossRef] [PubMed]
- Guimaraes, S.; Pruvost, M.; Daligault, J.; Stoetzel, E.; Bennett, E.A.; Cote, N.M.-L.; Nicolas, V.; Lalis, A.; Denys, C.; Geigi, E.-M.; et al. A cost-effective high-throughput metabarcoding approach powerful enough to genotype ~44,000 year-old rodent remains from Northern Africa. Mol. Ecol. Resour. 2016. [Google Scholar] [CrossRef]
- Avery, D. Holocene climatic change in southern Africa: The contribution of micromammals to its study. S. Afr. J. Sci. 1990, 86, 407–412. [Google Scholar]
- Willerslev, E.; Cooper, A. Ancient DNA. Proc. Biol. Sci. 2005, 272, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Kehlmaier, C.; Barlow, A.; Hastings, A.K.; Vamberger, M.; Paijmans, J.L.A.; Steadman, D.W.; Albury, N.A.; Franz, R.; Hofreiter, M.; Fritz, U. Tropical ancient DNA reveals relationships of the extinct Bahamian giant tortoise Chelonoidis alburyorum. Proc. R. Soc. B Biol. Sci. 2016, 21, 1–7. [Google Scholar]
- Gutiérrez-García, T.; Vázquez-Domínguez, E.; Arroyo-Cabrales, J.; Kuch, M.; Enk, J.; King, C.; Poinar, H.N. Ancient DNA and the tropics: A rodent’s tale. Biol. Lett. 2014, 10. [Google Scholar] [CrossRef] [PubMed]
- Avenant, N.L. Barn owl pellets: A useful tool for monitoring small mammal communities? Belgian J. Zool. 2005, 135, 39–43. [Google Scholar]
- Smoke, N.D.; Stahl, P.W. Post-burial fragmentation of microinvertebrate skeletons. J. Archaeol. Sci. 2004, 31, 1093–1100. [Google Scholar] [CrossRef]
- Oritz, P.E.; Madozzo Jaén, M.C.; Jayat, J.P. Micromammals and palaeoenvironments: Climatic oscillatons in the Monte desert of Catamarca (Argentina) during the last two millenia. J. Arid Environ. 2012, 77, 103–109. [Google Scholar] [CrossRef]
- McCrae, C. A comparative study of Late Holocene and Plio-Pleistocene-aged micromammalian owl accumulations from South Africa. Palaeontol. Afr. 2009, 44, 190–191. [Google Scholar]
- Taberlet, P.; Fumagalli, L. Owl pellets as a source of DNA for genetic studies of small mammals. Mol. Ecol. 1996, 5, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Poulakakis, N.; Lymberakis, P.; Paragamian, K.; Mylonas, M. Isolation and amplification of shrew DNA from barn owl pellets. Biol. J. Linn. Soc. 2005, 85, 331–340. [Google Scholar] [CrossRef]
- Guimaraes, S.; Fernandez-Jalvo, Y.; Stoetzel, E.; Gorgé, O.; Bennett, E.A.; Denys, C.; Grange, T.; Geigl, E.M. Owl pellets: A wise DNA source for small mammal genetics. J. Zool. 2016, 298, 64–74. [Google Scholar] [CrossRef]
- Brace, S.; Thomas, J.A.; Dalén, L.; Burger, J.; MacPhee, R.D.; Barnes, I.; Turvey, S.T. Evolutionary history of the Nesophontidae, the last unplaced Recent mammal family. Mol. Biol. Evol. 2016, 33, 3095–3103. [Google Scholar] [CrossRef] [PubMed]
- Pagès, M.; Chevret, P.; Gros-Balthazard, M.; Hughes, S.; Alcover, J.A.; Hutterer, R.; Rando, J.C.; Michaux, J.; Hänni, C. Paleogenetic analyses reveal unsuspected phylogenetic affinities between mice and the extinct Malpaisomys insularis, an endemic rodent of the Canaries. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Van, M.A.J. Middle Pleistocene Smaller Mammals from the Monte Peglia (Orvieto, Italy) with Special Reference to the Phylogeny of Microtus (Arvicolidae, Rodentia). Quaternaria 1973, 17, 1–144. [Google Scholar]
- Avery, D.M. An assessment of the Lower Pleistocene micromammalian fauna from Swartkrans members 1–3, Gauteng, South Africa. Geobios 1998, 31, 393–414. [Google Scholar] [CrossRef]
- Matthews, T.; Stynder, D.D. An analysis of two Myosorex species (Soricidae) from the Early Pliocene site of Langebaanweg (West coast, South Africa) using geometric morphometrics, linear measurements, and non-metric characters. Geobios 2011, 44, 87–99. [Google Scholar] [CrossRef]
- Adams, D.C.; Otárola-Castillo, E. Geomorph: An r package for the collection and analysis of geometric morphometric shape data. Methods Ecol. Evol. 2013, 4, 393–399. [Google Scholar] [CrossRef]
- Cornette, R.; Herrel, A.; Stoetzel, E.; Moulin, S.; Hutterer, R.; Denys, C.; Baylac, M. Specific information levels in relation to fragmentation patterns of shrew mandibles: Do fragments tell the same story? J. Archaeol. Sci. 2015, 53, 323–330. [Google Scholar] [CrossRef]
- Churakov, G.; Sadasivuni, M.K.; Rosenbloom, K.R.; Huchon, D.; Brosius, J.; Schmitz, J. Rodent evolution: Back to the root. Mol. Biol. Evol. 2010, 27, 1315–1326. [Google Scholar] [CrossRef] [PubMed]
- Markova, E.; Beeren, Z.; van Kolfschoten, T.; Strukova, T.; Vrieling, K. Differentiating sibling species in the Quaternary fossil record: A comparison of morphological and molecular methods to identify Microtus arvalis and M. rossiaemeridionalis (Arvicolinae, Rodentia). J. Syst. Palaeontol. 2012, 10, 585–597. [Google Scholar] [CrossRef]
- Fumagalli, L.; Taberlet, P.; Stewart, D.T.; Gielly, L.; Hausser, J.; Vogel, P. Molecular phylogeny and evolution of Sorex shrews (Soricidae: Insectivora) inferred from mitochondrial DNA sequence data. Mol. Phylogenet. Evol. 1999, 11, 222–235. [Google Scholar] [CrossRef] [PubMed]
- Chaline, J.; Graf, J.D. Phylogeny of the Arvicolidae (Rodentia): Biochemical and paleontological evidence. J. Mammal. 1988, 69, 22–33. [Google Scholar] [CrossRef]
- Hofreiter, M.; Paijmans, J.L.A.; Goodchild, H.; Speller, C.F.; Barlow, A.; Fortes, G.G.; Thomas, J.A.; Ludwig, A.; Collins, M.J. The future of ancient DNA: Technical advances and conceptual shifts. Bioessays 2015, 37, 284–293. [Google Scholar]
- Tegelström, H. Transfer of mitochondrial DNA from the northern red-backed vole (Clethrionomys rutilus) to the bank vole (C. glareolus). J. Mol. Evol. 1987, 24, 218–227. [Google Scholar]
- Runck, A.; Matocq, M.; Cook, J. Historic hybridization and persistence of a novel mito-nuclear combination in red-backed voles (genus Myodes). BMC Evol. Biol. 2009, 9. [Google Scholar] [CrossRef] [PubMed]
- Ballard, J.W.; Whitlock, M.C. The incomplete natural history of mitochondria. Mol. Ecol. 2004, 13, 729–744. [Google Scholar] [CrossRef] [PubMed]
- Haring, E.; Voyta, L.L.; Däubl, B.; Tiunov, M.P. Comparison of genetic and morphological characters in fossil teeth of grey voles from the Russian Far East (Rodentia: Cricetidae: Alexandromys). Mamm. Biol. 2015, 80, 496–504. [Google Scholar] [CrossRef]
- Rodrigues, M.; Bos, A.R.; Hoath, R.; Schembri, P.J.; Lymberakis, P.; Cento, M.; Ghawar, W.; Ozkurt, S.O.; Santos-Reis, M.; Merilä, J.; et al. Taxonomic status and origin of the Egyptian weasel (Mustela subpalmata) inferred from mitochondrial DNA. Genetica 2016, 144, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Amori, G.; Gippoliti, S.; Helgen, K.M. Diversity, distribution, and conservation of endemic island rodents. Quat. Int. 2008, 182, 6–15. [Google Scholar] [CrossRef]
- Ricklefs, R.; Bermingham, E. The West Indies as a laboratory of biogeography and evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008, 363, 2393–2413. [Google Scholar] [CrossRef] [PubMed]
- Brace, S.; Turvey, S.T.; Weksler, M.; Hoogland, M.L.P.; Barnes, I. Unexpected evolutionary diversity in a recently extinct Caribbean mammal radiation. Proc. R. Soc. B Biol. Sci. 2015, 282. [Google Scholar] [CrossRef] [PubMed]
- Fabre, P.H.; Vilstrup, J.T.; Raghavan, M.; der Sarkissian, C.; Willerslev, E.; Douzery, E.J.P.; Orlando, L. Rodents of the Caribbean: Origin and diversification of hutias unravelled by next-generation museomics. Biol. Lett. 2014, 10. [Google Scholar] [CrossRef] [PubMed]
- Barnes, S.S.; Matisoo-Smith, E.; Hunt, T.L. Ancient DNA of the Pacific rat (Rattus exulans) from Rapa Nui (Easter Island). J. Archaeol. Sci. 2006, 33, 1536–1540. [Google Scholar] [CrossRef]
- Matisoo-Smith, E.; Sutton, D.G.; Ladefoged, T.N.; Lambert, D.M.; Allen, J.S. Prehistoric mobility in Polynesia: MtDNA variation in Rattus exulans from the Chatham and Kermadec Islands. Asian Perspect. 1999, 38, 186–199. [Google Scholar]
- Wyatt, K.B.; Campos, P.F.; Gilbert, M.T.P.; Kolokotronis, S.; Hynes, W.H.; DeSalle, R.; Daszak, P.; MacPhee, R.D.E.; Greenwood, A.D. Historical mammal extinction on Christmas Island (Indian Ocean) correlates with introduced infectious disease. PLoS ONE 2008, 3. [Google Scholar] [CrossRef] [PubMed]
- Martinkova, N.; Barnett, R.; Cucchi, T.; Struchen, R.; Pascal, M.; Pascal, M.; Fischer, M.C.; Higham, T.; Brace, S.; Simon, Y.W.H.; et al. Divergent evolutionary processes associated with colonization of offshore islands. Mol. Ecol. 2013, 22, 5205–5220. [Google Scholar] [CrossRef] [PubMed]
- Pickrell, J.K.; Reich, D. Toward a new history and geography of human genes informed by ancient DNA. Trends Genet. 2014, 30, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Castle, W.E. The domestication of the rat. Proc. Natl. Acad. Sci. USA 1947, 33, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Jones, E.; Skirnisson, K.; McGovern, T.H.; Gilbert, M.T.; Willerslev, E.; Searle, J.B. Fellow travellers: A concordance of colonization patterns between mice and men in the North Atlantic region. BMC Evol. Biol. 2012, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Clout, M.N.; Russell, J.C. The invasion ecology of mammals: A global perspective. Wildl. Res. 2008, 35, 180–184. [Google Scholar] [CrossRef]
- Withers, P.; Cooper, E.; Maloney, K.; Bozinovic, F.; Cruz, A. Introduction to mammals. In Ecological and Enviromental Physiology of Mammals; Oxford University Press: Oxford, UK, 2016. [Google Scholar]
- Hadly, E.A.; Ramakrishnan, U.M.A.; Chan, Y.L.; Van Tuinen, M.; O’Keefe, K.; Spaeth, P.A.; Conroy, C.J. Genetic response to climatic change: Insights from ancient DNA and phylochronology. PLoS Biol. 2004, 2, e290. [Google Scholar] [CrossRef] [PubMed]
- Beaumont, M.A.; Zhang, W.; Balding, D.J. Approximate Bayesian computation in population genetics. Genetics 2002, 162, 2025–2035. [Google Scholar] [PubMed]
- Hadly, E.A.; van Tuinen, M.; Chan, Y.; Heiman, K. Ancient DNA evidence of prolonged population persistence with negligible genetic diversity in an endemic Tuco-Tuco (Ctenomys sociabilis). J. Mammal. 2003, 84, 403–417. [Google Scholar] [CrossRef]
- Chan, Y.L.; Lacey, E.A.; Pearson, O.P.; Hadly, E.A. Ancient DNA reveals Holocene loss of genetic diversity in a South American rodent. Biol. Lett. 2005, 1, 423–426. [Google Scholar] [CrossRef] [PubMed]
- Hofman, C.A.; Rick, T.C.; Fleischer, R.C.; Maldonado, J.E. Conservation archaeogenomics: Ancient DNA and biodiversity in the Anthropocene. Trends Ecol. Evol. 2015, 30, 540–549. [Google Scholar] [CrossRef] [PubMed]
- Prost, S.; Smirnov, N.; Fedorov, V.B.; Sommer, R.S.; Stiller, M.; Nagel, D.; Knapp, M.; Hofreiter, M. Influence of climate warming on arctic mammals? new insights from ancient DNA studies of the collared lemming Dicrostonyx torquatus. PLoS ONE 2010, 5. [Google Scholar] [CrossRef]
- Jaarola, M.; Martínková, N.; Gündüz, İ.; Brunhoff, C.; Zima, J.; Nadachowski, A.; Amori, G.; Bulatova, N.S.; Chondropoulos, B.; Fraguedakis-Tsolis, S.; et al. Molecular phylogeny of the speciose vole genus Microtus (Arvicolinae, Rodentia) inferred from mitochondrial DNA sequences. Mol. Phylogenet. Evol. 2004, 33, 647–663. [Google Scholar] [CrossRef] [PubMed]
- Catania, K.C. Evolution of sensory specializations in insectivores. Anat. Rec. Part A Discov. Mol. Cell. Evol. Biol. 2005, 287, 1038–1050. [Google Scholar] [CrossRef] [PubMed]
- Brace, S.; Barnes, I.; Powell, A.; Pearson, R.; Woolaver, L.G.; Thomas, M.G.; Turvey, S.T. Population history of the Hispaniolan hutia Plagiodontia aedium (Rodentia: Capromyidae): Testing the model of ancient differentiation on a geotectonically complex Caribbean island. Mol. Ecol. 2012, 21, 2239–2253. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, L.D.; Lambert, D.M. Ancient DNA and conservation: Lessons from the endangered kiwi of New Zealand. Mol. Ecol. 2008, 17, 2174–2184. [Google Scholar] [CrossRef] [PubMed]
- Bi, K.; Vanderpool, D.; Singhal, S.; Linderoth, T.; Moritz, C.; Good, J.M. Transcriptome-based exon capture enables highly cost-effective comparative genomic data collection at moderate evolutionary scales. BMC Genom. 2012, 13, 403. [Google Scholar] [CrossRef] [PubMed]
- Bi, K.; Linderoth, T.; Vanderpool, D.; Good, J.M.; Nielsen, R.; Moritz, C. Unlocking the vault: Next-generation museum population genomics. Mol. Ecol. 2013, 22, 6018–6032. [Google Scholar] [CrossRef] [PubMed]
- Smulders, M.J.M.; Snoek, L.B.; Booy, G.; Vosman, B. Complete loss of MHC genetic diversity in the Common Hamster (Cricetus cricetus) population in The Netherlands. Consequences for conservation strategies. Conserv. Genet. 2003, 4, 441–451. [Google Scholar] [CrossRef]
- Provan, J.; Bennett, K.D. Phylogeographic insights into cryptic glacial refugia. Trends Ecol. Evol. 2008, 23, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Brace, S.; Palkopoulou, E.; Dalén, L.; Lister, A.M.; Miller, R.; Otte, M.; Germonpré, M.; Blockley, S.P.; Stewart, J.R.; Barnes, I. Serial population extinctions in a small mammal indicate Late Pleistocene ecosystem instability. Proc. Natl. Acad. Sci. USA 2012, 109, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Ammerman, A.J.; Luigi, L.C. The Neolithic Transition and the Genetics of Populations in Europe; Princeton University Press: Princeton, NJ, USA, 2014. [Google Scholar]
- Thomas, W.K.; Pääbo, S.; Villablanca, F.X.; Wilson, A.C. Spatial and temporal continuity of kangaroo rat populations shown by sequencing mitochondrial DNA from museum specimens. J. Mol. Evol. 1990, 31, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Matisoo-Smith, E.; Allen, J.S.; Ladefoged, T.N.; Roberts, R.; Lambert, D.M. Ancient DNA from Polynesian rats: Extraction, amplification and sequence from single small bones. Electrophoresis 1997, 18, 1534–1537. [Google Scholar] [CrossRef] [PubMed]
- Matisoo-Smith, E.; Robins, J.H. Origins and dispersals of Pacific peoples: Evidence from mtDNA phylogenies of the Pacific rat. Proc. Natl. Acad. Sci. USA 2004, 101, 9167–9172. [Google Scholar] [CrossRef] [PubMed]
- Matisoo-Smith, E.; Robins, J. Mitochondrial DNA evidence for the spread of Pacific rats through Oceania. Biol. Invasions 2009, 11, 1521–1527. [Google Scholar] [CrossRef]
- Robins, J.H.; Tintinger, V.; Aplin, K.P.; Hingston, M.; Matisoo-Smith, E.; Penny, D.; Lavery, S.D. Phylogenetic species identification in Rattus highlights rapid radiation and morphological similarity of New Guinean species. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Orlando, L.; Mauffrey, J.F.; Cuisin, J.; Patton, J.L.; Hänni, C.; Catzeflis, F. Napoleon Bonaparte and the fate of an amazonian rat: New data on the taxonomy of Mesomys hispidus (Rodentia: Echimyidae). Mol. Phylogenet. Evol. 2003, 27, 113–120. [Google Scholar] [CrossRef]
- Demastes, J.W.; Butt, A.L.; Hafner, M.S.; Light, J.E. Systematics of a rare species of pocket gopher, Pappogeomys alcorni. J. Mammal. 2003, 84, 753–761. [Google Scholar] [CrossRef]
- Spaeth, P.A.; van Tuinen, M.; Chan, Y.L.; Terca, D.; Hadly, E.A. Phylogeography of Microtus. Longicaudus. in the Tectonically and Glacially Dynamic Central Rocky Mountains. J. Mammal. 2009, 90, 571–584. [Google Scholar] [CrossRef]
- Prost, S.; Guralnick, R.P.; Waltari, E.; Fedorov, V.B.; Kuzmina, E.; Smirnov, N.; Kolfschoten, T.; Hofreiter, M.; Vrieling, K. Losing ground: Past history and future fate of Arctic small mammals in a changing climate. Glob. Chang. Biol. 2013, 19, 1854–1864. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.L.; Hadly, E.A. Genetic variation over 10,000 years in Ctenomys: Comparative phylochronology provides a temporal perspective on rarity, environmental change and demography. Mol. Ecol. 2011, 20, 4592–4605. [Google Scholar] [CrossRef] [PubMed]
- Krystufek, B.; Buzan, E.V.; Hutchinson, W.F.; Hänfling, B. Phylogeography of the rare Balkan endemic Martino’s vole, Dinaromys bogdanovi, reveals strong differentiation within the western Balkan Peninsula. Mol. Ecol. 2007, 16, 1221–1232. [Google Scholar] [CrossRef] [PubMed]
- Kerhoulas, N.J.; Arbogast, B.S. Molecular systematics and Pleistocene biogeography of Mesoamerican flying squirrels. J. Mammal. 2010, 91, 654–667. [Google Scholar] [CrossRef]
- Fulton, T.L.; Norris, R.W.; Graham, R.W.; Semken, H.A.; Shapiro, B. Ancient DNA supports southern survival of Richardson’s collared lemming (Dicrostonyx richardsoni) during the last glacial maximum. Mol. Ecol. 2013, 22, 2540–2548. [Google Scholar] [CrossRef] [PubMed]
- Palkopoulou, E.; Baca, M.; Abramson, N.I.; Sablin, M.; Socha, P.; Nadachowski, A.; Prost, S.; Germonpré, M.; Kosintsev, P.; Smirnov, N.G.; et al. Synchronous genetic turnovers across Western Eurasia in Late Pleistocene collared lemmings. Glob. Chang. Biol. 2016, 22, 1710–1721. [Google Scholar] [CrossRef] [PubMed]
- Kalkvik, H.M.; Stout, I.J.; Doonan, T.J.; Parkinson, C.L. Investigating niche and lineage diversification in widely distributed taxa: Phylogeography and ecological niche modeling of the Peromyscus maniculatus species group. Ecography 2012, 35, 54–64. [Google Scholar] [CrossRef]
- Brace, S.; Ruddy, M.; Miller, R.; Schreve, D.C.; Stewart, J.R.; Barnes, I. The colonization history of British water vole (Arvicola amphibius (Linnaeus, 1758)): Origins and development of the Celtic fringe. Proc. R. Soc. B Biol. Sci. 2016, 283, 20160130. [Google Scholar] [CrossRef] [PubMed]
- Kuch, M.; Rohland, N.; Betancourt, J.L.; Latorre, C.; Steppan, S.; Poinar, H.N. Molecular analysis of an 11,700-year-old rodent midden from the Atacama Desert, Chile. Mol. Ecol. 2002, 11, 913–924. [Google Scholar] [CrossRef] [PubMed]
- Pacioni, C.; Hunt, H.; Allentoft, M.E.; Vaughan, T.G.; Wayne, A.F.; Baynes, A.; Haouchar, D.; Dortch, J.; Bunce, M. Genetic diversity loss in a biodiversity hotspot: Ancient DNA quantifies genetic decline and former connectivity in a critically endangered marsupial. Mol. Ecol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Prost, S.; Klietmann, J.; van Kolfschoten, T.; Guralnick, R.; Waltari, E.; Vrieling, K.; Stiller, M.; Nagel, D.; Rabeder, G.; Hofreiter, M.; et al. Effects of late quaternary climate change on Palearctic shrews. Glob. Chang. Biol. 2013, 19, 1865–1874. [Google Scholar] [CrossRef] [PubMed]
- Fraser, M.; Sten, S.; Götherström, A. Neolithic Hedgehogs (Erinaceus europaeus) from the Island of Gotland show early contacts with the Swedish mainland. J. Archaeol. Sci. 2012, 39, 229–233. [Google Scholar] [CrossRef]
- Tougard, C.; Renvoisé, E. Rodents and palaeogenetics: New perspectives. Comptes Rendus Palevol 2008, 7, 125–134. [Google Scholar] [CrossRef]
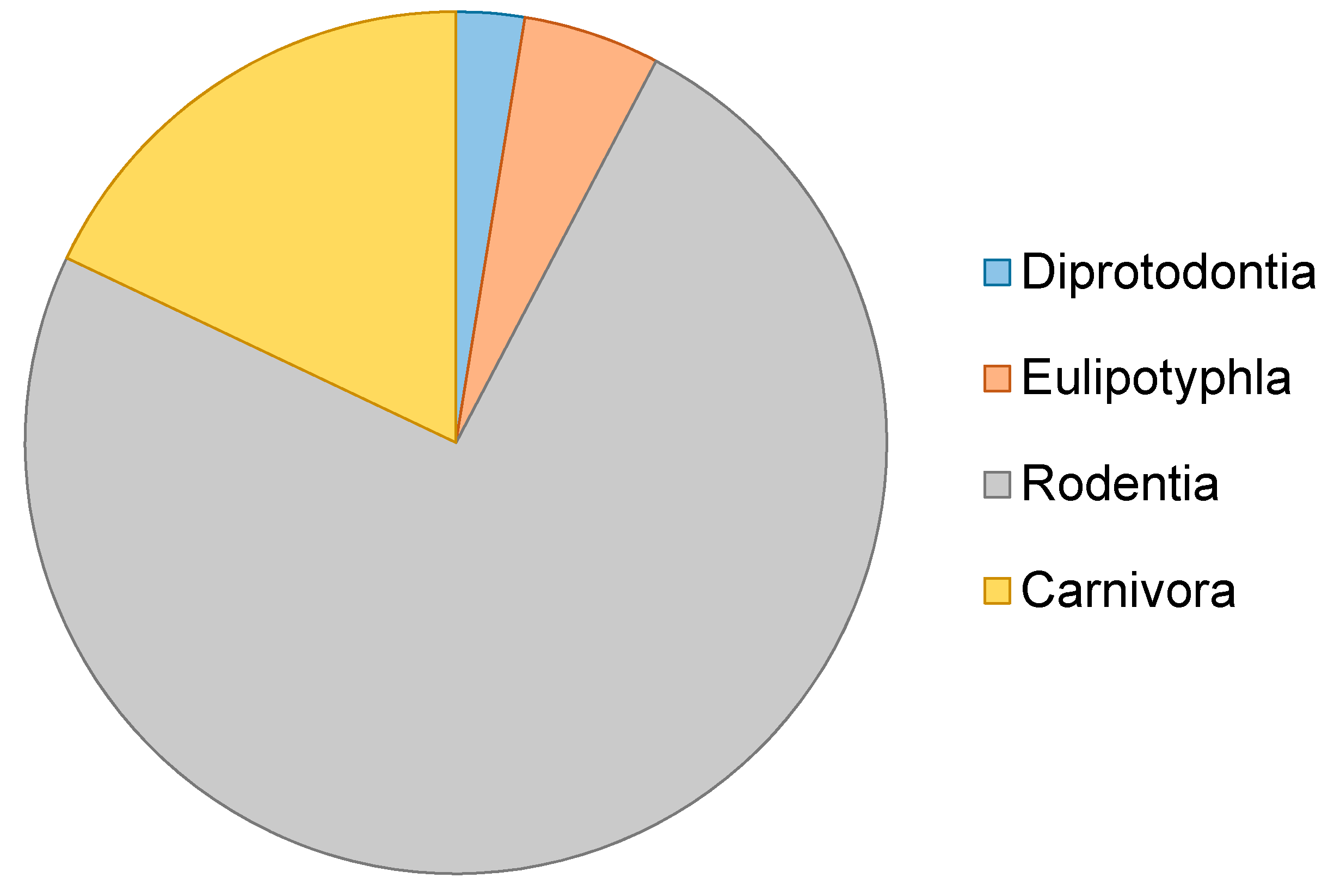
| Order | Genus | Species | Date | Author | Genetic Marker |
|---|---|---|---|---|---|
| Rodentia | Dipodomys | panamintinus | 1990 | Thomas et al. [93] | Control Region |
| Rodentia | Rattus | exulans | 1999 | Matisoo-Smith et al. [69] | Control Region |
| Rodentia | Rattus | exulans | 1997 | Matisoo-Smith et al. [94] | Control Region |
| Rodentia | Rattus | exulans | 2004 | Matisoo-Smith et al. [95] | mtDNA (~240 bp D-loop) |
| Rodentia | Rattus | exulans | 2006 | Barnes et al. [68] | Control Region |
| Rodentia | Rattus | exulans | 2009 | Matisoo-Smith & Robins [96] | Control region, Cytochrome b, Cytochrome Oxidase Subunit I |
| Rodentia | Rattus | - | 2014 | Robins et al. [97] | Control region, Cytochrome Oxidase Subunit I |
| Rodentia | Mesomys | hispidus | 2003 | Orlando et al. [98] | Cytochrome b |
| Rodentia | Pappogeomys | alcorni | 2003 | Demastes et al. [99] | Cytochrome b |
| Rodentia | Megalomys | georginae, desmarestii | 2015 | Brace et al. [66] | Cytochrome b |
| Rodentia | Microtus | montanus | 2004 | Hadly et al. [77] | Cytochrome b |
| Rodentia | Microtus | longicaudus | 2009 | Spaeth et al. [100] | Cytochrome b |
| Rodentia | Microtus | arvalis, rossiaemeridionalis | 2012 | Markova et al. [55] | Cytochrome b |
| Rodentia | Microtus | gregalis | 2013 | Prost et al. [101] | Cytochrome b |
| Rodentia | Ctenomys | sociabilis | 2003 | Hadly et al. [79] | |
| Rodentia | Ctenomys | sociabilis | 2005 | Chan et al. [80] | Cytochrome b |
| Rodentia | Ctenomys | sociabilis | 2006 | Chan et al. [72] | Cytochrome b |
| Rodentia | Ctenomys | haigi, sociabilis | 2011 | Chan & Hadley [102] | Cytochrome b |
| Rodentia | Dinaromys | bogdanovi | 2007 | Krystufek et al. [103] | Cytochrome b |
| Rodentia | Glaucomys | volans | 2010 | Kerhoulas & Arbogast [104] | Cytochrome b |
| Rodentia | Dicrostonyx | torquatus | 2010 | Prost et al. [82] | Control Region |
| Rodentia | Dicrostonyx | torquatus | 2012 | Brace et al. [91] | Cytochrome b |
| Rodentia | Dicrostonyx | richardsoni | 2013 | Fulton et al. [105] | Cytochrome b |
| Rodentia | Dicrostonyx | torquatus | 2013 | Prost et al. [101] | Cytochrome b |
| Rodentia | Dicrostonyx | groenlandicus, richardsoni, torquatus | 2016 | Palkopoulou et al. [106] | Cytochrome b |
| Rodentia | Peromyscus | polionotus | 2012 | Kalkvik et al. [107] | Cytochrome b |
| Rodentia | Plagiodontia | aedium | 2012 | Brace et al. [85] | Cytochrome b |
| Rodentia | Ototylomus | phyllotis | 2014 | Gutierrez-Garcia et al. [39] | Cytochrome b |
| Rodentia | Alexandromys | fortis, indet, maximowiczii, oeconomus | 2015 | Haring et al. [62] | Control Region |
| Rodentia | Pennatomys | nivalis, luciae | 2015 | Brace et al. [66] | Cytochrome b |
| Rodentia | Arvicola | amphibius | 2016 | Brace et al. [108] | Control Region |
| Rodentia | Malpaisomys, Mus, Murinae | - | 2012 | Pages et al. [48] | Cytochrome b, Interphotoreceptor Retinoid Binding Protein (IRBP) |
| Rodentia | Mus, Meriones, Apodemus, Lemniscomys, Rattus, Gerbillus, Cricetulus, Microtus, Eliomys | - | 2016 | Guimaraes et al. [35] | Single Nucleotide Polymorphisms (SNPs) |
| Rodentia | - | - | 2002 | Kuch et al. [109] | 12S & 16S rDNA, Cytochrome b |
| Diprotodontia | Bettongia | penicillata | 2015 | Pacioni et al. [110] | Control Region, Microsatellites |
| Eulipotyphla | Sorex | araneus, tundrensis | 2013 | Prost et al. [111] | Cytochrome b |
| Eulipotyphla | Erinaceus | europaeus | 2012 | Fraser et al. [112] | Cytochrome b |
| Eulipotyphla | Nesophontes | paramicrus | 2016 | Brace et al. [47] | Mitochondrial genome, ADORA3, ADRA2B, ADRB2, APOB, APP, ATP7A, BCHE, BDNF, BMI1, BMP4, BRCA1, CREM, EDG1, GHR, PLCB4, RAG1, RAG2, RHO, TTN, TYR, VWF |
| - | - | - | 2005 | Avenant et al. [40] | Cytochrome b |
| - | - | - | 2005 | Poulakakis et al. [45] | Cytochrome b |
| - | - | - | 2016 | Guimaraes et al. [46] | MtDNA |
| - | - | - | 2008 | Tougard & Renvoise [113] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Woods, R.; Marr, M.M.; Brace, S.; Barnes, I. The Small and the Dead: A Review of Ancient DNA Studies Analysing Micromammal Species. Genes 2017, 8, 312. https://doi.org/10.3390/genes8110312
Woods R, Marr MM, Brace S, Barnes I. The Small and the Dead: A Review of Ancient DNA Studies Analysing Micromammal Species. Genes. 2017; 8(11):312. https://doi.org/10.3390/genes8110312
Chicago/Turabian StyleWoods, Roseina, Melissa M. Marr, Selina Brace, and Ian Barnes. 2017. "The Small and the Dead: A Review of Ancient DNA Studies Analysing Micromammal Species" Genes 8, no. 11: 312. https://doi.org/10.3390/genes8110312
APA StyleWoods, R., Marr, M. M., Brace, S., & Barnes, I. (2017). The Small and the Dead: A Review of Ancient DNA Studies Analysing Micromammal Species. Genes, 8(11), 312. https://doi.org/10.3390/genes8110312





